Abstract
Sex determination systems in Animalia encompass a diverse array of genes, functioning in complex regulatory networks. This diversity is even pronounced within taxonomic ranks and the crustacean Order Decapoda is no exception. The commercial importance of the decapods and the ambition to develop their potential in aquaculture has resulted in the necessity to better understand the processes of sexual development. However, due to a lagging understanding of the regulation of sex determination, systems characterised in other model species often serve as the basis for these investigations. This work presents a collated summary of the current information of sex determination in Decapoda, including all determined chromosomal mechanisms and identified “sex-regulator” homologues, often focussing on genes characterised in the model arthropod Drosophila melanogaster (namely Sxl, Tra, Tra-2, Fru and Dsx), the nematode Caenorhabditis elegans (Fem-1 and Mab-3) and Mammalia (Sry, Sox9, Foxl2 and Dmrt1). Although homologue analyses such as these offer a good method to guide investigations in non-model species, the low conservation and variability of sex determination systems cautions against the assumption of conserved functionality. Thus, we propose a better suited approach to guide studies into sex determination in Decapoda, primarily relating to the functionally conserved sex regulators, the Dmrts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Sexual differentiation in Malacostraca
The integrated signalling cascades of sexual development make it difficult to clearly distinguish the processes of sex determination from sexual differentiation (Matson & Zarkower, 2012). However, in the decapods (Crustacea, Malacostraca), as members of the Malacostraca, the onset of male sexual differentiation is more clearly defined, due to the unique involvement of a male-specific endocrine gland known as the androgenic gland (AG). First characterised in the sister Order, Amphipoda (Crustacea, Malacostraca), the AG was determined to be the sole regulator of male primary and secondary sexual differentiation (Charniaux-Cotton, 1954). It was then in a second sister Order, Isopoda, that the specific regulatory hormone was first isolated, defined as an insulin-like peptide and so termed the insulin-like androgenic gland hormone (AGH) (Hasegawa et al., 1987; Martin et al., 1999; Okuno et al., 1999; Suzuki, 1999).
As a sex-differentiating factor shared by all malacostracans, an understanding of the regulation of male sexual differentiation in Decapoda soon followed, now known to be regulated through a unique developmental axis known as the X-organ–sinus-gland neuroendocrine complex (XO–SG)—AG—Testis axis. In brief, the XO–SG (located in the eyestalk) secretes an array of neuropeptides including the gonad-inhibiting hormone (GIH) (also known as vitellogenesis-inhibiting hormone in females) and moult-inhibiting hormone (MIH) (Rodríguez et al., 2007; Nagaraju, 2011), both belonging to the crustacean hyperglycaemic hormone (CHH) family. In males, these neurohormones do not act directly on the gonad but instead modulate the proliferation of the AG; in the absence of their inhibitory signal, the AG develops bilaterally on each of the posterior sections of the sperm ducts (Charniaux-Cotton, 1954; Charniaux-Cotton, 1958; Charniaux-Cotton et al., 1966; Sagi et al., 1997). Once established, the male-specific AG is solely responsible for the synthesis and secretion of the insulin-like hormone, named IAG in the decapods, which in turn stimulates both testicular differentiation (Rodríguez et al. 2007) and the broad tissue effects of male sexual dimorphism through to full reproductive capacity (Manor et al., 2007; Martin et al., 1999; Okuno et al., 1999; Rosen et al., 2010; Ventura et al., 2009; reviewed in Ventura et al. (2011a)).
The resulting sexual systems
Although the male sex-differentiating influence of IAG is a unifying feature of malacostracans, the hormone’s functional interaction with the broader networks of sexual development is not so conserved. While most decapods are gonochoristic, meaning that they develop into one distinct sex, multiple sexual systems have been described. First, there is a prevalence of complex gonochoristic species, where, although sex is genetically determined by a distinct genotype, simultaneous intersex (both male and female) phenotypes occur. This is particularly pronounced in certain Infraorders, such as the Astacidea (Grilo and Rosa, 2017); an example being the red-claw crayfish (Sagi et al., 1996), which can present as one of the seven distinct intersex phenotypes, all of which develop from a ZW (female) genotype (Parnes et al., 2003).
There are also examples of protandrous hermaphroditic species, where individuals are born as males and later develop into females. This appears to be common in the Caridea, such as the Peppermint shrimp (Lysmata wurdemanni) (Bortolini & Bauer, 2016; Zhang et al., 2017) and is a process regulated through the sex-differentiating effects of IAG (Zhang et al. 2017). Such hermaphroditism has also been documented in the Thalassinidae and Anomura (Subramoniam, 2017). To complicate things further, the shrimp species, L. wurdemanni, can also exist as a simultaneous hermaphrodite, where an individual can function as a male and a female at any one time (a protandrous simultaneous hermaphrodite) (Bauer & Holt, 1998).
More staggeringly, the Order also contains one documented parthenogenic species. The marbled crayfish (Procambarus fallax forma virginalis), only exists as female, of which all offspring is genetically identical to the mother and therefore, female (Scholtz et al., 2003; Vogt et al., 2015). Finally, to truly demonstrate the plasticity of the IAG-mediated system, one should consider the sister Order, Isopoda, where the influence of the AG is superseded by the feminisation effects of the bacterial endosymbiont, Wolbachia. This parasitism is thought to prevent the initial differentiation of the male-specific AG (Rigaud et al., 1997; Bouchon et al., 2008; Cordaux et al., 2011), allowing for female development. So dramatic are these AG-disruptive effects, not only do they result in total feminisation of ZZ males but ultimately, the loss and re-emergence of the W chromosome (Leclercq et al., 2016; Cordaux & Gilbert 2017).
How is this diversity in sexual systems achieved?
Based on our understanding of the conserved mode of sexual differentiation amongst these species, as well as the resulting diversity of sexual systems, the regulatory axis of the malacostracan AG, although ubiquitous, is certainly labile. This suggests that the preceding regulation of sex determination, which serves as the foundation of sexual development within each species, may be responsible for the diversity of sexual systems observed. Sex determination mechanisms are known to be highly divergent across Animalia, even across closely related species, particularly amongst those genes acting at the very top of the genetic cascades, the master sex determinants (Bachtrog et al., 2014). This variability is owing to the relaxed evolutionary restraint on sex determination genes (Meiklejohn et al., 2003; Parsch & Ellegren 2013), readily apparent in the increased rates of several fundamental evolutionary phenomena: (1) Gene duplication and neofunctionalization: examples are found in the master sex determinants of medaka (Oryzias latipes) (Matsuda et al., 2002; Nanda et al., 2002) and the African clawed frog (Xenopus laevis) (Yoshimoto et al., 2008), both of which have evolved through the sex-specific gene duplication of autosomal Dmrt1 genes. Neofunctionalization can result in the recruitment of entirely unrelated genes, such as the immune-related gene duplicate SdY, acting as the master sex determinant in the rainbow trout (Yano et al., 2012) and more staggeringly still, the Wolbachia bacterial element gained through horizontal gene transfer to become the master female sex determinant in the common pill bug (Armadillidium vulgare) (Leclercq et al., 2016). (2) Hierarchical rearrangement: resulting in the loss of function between seemingly well conserved genes across species, such as the master sex determinant Sxl in Drosophila melanogaster, a gene which has been expelled from the master regulatory role in the fly species Musca domestica and Ceratitis capitata (Meise et al., 1998), where it actually lacks any role in sex determination. (3) The increased occurrence of functional mutations: like the single nucleotide mutation in the male pufferfish (Takifugu rubripes) anti-Müllerian hormone receptor (Amhr2), a polymorphism which now acts as the master male sex-determining signal (Kamiya et al., 2012). (4) Altered translational and temporal gene expression patterns (Bachtrog et al., 2014; Beukeboom & Perrin 2014) and (5) epigenetic effects (Piferrer 2013). These factors culminate to make the characterisation of each sex determination system highly cryptic.
Thus, considering the diversity of sexual systems amongst Decapoda and the apparent plasticity of IAG endocrinology, it is apparent that to fully understand the regulation of sexual development, we must integrate the genetic networks that serve as the basis of each system. Furthermore (knowing the evolutionary characteristics and diversification of sex determination genes) to gain an accurate understanding of this highly complex regulatory process, this characterisation must be tackled on a species-by-species basis. In response, this review has collated the current research findings regarding the molecular basis of sex determination in Decapoda, in an attempt to serve both as a reference for preliminary investigations and to provide a critical evaluation of the current data. To provide evolutionary context, we have included the sister Order Isopoda (Crustacea, Malacostraca), which shares the commonality of the AG and the model crustacean species Daphnia pulex and D. magna (Crustacea, Branchiopoda) from the sister Class Branchiopoda. It is thought that Insecta evolved from the freshwater branchiopod crustaceans around 410 MYA (Glenner et al., 2006). Hence the Daphnia sp. are included as a phylogenetic link between the decapods and the insects (Hexapoda, Insecta), from which a significant proportion of the model sex determination genes have been characterised.
The genetic (chromosomal) modes of sex determination characterised to date are presented, followed by a list of candidate genes implicated in sex determination in Decapoda. These genes were primarily identified through transcriptomic homologue screening and therefore the list is heavily biased by systems characterised in the model species D. melanogaster, Caenorhabditis elegans and Mammalia. Given the high evolutionary rates of sex determination systems, this review highlights the significance of functional genomics to ensure that genes are assigned with appropriate functions. To reiterate such, we have included a brief functional description of the genes most commonly targeted for homologue analyses. It is of note that there is only a single gene family common to all three model systems: namely Drosophila-Dsx, C. elegans Mab-3 and Mammalia-Dmrt1 (denoted with an * in Table 2). These genes are from the DM domain transcription factor family, collectively known as the doublesex and male abnormal-3-related transcription factors (Dmrts).
Functional definitions of sex-regulator genes in model species
The fruit fly, Drosophila melanogaster (Arthropoda, Insecta)
Sex lethal: pre-mRNA splicing protein
Sex lethal (Sxl) is a splicing factor, defined by an RNA-binding domain. This gene is responsive to the higher X: Autosome chromosome ratio in females. The X chromosome gene products (e.g. Runt, Sisterless-A and Sisterless-B) bind to the Sxl promoter and induce its activation. Thus, Sxl is the master sex determinant for female development. Its primary target is Tra mRNA (Hashiyama et al., 2011; Beukeboom & Perrin, 2014).
Transformer: pre-mRNA splicing protein
Transformer (Tra) is an mRNA splicing factor, defined by an RNA-recognition motif and an arginine/serine-rich domain followed by a proline-rich region. The action of Sxl results in the active splice variant of Tra in females. Tra then acts as a downstream splicing factor, regulating female-specific splicing of target RNAs (Kulathinal et al., 2003).
Transformer-2: pre-mRNA splicing protein
Transformer-2 (Tra-2) is an mRNA splicing factor, defined by an RNA-recognition motif and arginine/serine-rich domain. Tra-2 is also a downstream splicing factor, which is constitutively produced (in both males and females) but only in females can it act in concert with Tra to regulate female-specific splicing of target RNAs. A primary target of Tra and Tra-2 is Dsx mRNA (Amrein et al., 1990).
Doublesex: DM domain, zinc finger protein (transcription factor)*
Doublesex (Dsx) is a transcription factor defined by a zinc finger domain, termed the DM domain. In the presence of the Tra–Tra-2 complex, the female splice variant of Dsx is produced, generating the DsxF protein. DsxF works in concert with an array of regulatory genes as the major effector of sexual development (Hoshijima et al., 1991; Beukeboom & Perrin 2014).
Fruitless: zinc finger protein (transcription factor)
Fruitless (Fru) is a transcription factor defined by its zinc fingers. Fru is a male-promoting gene, regulating development of the male central nervous system and male sexual behaviour. Fru is spliced in the absence of the female Tra–Tra-2 complex, carried out by non-sex-specific splicing machinery (Billeter et al., 2006).
The nematode, Caenorhabditis elegans (Nematoda, Chromadorea)
Feminization of XX and XO animals-1: Ankyrin repeat-containing protein
Feminization of XX and XO animals-1 (Fem-1) is a second messenger protein, defined by ankyrin repeats. It acts in male sex determination, as a signal-transducing regulator between the membrane receptor Tra-2 (not to be confused with the mRNA splicing factor, Tra-2 in Drosophila) and the transcription factor Tra-1. It works in concert with Fem-2 and Fem-3 (Yi et al., 2000; Haag 2005).
Male Abnormal-3: DM domain, zinc finger protein (transcription factor)*
Male Abnormal-3 (Mab-3) is a transcription factor defined by a novel zinc finger domain, termed the DM domain. In males, Her-1 binds the Tra-2 receptor, releasing Fem-1,2,3 which then sequester Tra-1. This in turn activates the major effector Mab-3. Mab-3 is vital for the male sexual differentiation of the peripheral nervous system and the intestine, causing the repression of vitellogenesis (Yi et al., 2000; Haag 2005).
Mammals (Chordata, Mammalia)
Sex-determining region Y: HMG-box DNA-binding protein (transcription factor)
Sex-determining region Y (Sry) is the master sex determinant in males. Sry, along with SF-1, binds and activates Sox9. Its function is transient, after its activation of Sox9, other major effector genes are responsible for the continuity of its signal (Sinclair et al., 1990; Matson et al., 2011).
Sex-determining region Y-box 9: HMG-box DNA-binding protein (transcription factor)
Sex-determining region Y-box 9 (Sox9) is a transcription factor defined by its Sry-related HMG-box. Once activated by Sry it is responsible for upregulating the expression of the male-promoting Dmrt1 via Fgf9. These genes then function in an auto-regulating loop, maintaining their own expression, stimulating (and maintaining) the male-specific programme of development (Jakob & Lovell-Badge 2011; Matson et al., 2011).
Doublesex and male abnormal-3-related transcription factor 1: DM domain, zinc finger protein (transcription factor)*
Doublesex and male abnormal-3-related transcription factor 1 (Dmrt1) is a transcription factor defined by a novel zinc finger domain, termed the DM domain. It is upregulated in response to Sox9 via Fgf9. Dmrt1 works in concert with an array of regulatory genes as the major effector of male sexual differentiation, specifically testicular development and maintenance. It also acts to inhibit the female developmental pathway (e.g. by suppressing Foxl2) (Matson et al., 2011).
Forkhead box L2: Forkhead box DNA-binding protein (transcription factor)
Forkhead box L2 (Foxl2) is a transcription factor that is defined by a unique DNA-binding domain. In the absence of the Sry-driven expression of Sox9, Foxl2 expression ensues and acts to inhibit the male pathway, whilst promoting the female pathway through the action of Rspo1, Wnt4 and B-catenin (Matson et al., 2011).
Acting at the top of the cascade: the sex chromosomes
The summary presented in Table 1 provides clear demonstration of the diversity of sex determination modes present in Crustacea. Both D. pulex (Crease et al., 1989; Chen et al., 2014) and D. magna (Kleiven et al., 1992; Kato et al., 2011) are subject to environmental sex determination, through cyclic parthenogenesis. This contrasts to the decapods where, thus far, genetic sex determination is common across the Order, although both XY/XX (heterogametic male) and ZW/ZZ (homogametic male) mechanisms exist sporadically across families (Chandler et al., 2016b). In Isopoda, male homogamety (ZZ) seems to be more common than male heterogamety (XY) (Becking et al., 2017). There is also strong evidence for frequent sex chromosome turnover, with an estimated three to thirteen heterogametic transitions occurring, accounting for the dramatic XY and ZW diversity observed within Genera (Becking et al., 2017).
This suggests that the disruptive effects of Wolbachia have had significant consequence on the evolution of sex determination mechanisms in isopods. Furthermore, the dramatic influence of Wolbachia has given rise to a third, distinct mode of sex determination: cytoplasmic. Genetic and environmental (including social) modes of sex determination are both extensively characterised across species (Beukeboom & Perrin 2014), but cytoplasmic sex determination appears to be specific to the Crustacea, resulting from the feminising effect of parasites (Rigaud et al., 1997). Hence, this mode of inheritance described in the isopods, accounts for a third mode of sex determination in the already diverse regulation of sexual development in Crustacea.
One unifying factor shared by the malacostracans, is the potential for sex to be manipulated, whether by parasitic infection, acting to prevent the differentiation of the AG in males (Isopoda) (Rigaud et al., 1997; Bouchon et al., 2008; Cordaux et al., 2011), or the RNAi-induced suppression of IAG expression (Decapoda) (Ventura et al., 2012); both causing fully functional feminisation. Taken together, this suggests that both sexes, irrelevant of their genetic background, have the genetic potential to develop as male or female, suggesting that the sex chromosomes must be predominantly homologous. This is further supported by the viability of WW females and YY males observed in both isopods (Rigaud et al., 1997; Becking et al., 2017) and decapods (Shpak et al., 2016), therefore suggesting that the sex chromosomes of these Orders are in the primary stages of differentiation (Charlesworth 1991; Rigaud et al., 1997), perhaps only differing in the one master sex-determining gene responsible for flipping the male/female switch.
The mechanism: evaluating the role of sex-regulator genes
The data presented in Table 2 clearly illustrate how transcriptomics (and genomics) have greatly advanced the identification of new gene families in the decapods. From an evolutionary perspective, this has enhanced our understanding of the divergence of these genes both within the Order and across Animalia. However, with regards to the functional context of sexual development, these data must be interpreted with a critical understanding of the evolutionary rates that define sex-determining genes.
The Tra homologues provide an apt example: the Tra genes are known to be highly divergent in sequence even amongst Drosophila sp. (Kulathinal et al., 2003); in other non-Drosophilid fly species such as the housefly (M. domestica) (Hediger et al., 2010) and medfly (C. capitata) (Pane et al., 2002) the Tra orthologues show functional divergence, having evolved up the hierarchy (expelling Sxl) to adopt the master sex-determining role; and in the distantly related decapods, thus far Tra homologues appear to be lacking.
There is some level of conservation within Insecta, as a functional Tra orthologue (termed Feminiser, Fem) also acts as the female master sex determinant in the honey bee (Apis mellifera) (Hasselmann et al., 2008). Fem lacks complete conservation of the sequence motifs described in Drosophila but does contain the same arginine/serine and proline-rich domain organisation and one conserved sequence motif, also described in C. capitata (Pane et al., 2002). Although diverged in both sequence and function across these three species (telling of significant independent evolution over the ~ 300 million years that separates the Orders), the conserved role of the three orthologues is indicative of a Tra-based ancestral pathway of sex determination in Insecta (Hasselmann et al., 2008). Furthermore, the discovery of Fem suggests that the complementary sex determiner (csd) gene may not be the universal master sex determinant across hymenopteran insects (bees, ants and wasps), as was previously thought (Hasselmann et al., 2008; Heimpel & Boer 2008).
It is interesting to note that the brachiopods D. pulex (Chen et al., 2014) and D. magna (Kato et al., 2010) do have Tra homologues; however, both show even greater divergence in domain organisation from that of Drosophila Tra. Furthermore, it is only in D. magna that a function in sexual development has been suggested, not in the female sex determination pathway (as with the insect sp.), but in the maintenance of male phenotype (Chen et al., 2014). This evolutionary emergence and divergence of Tra is in support of the proposed decapod—branchiopod—hexapod trajectory of the Pancrustacea (Glenner et al., 2006). A similar story is true of the highly conserved Sxl splice variants, which although seemingly well conserved in sequence throughout Insecta and Decapoda (Table 2), have not retained conserved functionality. These evolutionary patterns exemplify why the functional conservation of the genes summarised in this work need to be appropriately evaluated; a conserved function to that described in each model species should not be assumed a priori.
Dmrts: the functionally conserved sex regulators
There is however, a gene family that does display the functional conservation that appears to be somewhat assumed in other cases: the Dmrts. Indeed, the Dmrts are the only gene family with a conserved function in sex determination across Animalia (Kopp 2012; Beukeboom & Perrin 2014), being identified in all investigated species to date, with the only exception being the sponge Amphimedon queenslandica (Wexler et al., 2014) (which may well reflect its lack of tissue differentiation). There has been good progress in the identification of Dmrts in the decapods, with homologues identified in seven species comprising the Penaeidae (prawns), Palinuridae (lobsters), Palaemonidae (shrimp) and the Portunidae (crab). Although from our data collation, they appear yet to be identified in the isopods. Indeed, this work may suggest that the characterisation of the Dmrts in Isopoda is a promising avenue of further study regarding the mechanism by which the f element disrupts the native pathway of sexual development.
The functional conservation of the model genes characterised in D. melanogaster (Dsx), C. elegans (Mab-3) and Mammalia (Dmrt1) is readily illustrated in D. magna. In this branchiopod, a Dmrt homologue (Dsx1) was determined to be the master male sex determinant (Kato et al., 2011). It is of note however, that four additional Dmrts were also identified, Dmrt11E, Dmrt93B, Dmrt99B (Kato et al., 2008) and Dsx2 (Kato et al., 2011) and determined to have no clear function in sexual development. This highlights the critical importance of the functional analyses to reliably determine the explicit regulatory role of Dapma–Dsx1. The functional conservation of the Dmrts continues in the decapods, as recently a Y-chromosome-linked Dmrt was identified in the Eastern spiny lobster (S. verreauxi) (Chandler et al., 2017). This gene, termed as Sv–iDMY, offers the third example of a sex-linked Dmrt across Animalia (joining medaka DMY and frog DM-W) and is the first example in an invertebrate. As is the case in both medaka and frog, our functional analyses suggest that Sv-iDMY has evolved as the master sex determinant in this decapod species (Chandler et al., 2017).
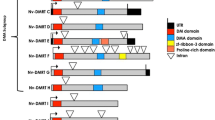
To better illustrate the evolutionary and mechanistic features of the Dmrt family, Fig. 1 presents the phylogeny of all the available decapod Dmrt sequences, as well as those from D. pulex and D. magna and the model Dsx, Mab-3 and Dmrt1. Figure 1A shows the full protein sequence, highlighting the clear clustering of Dmrt subclasses, but more significantly, the pronounced variation in domain organisation outside of the DM domain itself. In contrast, Fig. 1B shows the phylogeny of the DM domains only, emphasising the extreme conservation of the DNA-binding domain that defines the family. Taken together, Fig. 1 demonstrates how the mechanistic diversity of this family is achieved: routed in the sequence and domain variation outside of the DM domain itself. The only exception being the specialised repeat DM domain (defined as the Dmrt1 domain) of H. sapiens Dmrt1, which defines all vertebrate Dmrt1 homologues and is not present in any invertebrates thus far (Wexler et al., 2014).
Neighbour-joining phylogram and domain illustration of Dmrts in Decapoda, alongside model comparative species. A Displays phylogeny of full length Dmrt peptides, TAD predictions were conducted using the Nine Amino Acids Transactivation Domain (9aaTAD) Prediction Tool (Piskacek et al., 2007) as described in (Chandler et al., 2017). B Displays phylogeny of isolated DM domains. Throughout, the Dmrt11E cluster is highlighted in green, 99B cluster in purple, 93B cluster in blue and Dsx cluster in orange. Domain illustration and taxonomy keys are provided in figure. Bootstrap values are shown at each node and were performed with 1000 replicates to ensure reliability; scale bar indicates number of amino acid substitutions per site. All decapod GenBank Accession Numbers are given in Table 2, with the addition of H. sapiens Dmrt1 (Q9Y5R6.2), D. melanogaster Dmrt11E (AAF48261.2), Dmrt99B (AAF56919.1), Dmrt93B (AAF55843.1), DsxM (AAF54169.1), DsxF (AAN13385.1) and C. elegans Mab-3 (O18214.1)
The mechanistic diversity of the Dmrts
A pronounced example of the domain variation illustrated in Fig. 1A is the DMA domain. This domain has been identified in multiple animal taxa, including the decapods, cnidarians and the ctenophores and tends to be associated with the Dmrt93B and 99B clades, lacking from Dsx homologues (Wexler et al., 2014). However, a function for this domain is yet to be determined. The transactivation domain (TAD) however, has an explicit functional significance, responsible for the transcriptional activation of the bound Dmrt-DNA complex (Mapp & Ansari 2007; Piskacek et al., 2007; Beukeboom & Perrin 2014). Hence its occurrence has dramatic potential to shape the mechanistic action of each Dmrt. In cases where the TAD is absent, such as Drosophila DsxF (Fig. 1A), additional coactivators (hermaphrodite (HER) and intersex (IX)) are required to achieve transcriptional activity (Pultz & Baker 1995; Garrett-Engele et al., 2002). Or indeed, in the absence of such coactivators, there is potential for the Dmrt to exert suppressive mechanisms, such as the antagonistic mechanisms described for DM-W (Yoshimoto et al., 2006; Yoshimoto et al., 2008) and iDMY (Chandler et al., 2017). When one considers the functional significance of each additional domain, it becomes apparent how the seemingly well conserved Dmrts adopt the diversity of mechanisms observed across species. Moreover, this domain variation points towards the varied necessity to recruit additional genes to support functionality (explaining the diversity of described sex determination networks), from which the integration of co-regulatory genes can be better elucidated.
The DM domain that defines the Dmrts
In contrast, that depicted in Fig. 1B, reiterates the dramatic sequence conservation of the DNA-binding domain that defines the Dmrt family; the DM domain is that responsible for binding the target DNA. This commonality across species therefore suggests that DNA-binding motifs, which constitute the response elements targeted by each Dmrt, must also be relatively well conserved. Indeed, such conservation of the Dmrt-binding motif has already been demonstrated amongst Drosophila sp. (Luo et al., 2011). This conservation therefore offers a perfect method by which to bridge the gap, identifying the regulatory genes that act downstream of these key node regulators, through the identification of the conserved DM domain-binding motif. This has already been done in Mammalia (Murphy et al., 2010), X. laevis (Herpin et al., 2010) and Drosophila (Luo et al., 2011), informing of the auto and cross-regulatory effects of the Dmrt genes (Murphy et al., 2010), the mechanisms by which they coordinate their own expression (Herpin et al., 2010), as well as the extent of divergence of the DNA-binding motif itself (Luo et al., 2011). Similar promoter analysis in the decapods would facilitate the identification of genomic-binding motifs, from which candidate response genes can be elucidated and fully investigated through the more telling expression patterns gained through transcriptomics. Similar analyses have been conducted on a gene-by-gene basis, such as that conducted for the genomic regions of IAG in S. paramamosain (Zhang et al., 2014) and M. nipponense (Ma et al., 2016b), identifying a range of transcription factor binding sites in the 5′ promotor region (Table 2). Although informative, when one considers the interconnected nature of genetic networks, a genome-wide analysis would prove a far more powerful tool to accurately assemble the regulatory map of sex determination.
Future directions
In summary, this review serves as a detailed reference for those interested in sex determination in Decapoda, offering a list of putative candidates that (in most cases) can act to guide further functional investigations. However, we also intend for the critical assessment presented throughout this summary, to highlight the risks associated with the arbitrary identification of target genes without an appropriate consideration of functional conservation. Thus, we urge, that when evaluating each homologous candidate, one considers the divergence that defines the rapidly evolving genes of sex determination.
The spatial expression analyses that constitute the majority of the molecular studies presented, are a sound starting point for functional investigations; they are however indicative and not conclusive. This, considered with the fact that Decapoda is a non-model Order, advises that the best approach to advance current understanding should build on what is known in these species, rather than relying on that characterised in others. As described above, the well characterised (and conserved) function of IAG in sexual differentiation, offers an ideal functional basis with which to integrate putative candidates.
Following this, we advocate for a greater emphasis to be placed on the Dmrt genes, which of all the candidate homologues, are known to have the most significant functional conservation. Work should aim to thoroughly investigate the spatial and temporal expression of the Dmrts in Decapoda. The use of genome-guided promoter analyses would inform of Dmrt interconnectivity, as well as facilitate the identification of, as yet, unknown response genes. In conjunction with (or, as is often the case, in the absence of) genomic resources, the use of RNAi knockdown is a well-suited tool to begin to understand the transcriptional integration of each Dmrt with each other, with IAG and with the regulatory elements of IAG, such as the CHH hormones (e.g. GIH) and the TKIR receptor (Aizen et al., 2016; Sharabi et al., 2016). Yu et al., (2014), present a sound example of such, using RNAi approaches to determine that of two Dmrts (both of which showed notable testicular-biased expression), Dmrt11E but not Dmrt99B, was functionally involved in the regulation of IAG expression. We therefore conclude with Fig. 2, presenting our revised suggestion of the regulatory axis of sexual development in Decapoda, emphasising the network-like qualities of the system, rather than the linear ones described at present. In following these suggested avenues of future research, we can begin to identify and integrate associative genes, assembling a functionally valid understanding of sexual development in the decapods.
A Illustration of the evolutionary relationships of the focal taxa discussed in this work, emphasising the extreme evolutionary distance over which the Dmrts are conserved, highlighted by the blue bar (note that the Dmrts remain yet to be identified in the isopods). The shared role of IAG in the malacostracan Orders is highlighted in orange. B Revised depiction of the XO–SG–AG–TS axis of the Malacostraca (Decapoda), with inclusion of the Dmrts. Proven interactions are displayed in black line and those that hold a promising avenue for future work are shown in grey. The red-ended line indicates the inhibitory signal of GIH/MIH; all endocrine factors are shown in orange and tissues in green
References
Aizen, J., J. C. Chandler, Q. P. Fitzgibbon, A. Sagi, S. C. Battaglene, A. Elizur & T. Ventura, 2016. Production of recombinant insulin-like androgenic gland hormones from three decapod species: in vitro testicular phosphorylation and activation of a newly identified tyrosine kinase receptor from the Eastern spiny lobster, Sagmariasus verreauxi. General and Comparative Endocrinology 229: 8–18.
Amrein, H., T. Maniatis & R. Nothiger, 1990. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. The EMBO Journal 9: 3619–3629.
Bachtrog, D., J. E. Mank, C. L. Peichel, M. Kirkpatrick, S. P. Otto, T.-L. Ashman, M. W. Hahn, J. Kitano, I. Mayrose, R. Ming, N. Perrin, L. Ross, N. Valenzuela & J. C. Vamosi, 2014. Sex determination: why so many ways of doing it? PLoS Biology 12: e1001899.
Bauer, R. T. & G. J. Holt, 1998. Simultaneous hermaphroditism in the marine shrimp Lysmata wurdemanni (Caridea: Hippolytidae): an undescribed sexual system in the decapod Crustacea. Marine Biology 132: 223–235.
Becking, T., I. Giraud, M. Raimond, B. Moumen, C. Chandler, R. Cordaux & C. Gilbert, 2017. Diversity and evolution of sex determination systems in terrestrial isopods. Scientific Reports 7: 1084.
Benzie, J. A. H., M. Kenway & E. Ballment, 2001. Growth of Penaeus monodon x Penaeus esculentus tiger prawn hybrids relative to the parental species. Aquaculture 193: 227–237.
Beukeboom, L. & N. Perrin, 2014. Molecular mechanisms of sex determination. In Beukeboom, L. & N. Perrin (eds.), The Evolution of Sex Determination. Oxford University Press, Oxford: 37–76.
Billeter, J. C., A. Villella, J. B. Allendorfer, A. J. Dornan, M. Richardson, D. A. Gailey & S. F. Goodwin, 2006. Isoform-specific control of male neuronal differentiation and behavior in drosophila by the fruitless gene. Current Biology 16: 1063–1076.
Bortolini, J. L. & R. T. Bauer, 2016. Persistence of reduced androgenic glands after protandric sex change suggests a basis for simultaneous hermaphroditism in a caridean shrimp. The Biological Bulletin 230: 110–119.
Bouchon, D., R. Cordaux & P. Greve, 2008. Feminizing Wolbachia and the evolution of sex determination in isopods. In Bourtzis, K. & T. A. Miller (eds.), Insect Symbiosis. Taylor and Fransis, New York: 273–294.
Chandler, J. C., J. Aizen, A. Elizur, S. C. Battaglene & T. Ventura, 2016a. Male sexual development and the androgenic gland: novel insights through the de novo assembled transcriptome of the eastern spiny lobster, Sagmariasus verreauxi. Sexual Development 9: 338–354.
Chandler, J. C., J. Aizen, Q. P. Fitzgibbon, A. Elizur & T. Ventura, 2016b. Applying the power of transcriptomics: understanding male sexual development in decapod Crustacea. Integrative and Comparative Biology 56: 1144–1156.
Chandler, J. C., Q. P. Fitzgibbon, G. Smith, A. Elizur & T. Ventura, 2017. Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: the first invertebrate sex-linked Dmrt. Developmental Biology 430: 337–345.
Charlesworth, B., 1991. The evolution of sex chromosomes. Science 251: 1030–1033.
Charniaux-Cotton, H., 1954. Discovery in, an amphipod crustacean (Orchestia gammarella) of an endocrine gland responsible for the differentiation of primary and secondary male sex characteristics. Comptes rendus hebdomadaires des séances de l’Académie des sciences 239: 780–782.
Charniaux-Cotton, H., 1958. The androgenic gland of some decapodal crustaceans and particularly of Lysmata seticaudata, a species with functional protandrous hermaphroditism. Comptes rendus hebdomadaires des séances de l’Académie des sciences 246: 2814–2817.
Charniaux-Cotton, H., C. Zerbib & J. J. Meusy, 1966. Monographie de la glande androgène des crustacés supérieurs. Crustaceana 10: 113–136.
Chen, P., S.-L. Xu, W. Zhou, X.-G. Guo, C.-L. Wang, D.-L. Wang & Y.-L. Zhao, 2014. Cloning and expression analysis of a transformer gene in Daphnia pulex during different reproduction stages. Animal Reproduction Science 146: 227–237.
Colbourne, J. K., M. E. Pfrender, D. Gilbert, W. K. Thomas, A. Tucker, T. H. Oakley, S. Tokishita, A. Aerts, G. J. Arnold, M. K. Basu, D. J. Bauer, C. E. Cáceres, L. Carmel, C. Casola, J.-H. Choi, J. C. Detter, Q. Dong, S. Dusheyko, B. D. Eads, T. Fröhlich, K. A. Geiler-Samerotte, D. Gerlach, P. Hatcher, S. Jogdeo, J. Krijgsveld, E. V. Kriventseva, D. Kültz, C. Laforsch, E. Lindquist, J. Lopez, J. R. Manak, J. Muller, J. Pangilinan, R. P. Patwardhan, S. Pitluck, E. J. Pritham, A. Rechtsteiner, M. Rho, I. B. Rogozin, O. Sakarya, A. Salamov, S. Schaack, H. Shapiro, Y. Shiga, C. Skalitzky, Z. Smith, A. Souvorov, W. Sung, Z. Tang, D. Tsuchiya, H. Tu, H. Vos, M. Wang, Y. I. Wolf, H. Yamagata, T. Yamada, Y. Ye, J. R. Shaw, J. Andrews, T. J. Crease, H. Tang, S. M. Lucas, H. M. Robertson, P. Bork, E. V. Koonin, E. M. Zdobnov, I. V. Grigoriev, M. Lynch & J. L. Boore, 2011. The ecoresponsive genome of Daphnia pulex. Science 331: 555–561.
Coman, F. E., M. J. Sellars, B. J. Norris, G. J. Coman & N. P. Preston, 2008. The effects of triploidy on Penaeus (Marsupenaeus) japonicus (Bate) survival, growth and gender when compared to diploid siblings. Aquaculture 276: 50–59.
Cordaux, R. & C. Gilbert, 2017. Evolutionary significance of wolbachia-to-animal horizontal gene transfer: female sex determination and the f element in the isopod Armadillidium vulgare. Genes 8: 186.
Cordaux, R., D. Bouchon & P. Grève, 2011. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends in Genetics 27: 332–341.
Crease, T. J., D. J. Stanton & P. D. N. Hebert, 1989. Polyphyletic origins of asexuality in daphnia pulex. II. Mitochondrial-DNA variation. Evolution 43: 1016–1026.
Cui, Z., M. Hui, Y. Liu, C. Song, X. Li, Y. Li, L. Liu, G. Shi, S. Wang, F. Li, X. Zhang, C. Liu, J. Xiang & K. H. Chu, 2015. High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115: 206–215.
Gao, J., X. Wang, Z. Zou, X. Jia, Y. Wang & Z. Zhang, 2014. Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab (Scylla paramamosain). BMC Genomics 15: 585.
Garrett-Engele, C. M., M. L. Siegal, D. S. Manoli, B. C. Williams, H. Li & B. S. Baker, 2002. intersex, a gene required for female sexual development in drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development 129: 4661–4675.
Glenner, H., P. F. Thomsen, M. B. Hebsgaard, M. V. Sørensen & E. Willerslev, 2006. The origin of insects. Science 314: 1883–1884.
Gopal, C., G. Gopikrishna, G. Krishna, S. S. Jahageerdar, M. Rye, B. J. Hayes, S. Paulpandi, R. P. Kiran, S. M. Pillai & P. Ravichandran, 2010. Weight and time of onset of female-superior sexual dimorphism in pond reared Penaeus monodon. Aquaculture 300: 237–239.
Grilo, T. F. & R. Rosa, 2017. Intersexuality in aquatic invertebrates: prevalence and causes. Science of the Total Environment 592: 714–728.
Haag, E. S., 2005. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. In WormBook (ed), The C. elegans Research Community. WormBook
Hasegawa, Y., K. Haino-Fukushima & Y. Katakura, 1987. Isolation and properties of androgenic gland hormone from the terrestrial isopod, Armadillidium vulgare. General and Comparative Endocrinology 67: 101–110.
Hashiyama, K., Y. Hayashi & S. Kobayashi, 2011. Drosophila Sex lethal gene initiates female development in germline progenitors. Science 333: 885–888.
Hasselmann, M., T. Gempe, M. Schiott, C. G. Nunes-Silva, M. Otte & M. Beye, 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–522.
Hediger, M., C. Henggeler, N. Meier, R. Perez, G. Saccone & D. Bopp, 2010. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184: 155–170.
Heimpel, G. E. & J. G. D. Boer, 2008. Sex determination in the Hymenoptera. Annual Review of Entomology 53: 209–230.
Herpin, A., I. Braasch, M. Kraeussling, C. Schmidt, E. C. Thoma, S. Nakamura, M. Tanaka & M. Schartl, 2010. Transcriptional rewiring of the sex determining Dmrt1 gene duplicate by transposable elements. PLoS Genetics 6: e1000844.
Hoshijima, K., K. Inoue, I. Higuchi, H. Sakamoto & Y. Shimura, 1991. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252: 833–836.
Jakob, S. & R. Lovell-Badge, 2011. Sex determination and the control of Sox9 expression in mammals. FEBS Journal 278: 1002–1009.
Jiang, X.-H. & G.-F. Qiu, 2013. Female-only sex-linked amplified fragment length polymorphism markers support ZW/ZZ sex determination in the giant freshwater prawn Macrobrachium rosenbergii. Animal Genetics 44: 782–785.
Jin, S., H. Fu, Q. Zhou, S. Sun, S. Jiang, Y. Xiong, Y. Gong, H. Qiao & W. Zhang, 2013. Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, Using Illumina Hiseq 2000. PLoS ONE 8: e76840.
Juchault, P. & J.-J. Legrand, 1964. Démonstration de l’homogamétie femelle par croisement de deux femelles génétiques, chez les Oniscoïdes Porcellio dilatatus et Helleria brevicornis. Comptes Rendus de l’Académie des Sciences de Paris 258: 2685–2686.
Juchault, P. & J.-J. Legrand, 1972. Croisements de néo-mâles expérimentaux chez Armadillidium vulgare Latr. (Crustacé Isopode Oniscoïde). Mise en évidence d’une hétérogamétie femelle. Comptes Rendus de l’Académie des Sciences. 1387–1389: 274–276.
Juchault, P. & J.-J. Legrand, 1979. Analyse génétique et physiologique de la déermination du sexe dans une population du Crustacé Isopode Oniscoïde Armadillidium nasatum. Archives de Zoologie Expérimentale et Générale 120: 25–43.
Juchault, P. & T. Rigaud, 1995. Evidence for female heterogamety in two terrestrial crustaceans and the problem of sex chromosome evolution in isopods. Heredity 75: 466–471.
Kamiya, T., W. Kai, S. Tasumi, A. Oka, T. Matsunaga, N. Mizuno, M. Fujita, H. Suetake, S. Suzuki, S. Hosoya, S. Tohari, S. Brenner, T. Miyadai, B. Venkatesh, Y. Suzuki & K. Kikuchi, 2012. A trans-species missense snp in amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (Fugu). PLOS Genetics 8: e1002798.
Kato, Y., K. Kobayashi, H. Watanabe & T. Iguchi, 2011. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a doublesex gene in the sex-determining pathway. PLoS Genetics 7: e1001345.
Kato, Y., K. Kobayashi, S. Oda, J. K. Colbourn, N. Tatarazako, H. Watanabe & T. Iguchi, 2008. Molecular cloning and sexually dimorphic expression of DM-domain genes in Daphnia magna. Genomics 91: 94–101.
Kato, Y., K. Kobayashi, S. Oda, N. Tatarazako, H. Watanabe & T. Iguchi, 2010. Sequence divergence and expression of a transformer gene in the branchiopod crustacean, Daphnia magna. Genomics 95: 160–165.
Kleiven, O. T., P. Larsson & A. Hobæk, 1992. Sexual reproduction in Daphnia magna requires three stimuli. Oikos 65: 197–206.
Kopp, A., 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends in Genetics 28: 175–184.
Kulathinal, R. J., L. Skwarek, R. A. Morton & R. S. Singh, 2003. Rapid evolution of the sex-determining gene, transformer: structural diversity and rate heterogeneity among sibling species of Drosophila. Molecular Biology and Evolution 20: 441–452.
Leclercq, S., J. Thézé, M. A. Chebbi, I. Giraud, B. Moumen, L. Ernenwein, P. Grève, C. Gilbert & R. Cordaux, 2016. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proceedings of the National Academy of Sciences 113: 15036–15041.
Leelatanawit, R., K. Sittikankeaw, P. Yocawibun, S. Klinbunga, S. Roytrakul, T. Aoki, I. Hirono & P. Menasveta, 2009. Identification, characterization and expression of sex-related genes in testes of the giant tiger shrimp Penaeus monodon. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 152: 66–76.
Legrand, J. J., P. Juchault, J. C. Artault, J. P. Mocquard & J. L. Picaud, 1974. Le statut systématique de la « forme » petiti Vandel de Porcellio dilatatus Brandt, récoltée à l’île Saint-Honorat (Alpes-Maritimes). Critères morphologiques, génétiques et physiologiques. Bulletin de la Société Zoologique de France 99: 461–471.
Li, P., Y. Liu, D. Luo, C. Song & Z. Cui, 2017. Two spliced isoforms of the sex-determination gene fruitless in the Chinese mitten crab Eriocheir sinensis. Comparative Biochemistry and Physiology Part B: Biochemistry & Molecular Biology 208–209: 75–83.
Li, S., F. Li, R. Wen & J. Xiang, 2012. Identification and characterization of the sex-determiner transformer-2 homologue in Chinese shrimp, Fenneropenaeus chinensis. Sexual Development 6: 267–278.
Li, Y., K. Byrne, E. Miggiano, V. Whan, S. Moore, S. Keys, P. Crocos, N. Preston & S. Lehnert, 2003. Genetic mapping of the kuruma prawn Penaeus japonicus using AFLP markers. Aquaculture 219: 143–156.
Liu, Y., M. Hui, Z. Cui, D. Luo, C. Song, Y. Li & L. Liu, 2015. Comparative transcriptome analysis reveals sex-biased gene expression in juvenile chinese mitten crab Eriocheir sinensis. PLoS ONE 10: e0133068.
Luo, S. D., G. W. Shi & B. S. Baker, 2011. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138: 2761–2771.
Ma, K., G. Qiu, J. Feng & J. Li, 2012. Transcriptome analysis of the oriental river prawn, Macrobrachium nipponense using 454 pyrosequencing for discovery of genes and markers. PLoS ONE 7: e39727.
Ma, K. -Y., Z. Q. Liu, J. Y. Lin, J. L. Li & G. F. Qiu, 2016a. Molecular characterization of a novel ovary-specific gene fem-1 homolog from the oriental river prawn, Macrobrachium nipponense. Gene 575: 244–252.
Ma, K.-Y., J.-L. Li & G.-F. Qiu, 2016b. Identification of putative regulatory region of insulin-like androgenic gland hormone gene (IAG) in the prawn Macrobrachium nipponense and proteins that interact with IAG by using yeast two-hybrid system. General and Comparative Endocrinology 229: 112–118.
Malecha, S. R., P. A. Nevin, P. Ha, L. E. Barck, Y. Lamadrid-Rose, S. Masuno & D. Hedgecock, 1992. Sex-ratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii. Aquaculture 105: 201–218.
Manor, R., S. Weil, S. Oren, L. Glazer, E. D. Aflalo, T. Ventura, V. Chalifa-Caspi, M. Lapidot & A. Sagi, 2007. Insulin and gender: an insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. General and Comparative Endocrinology 150: 326–336.
Mapp, A. K. & A. Z. Ansari, 2007. A TAD further: exogenous control of gene activation. ACS Chemical Biology 2: 62–75.
Martin, G., O. Sorokine, M. Moniatte, P. Bulet, C. Hetru & A. Van Dorsselaer, 1999. The structure of a glycosylated protein hormone responsible for sex determination in the isopod, Armadillidium vulgare. European Journal of Biochemistry 262: 727–736.
Matson, C. K. & D. Zarkower, 2012. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nature Review Genetics 13: 163–174.
Matson, C. K., M. W. Murphy, A. L. Sarver, M. D. Griswold, V. J. Bardwell & D. Zarkower, 2011. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476: 101–104.
Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda, T. Kobayashi, C. E. Morrey, N. Shibata, S. Asakawa, N. Shimizu, H. Hori, S. Hamaguchi & M. Sakaizumi, 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563.
Meiklejohn, C. D., J. Parsch, J. M. Ranz & D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proceedings of the National Academy of Sciences 100: 9894–9899.
Meise, M., D. Hilfiker-Kleiner, A. Dubendorfer, C. Brunner, R. Nothiger & D. Bopp, 1998. Sex-lethal, the master sex-determining gene in drosophila, is not sex-specifically regulated in Musca domestica. Development 125: 1487–1494.
Mittal, O. P. & S. Pahwa, 1980. Cytogenetical studies on two terrestrial isopods (Crustacea), Philoscia iodnensis ramakrishna and Porcellio rathkei brandt. Cytobios 29: 17–24.
Mittal, O. P. & S. Pahwa, 1981. Germ cell chromosomes in two species of terrestrial isopods from India. Experientia 37: 343–345.
Mlinarec, J., I. Porupski, I. Maguire & G. Klobučar, 2016. Comparative karyotype investigations in the white-clawed crayfish (Lereboullet, 1858) species complex and stone crayfish (Schrank, 1803) (Decapoda: Astacidae). Journal of Crustacean Biology 36: 87–93.
Ishak, N. S. M., Q. D. Nong, T. Matsuura, Y. Kato & H. Watanabe, 2017. Co-option of the bZIP transcription factor Vrille as the activator of Doublesex1 in environmental sex determination of the crustacean Daphnia magna. PLOS Genetics 13: e1006953.
Murphy, M. W., A. L. Sarver, D. Rice, K. Hatzi, K. Ye, A. Melnick, L. L. Heckert, D. Zarkower & V. J. Bardwell, 2010. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proceedings of the National Academy of Sciences 107: 13360–13365.
Nagaraju, G. P. C., 2011. Reproductive regulators in decapod crustaceans: an overview. The Journal of Experimental Biology 214: 3–16.
Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler, A. Shimizu, Z. Shan, T. Haaf, N. Shimizu, A. Shima, M. Schmid & M. Schartl, 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proceedings of the National Academy of Sciences 99: 11778–11783.
Okuno, A., Y. Hasegawa, T. Ohira, Y. Katakura & H. Nagasawa, 1999. Characterization and cDNA cloning of androgenic gland hormone of the terrestrial isopod Armadillidium vulgare. Biochemical and Biophysical Research Communications. 264: 419–423.
Pane, A., M. Salvemini, P. Delli Bovi, C. Polito & G. Saccone, 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129: 3715–3725.
Parnes, S., I. Khalaila, G. Hulata & A. Sagi, 2003. Sex determination in crayfish: are intersex Cherax quadricarinatus (Decapoda, Parastacidae) genetically females? Genetical Research 82: 107–116.
Parsch, J. & H. Ellegren, 2013. The evolutionary causes and consequences of sex-biased gene expression. Nature Review Genetics 14: 83–87.
Peng, J., P. Wei, B. Zhang, Y. Zhao, D. Zeng, X. Chen, M. Li & X. Chen, 2015. Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC genomics 16: 1006.
Piferrer, F., 2013. Epigenetics of sex determination and gonadogenesis. Developmental Dynamics 242: 360–370.
Piskacek, S., M. Gregor, M. Nemethova, M. Grabner, P. Kovarik & M. Piskacek, 2007. Nine-amino-acid transactivation domain: establishment and prediction utilities. Genomics 89: 756–768.
Powell, D., W. Knibb, C. Remilton & A. Elizur, 2015. De-novo transcriptome analysis of the banana shrimp (Fenneropenaeus merguiensis) and identification of genes associated with reproduction and development. Marine genomics 22: 71–78.
Preechaphol, R., R. Leelatanawit, K. Sittikankeaw, S. Klinbunga, B. Khamnamtong, N. Puanglarp & P. Menasveta, 2007. Expressed sequence tag analysis for identification and characterization of sex-related genes in the giant tiger shrimp Penaeus monodon. Journal of biochemistry and molecular biology 40: 501–510.
Preston, N. P., P. J. Crocos, S. J. Keys, G. J. Coman & R. Koenig, 2004. Comparative growth of selected and non-selected Kuruma shrimp Penaeus (Marsupenaeus) japonicus in commercial farm ponds; implications for broodstock production. Aquaculture 231: 73–82.
Pultz, M. A. & B. S. Baker, 1995. The dual role of hermaphrodite in the Drosophila sex determination regulatory hierarchy. Development 121: 99–111.
Qiao, H., H. Fu, S. Jin, Y. Wu, S. Jiang, Y. Gong & Y. Xiong, 2012. Constructing and random sequencing analysis of normalized cDNA library of testis tissue from oriental river prawn (Macrobrachium nipponense). Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 7: 268–276.
Qiao, H., Y. W. Xiong, S. F. Jiang, H. T. Fu, S. M. Sun, S. B. Jin, Y. S. Gong & W. Y. Zhang, 2015. Gene expression profile analysis of testis and ovary of oriental river prawn, Macrobrachium nipponense, reveals candidate reproduction-related genes. Genetics and Molecular Research 14: 2041–2054.
Rahman, N. M., H. Fu, H. Qiao, S. Jin, H. Bai, W. Zhang, F. W. Jiang, G. Liang, S. Sun, Y. Gong, F. F. Jiang, Y. Xiong & Y. Wu, 2016. Molecular cloning and expression analysis of Fem1b from oriental river prawn Macrobrachium nipponense. Genetics and Molecular Research. https://doi.org/10.4238/gmr.15027950.
Rigaud, T., P. Juchault & J.-P. Mocquard, 1997. The evolution of sex determination in isopod crustaceans. BioEssays 19: 409–416.
Robinson, N. A., G. Gopikrishna, M. Baranski, V. K. Katneni, M. S. Shekhar, J. Shanmugakarthik, S. Jothivel, C. Gopal, P. Ravichandran, T. Gitterle & A. G. Ponniah, 2014. QTL for white spot syndrome virus resistance and the sex-determining locus in the Indian black tiger shrimp (Penaeus monodon). BMC genomics 15: 731.
Rocchi, A., G. Prantera, V. Lanza & M. D. Castro, 1984. Incipient sex chromosome differentiation in an isopod crustacean species, Asellus aquaticus. Chromosoma 89: 93–196.
Rodríguez, E. M., D. A. Medesani & M. Fingerman, 2007. Endocrine disruption in crustaceans due to pollutants: a review. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 146: 661–671.
Rosen, O., R. Manor, S. Weil, O. Gafni, A. Linial, E. D. Aflalo, T. Ventura & A. Sagi, 2010. A sexual shift induced by silencing of a single insulin-like gene in crayfish: ovarian upregulation and testicular degeneration. PLoS ONE 5: e15281.
Sagi, A. & D. Cohen, 1990. Growth, maturation and progeny of sex-reversed Macrobrachium rosenbergii males. World Aquaculture 21: 87–90.
Sagi, A., E. Snir & I. Khalaila, 1997. Sexual differentiation in decapod crustaceans: role of the androgenic gland. Invertebrate Reproductive and Development 31: 55–61.
Sagi, A., I. Khalaila, A. Barki, G. Hulata & I. Karplus, 1996. Intersex red claw crayfish, Cherax quadricarinatus (von Martens): functional males with pre-vitellogenic ovaries. Biological Bulletin 190: 16–23.
Scholtz, G., A. Braband, L. Tolley, A. Reimann, B. Mittmann, C. Lukhaup, F. Steuerwald & G. Vogt, 2003. Ecology: parthenogenesis in an outsider crayfish. Nature 421: 806.
Shaklee, J. B., 1983. Mannosephosphate isomerase in the Hawaiian spiny lobster Panulirus marginatus: a polymorphic, sex-linked locus useful in investigating embryonic and larval sex ratios. Marine Biology 73: 193–201.
Sharabi, O., R. Manor, S. Weil, E. D. Aflalo, Y. Lezer, T. Levy, J. Aizen, T. Ventura, P. B. Mather, I. Khalaila & A. Sagi, 2016. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 157: 928–941.
Shen, H., Y. Hu & X. Zhou, 2014. Sex-lethal gene of the Chinese mitten crab Eriocheir sinensis: cDNA cloning, induction by eyestalk ablation, and expression of two splice variants in males and females. Development Genes and Evolution 224: 97–105.
Shpak, N., R. Manor, E. D. Aflalo & A. Sagi, 2016. Three generations of cultured prawn without W chromosome. Aquaculture 467: 41–48.
Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths, M. J. Smith, J. W. Foster, A. M. Frischauf, R. Lovell-Badge & P. N. Goodfellow, 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244.
Song, C., Z. Cui, M. Hui, Y. Liu & Y. Li, 2015. Molecular characterization and expression profile of three Fem-1 genes in Eriocheir sinensis provide a new insight into crab sex-determining mechanism. Comparative Biochemistry and Physiology Part B: Biochemistry & Molecular Biology 189: 6–14.
Subramoniam, T., 2017. Sexual Systems Sexual Biology and Reproduction in Crustaceans. Elsevier, Amsterdam.
Suzuki, S., 1999. Androgenic gland hormone is a sex-reversing factor but cannot be a sex-determining factor in the female crustacean isopods Armadillidium vulgare. General and Comparative Endocrinology 115: 370–378.
Torrecilla, Z., A. Martínez-Lage, A. Perina, E. González-Ortegón & A. M. González-Tizón, 2017. Comparative cytogenetic analysis of marine Palaemon species reveals a X1X1X2X2/X1X2Y sex chromosome system in Palaemon elegans. Frontiers in Zoology 14: 47.
Trino, A., O. Millamena & C. Keenan, 1999. Commercial evaluation of monosex pond culture of the mud crab Scylla species at three stocking densities in the Philippines. Aquaculture 174: 109–118.
Ventura, T., O. Rosen & A. Sagi, 2011a. From the discovery of the crustacean androgenic gland to the insulin-like hormone in six decades. General and Comparative Endocrinology 173: 381–388.
Ventura, T., E. D. Aflalo, S. Weil, K. Kashkush & A. Sagi, 2011b. Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity 107: 456–461.
Ventura, T., R. Manor, E. D. Aflalo, S. Weil, S. Raviv, L. Glazer & A. Sagi, 2009. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 150: 1278–1286.
Ventura, T., R. Manor, E. D. Aflalo, S. Weil, O. Rosen & A. Sagi, 2012. Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biology of Reproduction 86: 1–6.
Vogt, G., C. Falckenhayn, A. Schrimpf, K. Schmid, K. Hanna, J. Panteleit, M. Helm, R. Schulz & F. Lyko, 2015. The marbled crayfish as a paradigm for saltational speciation by autopolyploidy and parthenogenesis in animals. Biology Open 4: 1583–1594.
Wexler, J. R., D. C. Plachetzki & A. Kopp, 2014. Pan-metazoan phylogeny of the DMRT gene family: a framework for functional studies. Development Genes and Evolution 224: 175–181.
Yano, A., R. Guyomard, B. Nicol, E. Jouanno, E. Quillet, C. Klopp, C. Cabau, O. Bouchez, A. Fostier & Y. Guiguen, 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Current Biology 22: 1423–1428.
Yi, W., J. M. Ross & D. Zarkower, 2000. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127: 4469–4480.
Yoshimoto, S., E. Okada, T. Oishi, R. Numagami, H. Umemoto, K. Tamura, H. Kanda, T. Shiba, N. Takamatsu & M. Ito, 2006. Expression and promoter analysis of Xenopus DMRT1 and functional characterization of the transactivation property of its protein. Development, Growth and Differentiation 48: 597–603.
Yoshimoto, S., E. Okada, H. Umemoto, K. Tamura, Y. Uno, C. Nishida-Umehara, Y. Matsuda, N. Takamatsu, T. Shiba & M. Ito, 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proceedings of the National Academy of Sciences 105: 2469–2474.
Yu, Y.-Q., W.-M. Ma, Q.-G. Zeng, Y.-Q. Qian, J.-S. Yang & W.-J. Yang, 2014. Molecular cloning and sexually dimorphic expression of two Dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agricultural Research 3: 181–191.
Zhang, D., M. Sun & X. Liu, 2017. Phase-specific expression of an insulin-like androgenic gland factor in a marine shrimp Lysmata wurdemanni: implication for maintaining protandric simultaneous hermaphroditism. PLoS ONE 12: e0172782.
Zhang, L., C. Yang, Y. Zhang, L. Li, X. Zhang, Q. Zhang & J. Xiang, 2007. A genetic linkage map of Pacific white shrimp (Litopenaeus vannamei): sex-linked microsatellite markers and high recombination rates. Genetica 131: 37–49.
Zhang, Y. P., H. Qiao, W. Y. Zhang, S. M. Sun, S. F. Jiang, Y. S. Gong, Y. W. Xiong, S. B. Jin & H. T. Fu, 2013. Molecular cloning and expression analysis of two sex-lethal homolog genes during development in the oriental river prawn, Macrobrachium nipponense. Genetics and Molecular Research 12: 4698–4711.
Zhang, Y., K. Qiao, S. Wang, H. Peng, Z. Shan & K. Wang, 2014. Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab, Scylla paramamosain. Aquaculture 433: 325–334.
Zhang, Y. P., S. M. Sun, H. T. Fu, X. P. Ge, H. Qiao, W. Y. Zhang, Y. W. Xiong, S. F. Jiang, Y. S. Gong & S. B. Jin, 2015. Characterization of the male-specific lethal 3 gene in the oriental river prawn, Macrobrachium nipponense. Genetics and Molecular Research 14: 3106–3120.
Acknowledgements
This work was supported by the Australian Research Council via a Discovery Early Career Research Award [DE130101089 to TV], a Discovery [DP160103320 to TV], a University of the Sunshine Coast (USC) Collaborative Research Networks (CRN) grant programme and a USC International Research Scholarship to JC.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Guest editors: Ferran Palero, Guiomar Rotllant, Peter Mather, Heather Bracken-Grissom & Begoña Santos / Crustacean Genomics
Rights and permissions
About this article
Cite this article
Chandler, J.C., Elizur, A. & Ventura, T. The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825, 61–80 (2018). https://doi.org/10.1007/s10750-017-3452-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3452-4