Abstract
This study explored the association of sepsis prognosis with dynamic changes in serum soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) and its polymorphisms. We enrolled 80 subjects with sepsis and 80 controls. Serum sTREM-1 was tested on days 1, 3, 5, 7, 10, and 14. PCR sequencing was performed to detect TREM-1 genetic variation on its four exons. sTREM-1 levels were significantly higher in the nonsurvivor than in the survivor group (p < 0.001), and those at each time point were the same (p ≤ 0.001). Of the three tested TREM-1 SNPs (rs144672509, rs2234237, and rs2234246), only rs2234237 (Ser25Thr) was significantly associated with sepsis prognosis in three inheritance models (p < 0.05). However, there was no relationship between TREM-1 polymorphism and dynamic concentration of sTREM-1. Logistic regression showed that sTREM-1, APACHE II, and rs2234237 polymorphism are risk factors for prognosis. Dynamic changes in serum sTREM-1 and rs2234237 polymorphism could be used in sepsis prognosis assessment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
More and more studies have confirmed that sepsis is a common disease caused by an interaction of environmental and genetic factors. There is a growing body of evidence that genetic factors play important roles in the occurrence and development of sepsis [1]. To further reveal the effect of genetic factors on sepsis, the 2001 International Sepsis Definition Conference recommended that individual genetic backgrounds be considered as an important part of the cause of sepsis [2]. Therefore, exploration of the pathogenesis of sepsis, especially for the genetic factors relevant to the development of severe sepsis, can improve the identification of susceptibility to sepsis, prediction capabilities of sepsis risk, monitoring, sepsis prognosis assessment, and early treatment intervention. These improvements will help to reduce the incidence of severe sepsis and mortality, improve patient outcomes, and lower medical costs. Meanwhile, based on individual genetic backgrounds, different treatments have been performed on different patients to achieve a real sense of future “individual treatment” [3].
In recent years, some studies that focused on gene polymorphism, susceptibility, and prognosis of sepsis found that several single nucleotide polymorphisms (SNPs) of some genes are relevant to the incidence, severity, and prognosis of sepsis [4]. Moreover, some genetic polymorphisms may be new indicators for sepsis susceptibility classification [2]. At present, research on sepsis and SNPs focuses on the pathogenesis of inflammation and some anti-inflammatory genes. More than 30 genes have had their respective polymorphisms studied in terms of relationships to sepsis and critical infection or inflammation [5]. Triggering receptor expressed on myeloid cells-1 (TREM-1) is a member of the immunoglobulin superfamily of receptors that is expressed on polymorphonuclear granulocytes and monocytes [6]. TREM-1 acts synergistically with Toll-like receptors (TLRs) and Nod-like receptors and activates downstream signaling pathways with DAP12 to induce and enhance the inflammatory reaction [7, 8]. This effect leads to synthesis and release of a large number of downstream inflammatory cytokines located in signaling pathways [7, 9]. Bacterial or fungal infections may induce the expression of TREM-1. sTREM-1 is a soluble form of TREM-1 that may be released into body fluids with upregulated expression of TREM-1 [10]. However, the association between genomic variations within the TREM-1 gene and sepsis remains unclear. In this study, we explored the dynamic changes in serum sTREM-1 and sequenced the genotype to reveal the clinical significance of sTREM-1 and its gene polymorphisms.
SUBJECTS AND METHODS
Subjects
All subjects were selected from inpatients who were hospitalized between May 2009 and July 2011 in the Respiratory ICU, Surgical ICU, and Emergency ICU of the Chinese People’s Liberation Army (CPLA) General Hospital. Sepsis met the criteria recommended by the 1991 ACCP/SCCM Joint Meeting [11] and the diagnostic criteria developed at the 2001 International Sepsis Definition Conference [2]. Based on 28-day survival, patients with sepsis were divided into a survivor group (≥28-day survival) and a nonsurvivor group (<28-day survival). Patients were excluded for (1) being <18 years of age, (2) having acquired immunodeficiency syndrome, (3) having a reduced polymorphonuclear granulocyte count (<500/μl), (4) dying within 24 h after admission into the ICU, (5) refusing to participate in the study, or (6) declining treatment during the period of observation. The control group was randomly selected from the healthy individuals who received a physical examination. Patients or their family members were fully informed and signed informed consent forms. This study was approved by the Ethics Committee of the CPLA General Hospital (project no. 20111013–009) and was registered with the US National Institutes of Health Clinical Trials Registry (NCT01490424).
Data Collection and Specimen Preparation
Upon admission into the ICU, the following items were recorded for each patient: source of patient, age, gender, chief complaints upon admission, symptoms, temperature, APACHE II scores, SOFA scores, and underlying diseases. Records were also kept for the 28-day survival patients.
Intravenous blood samples were obtained within 24 h (first day of study) after ICU admission. The same blood draw was used to collect both cells for DNA and serum for sTREM analysis. Blood was centrifuged at 3000 rpm for 15 min. The supernatants were transferred to Eppendorf tubes and stored at −80 °C for ELISA. Blood cells on the bottom of the tube were used to extract DNA. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The DNA was quantified in a spectrophotometer (GeneQuant Pro RNA/DNA Calculator; Amersham Biosciences, Piscataway, NJ, USA) and stored at −80 °C for sequencing.
ELISA Assays
All specimens were renumbered before the experiment to assure that the subsequent steps for researchers were relatively blind. sTREM-1 was determined with a double antibody sandwich ELISA (Quantikine Human TREM-1 Immunoassay ELISA Kit, product number DTRM10B; R&D Systems, Minneapolis, MN, USA). ELISA was performed in duplicate and in strict accordance with the manufacturer’s instructions.
Genotyping of Single Nucleotide Polymorphisms
We first sequenced 30 patients with sepsis and 30 controls with four exons of the TREM-1 gene by PCR sequencing. The sequencing results showed three gene polymorphisms located in exon-2 and exon-4. Compared with the NCBI dbSNP and Hapmap database, the two polymorphisms located in exon-2 were nonsynonymous variation rs2234237 (Ser25Thr) and rs144672509 (Arg97Cys), and the other located in exon-4 was synonymous variation rs2234246. Another 50 samples underwent PCR sequencing of exon-2 and exon-4 to explore polymorphisms of loci rs2234237, rs144672509, and rs2234246. The primers used for exon-2 were as follows: forward primer, 5′-GGGTGGTTGGACAAGAAA-3′; reverse primer, 5′-GAGAACAAAGAATGGGACTAAA-3′; and sequencing primer, GACTGCTGGGAATCCTGAAC. The primers for PCR amplification of exon-4 were designed as follows: forward primer, 5′-CAGGCAGTGCGGGAAAG-3′; reverse primer, 5′-CACCCAAGATTATTAGAGGAACG-3′; and sequencing primer, CAGGCAGTGCGGGAAAG. PCR amplification of genomic DNA was carried out in 25-μl volumes containing 50 ng of genomic DNA, 0.5 μl of each primer (10 mmol/l), 2 μl of dNTP (2.5 mmol/l), 2.5 μl of Takara HotStart Taq polymerase, and 2.5 μl of 10× ExTaq buffer (Takara, Dalian, China). The PCR reaction was performed in a thermal cycler (DNA Engine Peltier Thermal Cycler 200; Bio-Rad, Hercules, CA, USA). Amplification conditions were as follows: denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 40 s, annealing at 57.7 °C for 40 s, and extension at 72 °C for 60 s; an additional extension cycle at 72 °C for 10 min; and storage at −20 °C. The product was purified using Montage PCRμ96 (Millipore Corporation, Billerica, MA, USA) and loaded onto an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The chromatograms were generated on the basis of sequencing data obtained with the Applied Biosystems model ABI3730 DNA sequencing instrument and analyzed using Chromas1.56 (Technelysium Pty Ltd., Queensland, Australia). Beijing Genomics Institute at Shenzhen (Shenzhen, China) completed this experiment.
Statistical Analysis
Results for continuous variables with normal distributions are given as means ± standard deviations (SD). Student’s t-test was used to compare means between two groups. The logarithmic transformation was performed for the continuous variables that were not normally distributed (serum sTREM-1 concentration). Results for qualitative variables were expressed as percentages and compared between groups using a chi-square test. Repeated measure analysis of variance (ANOVA) was used to describe the dynamic changes in sTREM-1 levels between survivors and nonsurvivors at different time points of the study observation period (days 1, 3, 5, 7, 10, and 14). ANOVA was used to study the relationship between TREM-1 gene polymorphism and serum sTREM-1 concentration. All SNP data were evaluated for the Hardy–Weinberg equilibrium using the goodness-of-fit test (http://analysis.bio-x.cn) [12, 13]. Allele frequency and genotype frequency between patients with sepsis and control subjects or between survivors and nonsurvivors among sepsis patients were compared using Fisher’s exact test or the χ 2 test. Different models of inheritance were tested using SNPStats software (http://bioinfo.iconcologia.net/index.php?module=Snpstats) [14]. Linkage disequilibrium among genotyped SNPs was obtained, and haplotypes were estimated using the Haploview 4.2 program [15]. Logistic regression was used to evaluate the influence of the polymorphisms on the outcome of patients with sepsis, both of which included most clinical and biological data at admission as covariates, such as gender, age, sepsis severity, temperature, WBC counts, serum sTREM-1, APACHE II score, SOFA score, and TREM-1 polymorphism. Discrimination was assessed using the area under the receiver operating characteristic curve to evaluate how well the model distinguished patients who lived from those with a poor prognosis. Values of p < 0.05 were considered statistically significant. The statistical analysis was performed with SPSS version 16.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Subject Descriptions
We enrolled 160 subjects into this study, including 80 in the control group and 80 in the sepsis group. The patients with sepsis eventually comprised 79 Han Chinese people (one sample failed sequencing) who came from all regions of the Chinese Mainland. Patients from North China accounted for 53.2 %, followed by East China (16.5 %), Northeast China (11.4 %), Central China (10.1 %), Northwest China (3.8 %), Southwest China (3.8 %), and South China (1.3 %). Characteristics of patients with sepsis (divided into survivors and nonsurvivors) at admission to ICU are given in Table 1. The sepsis severity of the nonsurvivor group was higher than that of the survivor group (p < 0.001). The nonsurvivor group demonstrated higher serum sTREM-1 levels (p < 0.001). APACHE II and SOFA scores were significantly different between survivors and nonsurvivors (p < 0.001). In terms of age, gender, temperature, WBC counts, and underlying diseases, a cross-group comparison upon admission was devoid of statistical significance.
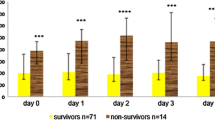
Values for Serum sTREM-1 Dynamic Assessment of Sepsis Prognosis
Table 2 compares the survivor and nonsurvivor groups in terms of dynamic changes in serum sTREM-1 levels on days 1, 3, 5, 7, 10, and 14. A polynomial test was used to analyze the trend curve (Fig. 1). The results showed that the sTREM-1 level of the dynamic change trend was significantly different between the survivor and nonsurvivor groups in the observation period (p < 0.05), which was reflected by a linear curve. The curves show that the nonsurvivor group had higher serum sTREM-1 levels initially and that they increased with the passage of time. In contrast, sTREM-1 of the survivor group revealed a tendency of declination. The sTREM-1 levels of the nonsurvivor group were significantly higher than those of the survivor group (F = 22.28, p < 0.001), and at each time point they were the same (p ≤ 0.001). There were no significant differences in the sTREM-1 level among patients with sepsis admitted to the ICU at different time points (F = 0.221, p = 0.954); the survivor and nonsurvivor groups were the same (F values were 0.714 and 1.908, and p values were 0.565 and 0.131, respectively). In addition, there was no interaction effect between the dynamics of sTREM-1 levels and the two groups of different patient prognoses (F = 2.332, p = 0.06).
Association Between TREM-1 Gene Polymorphisms and Sepsis
Only one patient of 79 individuals had rs144672509 polymorphism, so we analyzed rs2234237 and rs2234246. The genotype distribution for the assayed loci followed the Hardy–Weinberg equilibrium in patients with sepsis and controls (Table 3). All pairwise r 2 values for linkage disequilibrium among SNPs rs2234237 and rs2234246 in both of the groups were analyzed. Control group D′ was 0.615, and r 2 was 0.038; sepsis group D′ was 0.3, and r 2 was 0.064. Based on high LD values defined as r 2 > 1/3 as suggested by Ardlie et al. [16] or D′ > 0.7 as suggested by Gabriel et al. [17], there was no linkage disequilibrium between rs2234237 and rs2234246. Different models of inheritance were tested for the impact of rs2234237 and rs2234246 on susceptibility and prognosis. None of the assayed loci were significantly associated with susceptibility to sepsis in three inheritance models (dominant model, recessive model, and codominant model) (Table 4). However, the assayed locus of rs2234237 was significantly associated with prognosis of patients with sepsis in the three inheritance models (Table 5). Therefore, all patients with sepsis (79 cases) were divided into three groups according to genotype. Based on a log-rank test for trend, patients with the AA genotype had increased survival compared with patients with the AT or TT genotypes (p = 0.037) (Fig. 2).
Relationship Between rs2234237 Polymorphism and Serum sTREM-1 Levels
The assayed locus of rs2234237 was significantly associated with prognosis of sepsis in the three inheritance models. Nonsynonymous variation Ser25Thr (rs2234237) in exon-2 of the TREM-1 gene may influence the biologic function of TREM-1. We speculated whether different genotypes could affect sTREM-1 expression. Therefore, all patients with sepsis (79 cases) were divided into three groups according to genotype. Figure 3 shows the different genotypes in terms of dynamic changes in serum sTREM-1 levels on days 1, 3, 5, 7, 10, and 14. The serum sTREM-1 concentration in patients with the TT genotype was consistently higher than that in patients with the AT genotype. Patients with the AA genotype showed a trend toward increased serum sTREM-1 concentrations. However, there was no significant difference in sTREM-1 levels among patients with different genotypes at different time points.
Relationship between TREM-1 gene polymorphism and serum sTREM-1 dynamic concentration. Based on analysis of variance, serum sTREM-1 levels of rs2234237 genotypes AA, AT, and TT showed no significant differences at different time points. AA vs. AT vs. TT: day 1, 2.19 ± 0.32 vs. 2.12 ± 0.34 vs. 2.24 ± 0.27, F = 1.122, p = 0.301; day 3, 2.19 ± 0.32 vs. 2.11 ± 0.36 vs. 2.24 ± 0.31, F = 1.259, p = 0.29; day 5, 2.27 ± 0.31 vs. 2.14 ± 0.41 vs. 2.22 ± 0.32, F = 0.704, p = 0.498; day 7, 2.3 ± 0.38 vs. 2.16 ± 0.44 vs. 2.23 ± 0.32, F = 0.577, p = 0.564; day 10, 2.24 ± 0.36 vs. 2.16 ± 0.44 vs. 2.23 ± 0.33, F = 0.377, p = 0.687; day 14, 2.24 ± 0.4 vs. 2.19 ± 0.41 vs. 2.21 ± 0.36, F = 0.08, p = 0.923.
Best Values for Early Identification of Sepsis Prognosis
Univariate logistic regression was employed to assess possible risk factors for prognosis. The variables taken into account included gender, age, sepsis severity, temperature, WBC counts, serum sTREM-1, APACHE II score, SOFA score, and TREM-1 rs2234237 genetic variation. Five variables, namely sepsis severity, TREM-1 rs2234237 genetic variation, sTREM-1 concentration, APACHE II score, and SOFA score, were selected for multivariate regression. Finally, serum sTREM-1 level, APACHE II score, and TREM-1 rs2234237 genetic variation were able to enter the multivariable regression equation (Table 6). The ROC curve denoting the two risk parameters, serum sTREM-1 level and APACHE II score, and their combination was drawn on the ICU admission day. The AUC demonstrated that serum sTREM-1 + APACHE II measured 0.807 (95 % CI, 0.709–0.904). With 0.467 as the cutoff point, sensitivity measured 0.854 (Table 7, Fig. 4).
DISCUSSION
During the 14 days of observation, the nonsurvivors had a higher serum sTREM-1 level that continued to climb with the passage of time, while the serum sTREM-1 level of the survivors remained relatively lower and declined (Fig. 1). These serum sTREM-1 trends are similar to those of our previous studies, which focused on serum and urine, respectively [18, 19]. The continuous rise in the sTREM-1 level was possibly due to positive feedback provided by downstream inflammatory factors [20, 21]. Because of extracellular partial exfoliation brought about by metalloproteinase proteolysis, TREM-1 gives rise to its soluble protein sTREM-1 [22], which suggests the release of additional proinflammatory cytokines and mediators as well as a continuous or progressive overactive inflammatory response and a poor prognosis. Therefore, dynamic changes in serum sTREM-1 may well reflect the body’s inflammatory responses and the prognosis of sepsis: relatively lower expression indicates that the inflammatory response has been brought under control and that the prognosis might be better. sTREM-1 is an ideal medical biomarker because the progressive increase in serum sTREM-1 levels may signify a poor prognosis.
We examined the relationships between TREM-1 genetic polymorphisms and susceptibility to sepsis and prognosis. Three SNPs were detected in the exon sequencing. We discarded the rs144672509 polymorphism analysis because it was present in only one case. The assayed locus of rs2234237 was significantly associated with prognosis of patients with sepsis in three inheritance models: the codominant model (A/T: OR 0.51 and 95 % CI 0.19–1.38; and A/A: OR 0.07 and 95 % CI 0.01–0.61; p = 0.01), the dominant model (A/T-A/A: OR 0.36; 95 % CI 0.14–0.94; p = 0.033), and the recessive model (A/A: OR 0.09; 95 % CI 0.01–0.80; p = 0.0065). Therefore, the A allele may be a protective factor for sepsis prognosis, and the T allele may be a risk factor. The survival analysis showed that patients with the TT genotype had increased mortality over 28 days compared with patients with the AT or AA genotype (Fig. 2). According to r 2 and D′ values, we did not find linkage disequilibrium between the two measured SNP loci. However, a study investigating an association between TREM-1 gene polymorphisms and severe sepsis conducted in a Chinese cohort came to negative conclusions [23]. Three studied common polymorphisms within the TREM-1 gene (rs7768162, rs9471535, and rs2234237) may not play a major role in the predisposition to severe sepsis in the studied Chinese Han cohort. The reason for different conclusions in a similar study, in our opinion, may be that differences in the study population led to different results. Patients involved in our study came from all regions of the Chinese Mainland rather than being limited to one or two regions. Jung et al. [24] recently proved that TREM-1 SNPs (rs7768162, rs9471535, and rs2234237) may play a significant role in the development of intestinal Behçet’s disease and may have modest effects on disease severity. Therefore, TREM-1 gene polymorphism is still a problem and is worthy of investigation in further studies.
rs2234237 is a nonsynonymous variation (Ser25Thr) in exon-2 of the TREM-1 gene. Therefore, it may influence the biologic function of TREM-1. However, there is no relationship between TREM-1 gene polymorphism and the serum sTREM-1 dynamic concentration (Fig. 3). That is to say, we did not find a particular genotype that could influence sTREM-1 expression to be either too high or too low. The reason may involve the sample size of this study, which included only nine patients with genotype A/A. Logistic regression analysis showed that serum sTREM-1 level, APACHE II score, and TREM-1 rs2234237 genetic variation are risk factors for the prognosis of sepsis. Septic patients with the TREM-1 rs2234237 genetic variation had a 2.243-fold higher risk of death than did patients without this variation based on regression analysis. Therefore, this characteristic of septic patients should be given attention, and early intervention must occur. At the same time, we can observe the dynamics in the sTREM-1 levels to detect changes in patients’ conditions and determine whether treatment is effective. Through logistic regression, sTREM-1, APACHE II, and sTREM-1 + APACHE II were worked into an ROC curve. The sTREM-1 + APACHE II combination proved to be the most reliable for prognostic assessment. With 0.5467 as the cutoff point, sensitivity was 0.854, and specificity was 0.711. The sTREM-1 + APACHE II combination may improve the diagnostic performance. It is believed that sTREM-1 may be a good biomarker of sepsis.
The present study has some limitations. (1) The sample size was not sufficiently large to demonstrate associations between TREM-1 genetic polymorphisms and sepsis susceptibility and its prognosis, including only 79 patients with sepsis and 80 healthy individuals who received a physical examination. It is necessary to continue recruiting larger sample sizes to validate our conclusions. (2) The significant SNP rs2234237 associated with prognosis of sepsis may not be causal, and the functional relation between the genomic region and prognosis of sepsis is unknown; further functional studies, such as fine mapping and expression quantitative trait loci (eQTLs), are needed to explore the relationship between TREM-1 gene polymorphisms and its actual expression in serum. (3) Only three SNP loci were detected among the four exons of TREM-1. We speculate that these may be the only three SNPs relevant to this Chinese cohort with sepsis. Further study is necessary to identify new SNPs that might be further related to sepsis susceptibility and prognosis through the full sequencing of TREM-1. Therefore, polymorphisms located in promoter regions, intron sites, or intron–exon splice junctions are also important [25]. (4) TREM-1 is a key signaling molecule in inflammatory pathways and is closely linked to other inflammatory molecules. It would be necessary to search new genes that have close relationship with TREM-1 and then analyze the gene-to-gene interaction and functional relevance of these genes [26].
In summary, the dynamic change in serum sTREM-1 may be a more accurate and valuable parameter for the dynamic assessment of prognosis. There is no association between the variations rs2234246 and rs2234237 within the TREM-1 gene and susceptibility to sepsis. However, the TREM-1 rs2234237 polymorphism is associated with increased 28-day mortality in patients with sepsis. Because of the limitations of our sample size, further studies are to be expected on sTREM-1 polymorphism. TREM-1 may be a biomarker of sepsis prognosis.
References
Holmes, C.L., J.A. Russell, and K.R. Walley. 2003. Genetic polymorphisms in sepsis and septic shock: Role in prognosis and potential for therapy. Chest 124: 1103–1115.
Levy, M.M., M.P. Fink, J.C. Marshall, E. Abraham, D. Angus, D. Cook, J. Cohen, S.M. Opal, J.L. Vincent, and G. Ramsay. 2003. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Critical Care Medicine 31: 1250–1256.
Hotchkiss, R.S., and I.E. Karl. 2003. The pathophysiology and treatment of sepsis. The New England Journal of Medicine 348: 138–150.
Lin, M.T., and T.E. Albertson. 2004. Genomic polymorphisms in sepsis. Critical Care Medicine 32: 569–579.
Namath, A., and A.J. Patterson. 2009. Genetic polymorphisms in sepsis. Critical Care Clinics 25: 835–856. x.
Bouchon, A., J. Dietrich, and M. Colonna. 2000. Cutting edge: Inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. Journal of Immunology 164: 4991–4995.
Klesney-Tait, J., I.R. Turnbull, and M. Colonna. 2006. The TREM receptor family and signal integration. Nature Immunology 7: 1266–1273.
Tessarz, A.S., and A. Cerwenka. 2008. The TREM-1/DAP12 pathway. Immunology Letters 116: 111–116.
Sharif, O., and S. Knapp. 2008. From expression to signaling: Roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology 213: 701–713.
Bouchon, A., F. Facchetti, M.A. Weigand, and M. Colonna. 2001. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410: 1103–1107.
Bone, R.C., R.A. Balk, F.B. Cerra, R.P. Dellinger, A.M. Fein, W.A. Knaus, R.M. Schein, and W.J. Sibbald. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 101: 1644–1655.
Shi, Y.Y., and L. He. 2005. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research 15: 97–98.
Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, He L, Shi Y. 2009 A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio-x.cn). Cell Res 19: 519–523.
Sole, X., E. Guino, J. Valls, R. Iniesta, and V. Moreno. 2006. SNPStats: A web tool for the analysis of association studies. Bioinformatics 22: 1928–1929.
Barrett, J.C., B. Fry, J. Maller, and M.J. Daly. 2005. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265.
Ardlie, K.G., L. Kruglyak, and M. Seielstad. 2002. Patterns of linkage disequilibrium in the human genome. Nature Reviews Genetics 3: 299–309.
Gabriel, S.B., S.F. Schaffner, H. Nguyen, J.M. Moore, J. Roy, B. Blumenstiel, J. Higgins, M. DeFelice, A. Lochner, M. Faggart, S.N. Liu-Cordero, C. Rotimi, A. Adeyemo, R. Cooper, R. Ward, E.S. Lander, M.J. Daly, and D. Altshuler. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229.
Zhang, J., D. She, D. Feng, Y. Jia, and L. Xie. 2011. Dynamic changes of serum soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) reflect sepsis severity and can predict prognosis: A prospective study. BMC Infectious Diseases 11: 53.
Su, L.X., L. Feng, J. Zhang, Y.J. Xiao, Y.H. Jia, P. Yan, D. Feng, and L.X. Xie. 2011. Diagnostic value of urine sTREM-1 for sepsis and relevant acute kidney injuries: A prospective study. Critical Care 15: R250.
Colonna, M. 2003. TREMs in the immune system and beyond. Nature Reviews Immunology 3: 445–453.
Ornatowska, M., A.C. Azim, X. Wang, J.W. Christman, L. Xiao, M. Joo, and R.T. Sadikot. 2007. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. American Journal of Physiology. Lung Cellular and Molecular Physiology 293: L1377–L1384.
Gomez-Pina, V., A. Soares-Schanoski, A. Rodriguez-Rojas, C. Del Fresno, F. Garcia, M.T. Vallejo-Cremades, I. Fernandez-Ruiz, F. Arnalich, P. Fuentes-Prior, and E. Lopez-Collazo. 2007. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. Journal of Immunology 179: 4065–4073.
Chen, Q., H. Zhou, S. Wu, H. Wang, C. Lv, B. Cheng, G. Xie, and X. Fang. 2008. Lack of association between TREM-1 gene polymorphisms and severe sepsis in a Chinese Han population. Human Immunology 69: 220–226.
Jung, E.S., S.W. Kim, C.M. Moon, D.J. Shin, N.H. Son, E.S. Kim, H.J. Lee, S.P. Hong, T.I. Kim, W.H. Kim, and J.H. Cheon. 2011. Relationships between genetic polymorphisms of triggering receptor expressed on myeloid cells-1 and inflammatory bowel diseases in the Korean population. Life Sciences 89: 289–294.
Tabor, H.K., N.J. Risch, and R.M. Myers. 2002. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nature Reviews Genetics 3: 391–397.
Cordell, H.J. 2009. Detecting gene–gene interactions that underlie human diseases. Nature Reviews Genetics 10: 392–404.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dr. Longxiang Su and Dr. Changting Liu have contributed equally to this work.
The trial was registered under ClinicalTrial.gov identifier NCT01490424.
Rights and permissions
About this article
Cite this article
Su, L., Liu, C., Li, C. et al. Dynamic Changes in Serum Soluble Triggering Receptor Expressed on Myeloid Cells-1 (sTREM-1) and its Gene Polymorphisms are Associated with Sepsis Prognosis. Inflammation 35, 1833–1843 (2012). https://doi.org/10.1007/s10753-012-9504-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-012-9504-z