Abstract
Protists thrive in polar oceans, where they represent a major driving force for globally important biogeochemical cycles and a key food-web component. Their biogeography is frequently associated to bipolar patterns of distribution. Although conceptually well supported by apparently unrestricted migration rates, the experimental certification of these patterns copes with the protist paucity of morphological characters with taxonomic value and difficulties in applying conventional species concepts. We studied three marine species of the ciliate Euplotes, E. euryhalinus, E. nobilii, and E. petzi, for their bipolar distribution by comparing the SSU-rRNA gene sequences and mating interactions of Antarctic, Patagonian, and Arctic strains. Each species was analogously found not to carry significantly varied SSU-rRNA gene sequences, implying a common occurrence of trans-equatorial genetic mixing. However, mating analyses revealed significant inter-species differences. Scarce Antarctic × Arctic strain mating compatibility distinguished E. petzi from E. euryhalinus and E. nobilii, in which mating pairs between Antarctic and Arctic strains were successfully induced. Yet, E. nobilii was the only one of the two species to show cross-fertilizing and fertile mating pairs. Taking the biological concept of species as discriminatory, it was thus concluded that only E. nobilii warrants the definition of genuine bipolar species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An amazingly diverse wealth of single-cell eukaryotic microorganisms (protists) thrives in the Antarctic and Arctic marine environment (Vincent, 1988; Sorokin, 1999; Scott & Marchant, 2005; Lovejoy et al., 2006; Lovejoy, 2014). Their metabolisms play a vital role in the polar ecosystems’ maintenance of global biogeochemical cycles and, hence, the well-being of our planet. Since their first description by Ehrenberg (1844) in samples of Antarctic phytoplankton, diatoms have received more attention in polar biology than any other microbial group (Knox, 1994). This attention is substantially due to their major photosynthetic role in the biological pump which converts inorganic carbon to organic carbon (Falkowski et al., 2008; Armbrust, 2009), as well as to their value as fossil records in paleo-oceanographic and paleo-climatic reconstructions (Gersonde, 1990; Finkelstein et al., 2014), matched only by foraminifera (de Vargas et al., 1997) and dinoflagellates (Fensome et al., 1996; Taylor et al., 2008). Ciliates, while less remarkable than diatoms, foraminifera, and dinoflagellates on ecological and paleo-environmental grounds, certainly parallel these taxa in biodiversity and capacity for global dispersal up to the highest latitudes of the globe. This makes them equally valuable models for gaining insights into the microbial biogeographical patterns of bipolar (or anti-polar) distribution, of great interest to marine ecology for the information on contemporary and historical forces and mechanisms that regulate genetic drift, gene flow, and migration in the oceans that we can retrieve from studying them (Darling et al., 2000; Montresor et al., 2003; Pawlowski et al., 2007; Darling & Wade, 2008).
Among the roughly 400 species of ciliates from Antarctic and Arctic polar habitats that have been identified on a merely morphological basis (Valbonesi & Luporini, 1990a, b; Agatha et al., 1993; Petz et al., 1995, 2007; Petz & Foissner, 1997; Kepner et al., 1999; Petz, 2004, 2005; Wilbert & Song, 2005, 2008; Dolan et al., 2013; Mieczan et al., 2013), our attention has mostly been focused on the Euplotes species. These species can be readily identified in vivo, isolated and expanded into clonal cultures that, usually being able to reproduce true-to-type over many generations, guarantee virtually unlimited experimental material. In addition, they regulate intercellular genetic exchange through a sexual phenomenon of conjugation that is genetically controlled by highly multiple (virtually open) mating-type systems (Génermont et al., 1976; Nobili et al., 1978; Valbonesi et al., 1992; Dini & Nyberg, 1993). Compared to species endowed with binary or low-multiple systems, it is therefore relatively easier to collect higher numbers of genetically distinct strains of Euplotes from different localities and gain insights into the genetic relationships of their natural populations from an analysis of their mating interactions.
Over more than twenty years, we have had the opportunity to set up a vast collection of Euplotes strains isolated from various littoral sea sites located in the surroundings of the Italian “Mario Zucchelli” Research Station in Terra Nova Bay (Ross Sea), and scattered in sub-Antarctic (Patagonia) and Arctic (Alaska, Greenland, Russia, and Svalbard) regions. This collection was for a long time essentially used to investigate molecular mechanisms underlying cold adaptation (Pedrini et al., 2007; Alimenti et al., 2009; Vallesi et al., 2010, 2012; Chiappori et al., 2012; Candelori et al., 2013; Geralt et al., 2013). Only more recently has it turned out to be of research interest also within a biogeographic and ecological perspective, in relation to morphological and genetic evidence that co-specificity exists among strains inhabiting opposite sides of the globe (Di Giuseppe et al., 2011, 2013a, b).
Here we update and describe new data on the genetic relationships of high-latitude populations of three Euplotes species, namely E. euryhalinus, E. nobilii, and E. petzi, which are well distinct on morphological, eco-physiological, and phylogenetic grounds (Valbonesi & Luporini, 1990a; Achilles-Day et al., 2008; Chen et al., 2013; Di Giuseppe et al., 2014). These relationships were deduced from comparisons of small-subunit ribosomal RNA (SSU-rRNA) gene sequences and analyses of mating interactions.
Materials and methods
Collection sites, strain origin, and cultivation
A chart of the collection sites is shown in Fig. 1, while Table 1 lists the strains analyzed for each species and provides information on basic environmental parameters of each site. Each of the strains is monoclonal, as it was expanded starting from a single isolate. To minimize the possibility of growing laboratory cultures expanded from products of divisions of the same isolate, seawater and sediment samples (usually from 100 to 500 ml of volume) were as a rule inspected for the isolation of Euplotes specimens not later than 1–2 days after their collection. Whenever this inspection took longer time and multiple cultures were initially raised from specimens isolated from the same sample, only one culture for each sample was eventually stably cultivated. Cultivation was carried out in cold rooms adjusted with a daily cycle of 12 h of dark and 12 h of very weak light, and temperatures of 4–6°C. The green alga Dunaliella tertiolecta, grown either in natural or artificial seawater (PSU, 32–33) enriched with Walne medium, was used as standard food source. Replicated sets of cultures were also continuously maintained in the dark and fed with the bacterium Enterobacter aerogenes.
Mating pair induction and analysis
Mating mixtures were carried out between 1-ml cell samples taken from cultures maintained at a growth stage for 1–2 weeks and then re-suspended in fresh seawater for 2–3 days at adjusted concentrations of about 3 × 103 cells/ml. At the peak of the mating reaction (usually coinciding with the second day following cell mixing), stable mating pairs were individually isolated in a few drops of supernatant of their original cultures and each pair was left to separate. During 1 week without food, the two ex-partner cells of each pair were allowed to reorganize and develop their new nuclear apparatuses. Ex-partner cells surviving from the same mating pair were then fed and grown into couples of sister progeny clones (synclones). Ten of these synclones were chosen for each strain mixture to determine the homo- or heterotypic nature of their parental mating pairs through comparative analyses of the profiles of their SSU-rRNA nuclear gene sequences. Sequence diversity between the two sister clones of each synclone was taken as indication of heterotypic pairs destined to carry out cross-fertilization, being formed by genetically distinct partners provided by both of the strains mixed; sequence identity, as indication of homotypic pairs destined to carry out self-fertilization, being formed by genetically identical partners both provided by only either one of the two strains mixed.
DNA extraction and polymerase chain reaction (PCR)
SSU-rRNA nuclear gene sequences were PCR amplified from DNA prepared from cells of cultures that had been starved for several days, using the QIAamp® DNA Micro Kit (Qiagen, Milan, Italy) in accord with the manufacturer’s instructions. The universal eukaryotic forward primer 5′-CTGGTTGATCCTGCCAG-3′ (Medlin et al., 1988) and the ciliate hypotrich-specific 18S reverse primer 5′-TGATCCTTCYGCAGGTTC-3′ (Petroni et al., 2002) were used in PCR amplification of SSU-rRNA genes. Amplifications were performed by adding DNA aliquots (100 ng) to 50 μl of reaction mixture containing 2 mM MgCl2, 250 μM of dNTP, one unit of Taq DNA polymerase (Polymed, Florence, Italy), and 0.2 μM of each primer. They were run in a GenAmp PCR system 2400 (Applied Biosystems, Foster City, CA, USA), following standard programs. Each amplified product was purified using Quantum Prep PCR Kleen Spin columns (Bio-Rad, Hercules, CA, USA) and sequenced in both directions with an ABI Prism 310 automated DNA sequencer (Applied Biosystems). To minimize amplification errors, sequences of two distinct amplicons were compared for each strain.
Sequence availability and phylogenetic analyses
All the new nuclear SSU-rRNA gene sequences were deposited in the GenBank/EMBL databases and their accession numbers are listed in Table 2. Sequence alignment was carried out using the CLUSTAL X program (version 1.81) (Thompson et al., 1997) and the default parameter settings. The alignments, resulting in 2021 positions, were edited using the BIOEDIT program (version 7.0.0) (Hall, 1999). Maximum Likelihood (ML), Maximum Parsimony (MP), and Bayesian Inference (BI) analyses were performed with the programs Tree-Puzzle (version 5.0) (Schmidt et al., 2002), PAUP (version 4.b10) (Swofford, 2003), and MrBayes (version 3.1) (Ronquist & Huelsenbeck, 2003), respectively, using the GTR + G + I model selected under the AIC criterion by the Modeltest program (version 3.7) (Posada & Crandall, 1998). The reliability of the internal branches of the phylogenetic trees was evaluated with the bootstrap method (Felsenstein, 1988), assessing 1000 replicates in the ML and MP tree, and posterior probabilities in the BI tree.
Results
Morphological, eco-physiological, and phylogenetic specificities
The taxonomic distinction of the three species considered in this study, E. nobilii, E. petzi, and E. euryhalinus, has been originally based on significant variations in major diagnostic traits of Euplotes, in particular cell body shape and dimensions, number of dorso-lateral ciliary rows (kineties), geometrical pattern of the dorsal silver-line system (argyrome), and number of ciliary membranelles surrounding the adoral zone. However, also remarkable eco-physiological and phylogenetic aspects distinguish these species. With regard to the eco-physiological context, E. nobilii and E. petzi are genuine seawater species. All their strains come from locations of the marine littoral in which salinity was minimum 30 PSU, and none of their strains have been observed to be capable of forming resistant stages (cysts). In contrast, all E. euryhalinus strains have been isolated from seashore pools subject to rapid variations in temperature (from −1 up to 6°C) and, more significant, in salinity (from practically zero to 27 PSU) due to meltwater dilution, evaporation, and brine accumulation from surface freezing. Consistently, with its brackish habitat, and as its species denomination indicates, E. euryhalinus tolerates a wide range of salinity to the point that its strains have been adapted to grow in laboratory at a salinity of 32–33 PSU (like those of E. nobilii and E. petzi), and have been shown to be able to form cysts. The specific causes that induce E. euryhalinus to encyst have not been accurately studied; preliminary (unpublished) observations have identified two possible causes in prolonged starvation and exposure to PSU values over than 35_40.
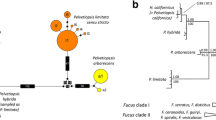
Phylogenetically, E. nobilii, E. petzi, and E. euryhalinus occupy significantly distant positions in the Euplotes phylogenetic tree. The tree shown in Fig. 2 was constructed from comparative alignments of only those Euplotes SSU-rRNA gene sequences available from the GenBank/EMBL databases that we judged to be of unequivocal species assignment, i.e., not taking into account those sequences of strains that were of dubious taxonomy. It appears that E. nobilii and E. petzi both fall into statistically well-supported clades (ML and MP, 100; BI, 1.0) that are coincident with two widely separated early branches of the tree, one of which (including E. nobilii) has even been regarded (Achilles-Day et al., 2008) as representative of a genus (Neteuplotes) distinct from Euplotes. Closer relationships link E. nobilii to E. raikovi and E. elegans which are species of temperate marine and brackish waters, respectively, while E. petzi appears to be more closely related to E. sinicus, a seawater species recently isolated (Jiang et al., 2010a) from coastal sites in Qingdao (China). The phylogenetic position taken by E. euryhalinus, yet appearing as more equivocal, sets quite apart from those of E. nobilii and E. petzi. Together with E. magnicirratus and E. trisulcatus, both of which are marine species from temperate waters, E. euryhalinus is included in a late-branching clade that receives weak statistical support (ML, <50; MP, 65; BI, 0.71) and may even be regarded as an unresolved clade.
Phylogenetic tree of Euplotes, based on SSU-rRNA nuclear gene sequences, highlighting (in bold) the positions of E. petzi, E. nobilii, and E. euryhalinus. Nodes are numbered progressively, and bootstrap and posterior probability values (≥50% and ≥0.50, respectively) are provided for each node. ML, maximum likelihood support; MP, maximum parsimony support; and BI, Bayesian inference support. Fully supported (100%, 1.00) branches are marked with solid circles. The scale bar corresponds to two substitutions per 100 nucleotide positions. The accession numbers of the sequences used to generate the tree are indicated for each species with the exception of E. nobilii, E. petzi, and E. euryhalinus, for which the accession numbers of the multiple sequences that have been used are reported in Di Giuseppe et al. (2011, 2014) and Table 2 (this article), respectively. In accord with Jiang et al. (2010a, b) and Chen et al. (2013), Certesia quadrinucleata and Aspidisca steini (species of a sister clade of Euplotes) were used as outgroups
Genetic relationships
The genetic relationships between Antarctic and non-Antarctic Euplotes populations have been originally studied on a collection of nine Antarctic, four Patagonian, nine Greenland, and three Svalbard E. nobilii strains, which were compared for their nuclear (18S and ITS) and mitochondrial (16S) rRNA gene sequences and mating interactions (Di Giuseppe et al., 2013a). Although genetically distinct by some nucleotide mutations in their gene sequences, one Greenland and three Antarctic strains were found to be mutually mating compatible to the point of forming mating pairs of heterotypic nature fully capable of generating viable offspring from cross-fertilization, thus implying that a gene flow actually bridges E. nobilii populations living geographically separated at the opposite poles of the globe.
This implication is now further supported by genetic and mating analyses of four new E. nobilii strains that we recently had the opportunity to isolate from the Alaskan coastal waters of Barrow. These strains were first analyzed for their SSU-rRNA gene sequences, which were determined for an extension of 916–918 bp and compared with all the other SSU-rRNA gene sequences previously determined for the E. nobilii strains of the original collection. Then, we assessed the spectrum of their mating interactions with some of the Patagonian and Antarctic strains that have remained in vigorous health throughout the many years passed since the time of their collection from the wild. Three of the new Alaskan strains revealed much closer sequence correlations with the Greenland and Svalbard strains than with any Patagonian or Antarctic strain (Fig. 3a), with which they consistently showed no mating compatibility. However, the fourth (ABY7-3) Alaskan strain not only revealed a SSU-rRNA gene sequence fully overlapping with those of the Antarctic and Patagonian strains but it also manifested full mating compatibility with the Patagonian strain IsH3-2 with which it formed stable mating pairs. These pairs were isolated and systematically observed to be able to complete the mating process and generate viable offspring clones. Yet, due to the full SSU-rRNA gene sequence matching shown by the ABY7-3 and IsH3-2 strains, we were so far unable to definitively assess the heterotypic, or homotypic nature of the mating pairs formed in the mixtures between these two strains.
Information on the intra-specific relationships of E. petzi has so far remained limited to the study of only three Antarctic and three Arctic strains (two from Greenland and one from Chupa, Russia), whose SSU-rRNA nuclear gene sequences have been determined for their entire extensions of 1770–1772 bp. No nucleotide substitution was identified within either of the two sets of strains, while four substitutions were detected between the two sets of strains, determining their consequent separation into two distinct clusters (Fig. 3b). Consistently with these nucleotide sequence variations between the Antarctic and Arctic strains, the Antarctic × Antarctic and Arctic × Arctic strain mixtures produced quite different results with respect to the Antarctic × Arctic ones. In the former case, it was systematically possible to obtain good mating reactions and the formation of stable and viable heterotypic mating pairs. In the latter case, only loose cell–cell ciliary sticking was observed between mixed cells, but no stable Antarctic × Arctic cell mating unions could be induced implying that a significant degree of genetic discontinuity is likely to have ensued between Antarctic and Arctic E. petzi populations.
The species that has most recently been studied for its trans-equatorial genetic relationships is E. euryhalinus, whose major diagnostic morphological traits are illustrated in Fig. 4. For this species, we determined the SSU-rRNA nuclear gene sequences for an extension of 1828_1832 bp in 18 Antarctic and 12 Arctic strains, half of Greenland and half of Svalbard origin. Complete sequence identity was equally observed within the Antarctic strains (except for one strain carrying a single nucleotide mutation) as well as within the Greenland strains, while the two sets of strains differed from one another in seven nucleotide mutations. Similarly, six nucleotide mutations distinguished the Svalbard strains into two sub-groups. One of these included four strains with sequences identical to those of the Antarctic strains, while the second group included two strains with sequences identical to those of the Greenland strains. In practice (Fig. 3c), the genetic structure of E. euryhalinus revealed a pattern mimicking that previously determined in E. nobilii, with a significant number of Svalbard strains that clustered together with the Antarctic strains, and not with their population members.
Morphological traits of E. euryhalinus, as they appear in SEM micrographs (a, c, dorsal and ventral surface, respectively) and silver-stained specimens (b, d, dorsal and ventral surface, respectively). DA dorsal alveoli, DK dorsal kinety, AM adoral membranelles, FVC fronto-ventral cirri, TC transverse cirri, CC caudal cirri, LK lateral kinety. After Valbonesi & Luporini (1990a), modified
However, a significant difference emerged in assessing whether E. euryhalinus mimics E. nobilii also with regard to the pattern of inter-polar mating reactions. As was the case for E. nobilii, within the E. euryhalinus strains too it was possible to identify three Antarctic (BTN4, CAD1, and TNK1) and two Arctic strains (1-3ILb1 and 1bILb3) from Greenland that were able to interact for mating in every pair-wise mixture (Fig. 5). Mating pairs were then isolated from each mixture between these Antarctic and Arctic strains and examined to determine their heterotypic, or homotypic nature. This determination could not be carried out through traditional Mendelian analyses of mating-type inheritance that are usually applied to temperate water ciliates, because in polar ciliates these analyses are impracticable due to the too long-lasting (months) stage of cell incompetence to mate (sexual immaturity) that ensues after the completion of every mating event (Valbonesi & Luporini, 1993). Therefore, as originally devised in E. nobilii (Di Giuseppe et al., 2011), we used the SSU-rRNA nuclear gene sequences, previously determined in relation to the phylogenetic analysis of the E. euryhalinus strains, as cell-specific and bi-parentally inheritable nuclear markers. Mating pairs were isolated from each reactive mating Antarctic × Arctic E. euryhalinus strain combination, and systematically found to be fully able to complete the mating process and generate viable offspring clones. These clones, however, all showed SSU-rRNA nuclear gene sequences identical to the sequence of the Antarctic strain involved in the mating mixture, thus providing evidence that they were descendant of homotypic, self-fertilizing pairs formed only between Antarctic mating partners.
Mating interactions between Antarctic and Arctic E. euryhalinus strains. For each strain combination the corresponding box indicates the intensity of the mating reactions, the strain involved in the formation of the homotypic (selfing) mating pairs, and the mating pair viability computed as percentage of ex-conjugant cells that were able to generate viable progeny clones
Discussion
Microbial species are commonly credited with bipolar biogeographic patterns essentially on the basis of criteria resulting from comparative analyses of morphological traits and molecular data of isolates from high-latitude populations. Based only on these criteria, E. nobilii, E. petzi, and E. euryhalinus would equally deserve the definition of bipolar species, their Antarctic and Arctic populations having revealed overlapping morphotypes and not significantly mutated SSU-rRNA nuclear gene sequences. However, the most objective principle to unequivocally establish whether populations belong to the same biological species, or are genetically separated to the point of representing incipient new species, is widely recognized to be provided by inter-population breeding analyses. Taking the results of these analyses as discriminatory, it would then be more appropriate to regard only E. nobilii as a genuine bipolar species. In effect, only in this case was unequivocal evidence obtained for the occurrence of effective cross-fertilization and, hence, outbreeding and trans-tropical gene flow between Antarctic and Arctic populations. The designation of E. petzi and E. euryhalinus as bipolar species would appear as less legitimate. In E. petzi, Antarctic and Arctic strains showed little, or no mutual mating compatibility, while mixing between Antarctic and Arctic E. euryhalinus strains resulted in the formation only of homotypic mating pairs destined to generate population inbreeding and, hence, genetic discontinuity through self-fertilization. In any case, these observations do not appear as yet sufficient to conclusively state that these two species did actually evolve physiological and/or genetic obstacles strong enough to definitively preclude a mutual mating compatibility between their Antarctic and Arctic populations. This conclusion would require to be further substantiated by analyses extended to mating interactions of new sets of strains representative of a wider array of geographically distinct Antarctic and Arctic populations. The fact, in E. euryhalinus in particular, that mixtures between Antarctic and Arctic strains form functional mating pairs, yet of homotypic nature, implies that these strains share and mutually recognize structurally homologous mating-inducing signaling pheromones and, hence, that they still have the potential to form also heterotypic mating pairs capable of cross-fertilization and mutual gene exchange.
As pointed out in the Introduction, Euplotes species modulate their gene exchange through high-multiple mating-type systems responsible for wide intra-specific genetic polymorphisms (Génermont et al., 1976; Nobili et al., 1978; Valbonesi et al., 1992). Although these polymorphisms demand caution in extrapolating genetic structures from the inevitably limited numbers of wild-type strains that one can analyze in the laboratory for their mating interactions, the breeding differences detected among E. nobilii, E. petzi, and E. euryhalinus find a plausible explanation in the distances that separate these species in the Euplotes phylogenetic tree. This separation implies that their evolutionary Antarctic and Arctic colonization took place independently from one another, with the consequent adoption of species-specific ecological strategies and genetic mechanisms. Unfortunately, unlike diatoms, foraminifera and dinoflagellates in which fossil records provide solid parameters to correlate inter-species phylogenetic distances with evolutionary times (Sims et al., 2006; Pawlowski et al., 2007; Souffreau et al., 2011), ciliates left no reliable fossil record. This absence thus prevents any reference to a definite molecular clock for connecting the phylogenetic distances of E. nobilii, E. petzi, and E. euryhalinus with the geological times that have seen the migration of these species into the Antarctic and Arctic oceans.
References
Achilles-Day, U. E. M., T. Pröschold & J. G. Day, 2008. Phylogenetic position of the freshwater ciliate Euplotes daidaleos within the family of Euplotidae, obtained from small subunit rDNA gene sequence. Denisia 23: 411–416.
Agatha, S., M. Spindler & N. Wilbert, 1993. Ciliated protozoa (Ciliophora) from Arctic sea ice. Acta Protozoologica 32: 261–268.
Alimenti, C., A. Vallesi, B. Pedrini, K. Wüthrich & P. Luporini, 2009. Molecular cold-adaptation: comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes. IUBMB Life 61: 838–845.
Armbrust, E. V., 2009. The life of the diatoms in the world’s oceans. Nature 459: 185–192.
Candelori, A., P. Luporini, C. Alimenti & A. Vallesi, 2013. Characterization and expression of the gene encoding En-MAPK1, an intestinal cell kinase (ICK)-like kinase activated by the autocrine pheromone-signaling loop in the polar ciliate, Euplotes nobilii. International Journal of Molecular Sciences 14: 7457–7467.
Chen, X., Y. Zhao, S. A. Al-Farraj, S. A. Al-Quraishy, H. A. El-Serehy, C. Shao & K. A. S. Al-Rasheid, 2013. Taxonomic descriptions of two marine ciliates, Euplotes dammamensis n. sp. and Euplotes balteatus (Dujardin, 1841) Kahl, 1932 (Ciliophora Spirotrichea, Euplotida), collected from the Arabian Gulf, Saudi Arabia. Acta Protozoologica 52: 73–89.
Chiappori, F., S. Pucciarelli, I. Merelli, P. Ballarini, C. Miceli & L. Milanesi, 2012. Structural thermal adaptation of β-tubulins from the Antarctic psychrophilic protozoan Euplotes focardii. Proteins 80: 1154–1166.
Darling, K. F. & C. M. Wade, 2008. The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Marine Micropaleontology 67: 216–238.
Darling, K. F., C. M. Wade, I. A. Steward, D. Kroon, R. Dingle & A. J. Leigh Brown, 2000. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature 405: 43–47.
de Vargas, C., L. Zaninetti, H. Hilbrecht & J. Pawlowski, 1997. Phylogeny and rates of molecular evolution of planktonic foraminifera: SSU rDNA sequences compared to fossil records. Journal of Molecular Evolution 45: 285–294.
Di Giuseppe, G., M. Barbieri, A. Vallesi, P. Luporini & F. Dini, 2013a. Phylogeographical pattern of Euplotes nobilii, a protist ciliate with a bipolar biogeographical distribution. Molecular Ecology 22: 4029–4037.
Di Giuseppe, G., F. Dini, C. Alimenti, A. Vallesi & P. Luporini, 2013b. Pole to pole gene flow in protozoan ciliates. In Di Prisco, G. & C. Verde (eds), Adaptation and Evolution in Marine Environments, Vol. 2. Springer, Berlin: 55–66.
Di Giuseppe, G., F. Erra, F. Dini, A. Alimenti, A. Vallesi, B. Pedrini, K. Wüthrich & P. Luporini, 2011. Antarctic and Arctic populations of the ciliate Euplotes nobilii show common pheromone mediated cell-cell signaling and cross-mating. Proceedings of the National Academy of Sciences of the United States of America 108: 3181–3186.
Di Giuseppe, G., F. Erra, F. P. Frontini, F. Dini, A. Vallesi & P. Luporini, 2014. Improved description of the bipolar ciliate, Euplotes petzi, and definition of its basal position in the Euplotes phylogenetic tree. European Journal of Protistology 50: 402–411.
Dini, F. & D. Nyberg, 1993. Sex in ciliates. In Jones, J. G. (ed.), Advances in Microbial Ecology. Plenum Press, New York: 85–153.
Dolan, J. R., E. J. Yang, S. H. Lee & S. Y. Kim, 2013. Tintinnid ciliates of Amundsen Sea (Antarctica) plankton communities. Polar Research 32: 19784.
Ehrenberg, C. G., 1844. Einige vorläufige Resultate seiner Untersuchungen der ihm von der Südpol reise des Captain Ross, so wie von den Herren Schayer und Darwin zugekommenen Materialien über das Verhalten des kleinsten Lebens in den Oceanen und den grössten bisher zugänglichen Tiefen des Weltmeeres. Bericht über die zur Bekanntmachung Geeigneten Verhandlungen Der Königl. Preuss. Akademie Der Wissenschaften zu Berlin 1844: 182–207
Falkowski, P. G., M. E. Katz, A. H. Knoll, A. Quigg, J. A. Raven, O. Schofield & F. J. R. Taylor, 2008. The evolution of modern eukaryotic phytoplankton. Science 305: 354–360.
Felsenstein, J., 1988. Phylogenies from molecular sequences: inference and reliability. Annual Review of Genetics 22: 521–565.
Fensome, R. A., R. A. MacRae, J. M. Moldowan, F. J. R. Taylor & G. L. Williams, 1996. The early Mesozoic radiation of dinoflagellates. Paleobiology 22: 329–338.
Finkelstein, S. A., J. Bunbury, K. Gajewski, A. P. Wolfe, J. K. Adams & J. E. Devlin, 2014. Evaluating diatom-derived Holocene pH reconstructions for Arctic lakes using an expanded 171-lake training set. Journal of Quaternary Science 29: 249–260.
Génermont, J., V. Machelon & M. Tuffrau, 1976. Données expérimentales relatives au problème de l’espèce dans le genre Euplotes (Ciliés hypotriches). Protistologica 12: 239–248.
Geralt, M., C. Alimenti, A. Vallesi, P. Luporini & K. Wüthrich, 2013. Thermodynamic stability of psychrophilic and mesophilic pheromones of the protozoan ciliate Euplotes. Biology 2: 142–150.
Gersonde, R., 1990. The paleontological significance of fossil diatoms from high latitude oceans. In Medlin, L. K. & J. Priddle (eds), Polar Marine Diatoms. British Antarctic Survey, Cambridge: 57–63.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium 41: 95–98.
Jiang, J., Q. Zhang, X. Hu, C. Shao, K. A. S. Al-Rasheid & W. Song, 2010a. Two new marine ciliates, Euplotes sinicus sp. nov. and Euplotes parabalteatus sp. nov., and a new small subunit rRNA gene sequence of Euplotes rariseta (Ciliophora, Spirotrichea, Euplotida). International Journal of Systematic and Evolutionary Microbiology 60: 1241–1251.
Jiang, J., Q. Zhang, A. Warren, K. A. S. Al-Rasheid & W. Song, 2010b. Morphology and SSU rRNA gene-based phylogeny of two marine Euplotes species, E. orientalis spec. nov. and E. raikovi Agamaliev, 1966 (Ciliophora, Euplotida). European Journal of Protistology 46: 121–132.
Kepner Jr, R. L., R. A. Jr Wharton & D. W. Coats, 1999. Ciliated protozoa of two Antarctic lakes: analysis by quantitative protargol staining and examination of artificial substrates. Polar Biology 21: 285–294.
Knox, G., 1994. The Biology of Southern Ocean. Cambridge University Press, Cambridge.
Lovejoy, C., 2014. Changing views of Arctic protists (marine microbial eukaryotes) in a changing Arctic. Acta Protozoologica 53: 91–100.
Lovejoy, C., R. Massana & C. Pedros-Alio, 2006. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Applied and Environmental Microbiology 72: 3085–3095.
Medlin, L., H. J. Elwood, S. Stickel & M. L. Sogin, 1988. The characterization of enzymatically amplified eukaryotic 16S-liker RNA-coding regions. Gene 71: 491–499.
Mieczan, T., D. Gorniek, A. Swiatecki, M. Zdanowski & M. Tarkowska-Kukuryk, 2013. The distribution of ciliates on Ecology Glacier (King George Island, Antarctica): relationships between species assemblages and environmental parameters. Polar Biology 36: 249–258.
Montresor, M., C. Lovejoy, L. Orsini, G. Procaccini & S. Roy, 2003. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biology 26: 186–194.
Nobili, R., P. Luporini & F. Dini, 1978. Breeding systems, species relationships and evolutionary trends in some marine species of Euplotidae. In Battaglia, B. & J. A. Beardmore (eds), Marine Organisms: Genetics, Eecology and Evolution. Plenum Press, New York: 591–616.
Pawlowski, J., J. Fahrni, B. Lecroq, D. Longet, N. Cornelius, L. Excoffier, T. Cedhagen & A. J. Gooday, 2007. Bipolar gene flow in deep-sea benthic foraminifera. Molecular Ecology 16: 4089–4096.
Pedrini, B., W. J. Placzek, E. Koculi, C. Alimenti, A. La Terza, P. Luporini & K. Wüthrich, 2007. Cold-adaptation in sea-water-borne signal proteins: sequence and NMR structure of the pheromone En-6 from the Antarctic ciliate Euplotes nobilii. Journal of Molecular Biology 372: 277–286.
Petroni, G., F. Dini, F. Verni & G. Rosati, 2002. A molecular approach to the tangled intrageneric relationships underlying phylogeny in Euplotes (Ciliophora, Spirotrichea). Molecular Phylogenetics and Evolution 22: 118–130.
Petz, W., 2004. Ciliate biodiversity in Antarctic and Arctic freshwater habitats – a bipolar comparison. European Journal of Protistology 39: 491–494.
Petz, W., 2005. Ciliates. In Scott, F. J. & H. J. Marchant (eds), Antarctic Marine Protists. Australian Biological Resources Study, Camberra: 347–448.
Petz, W. & W. Foissner, 1997. Morphology and infraciliature of some soil ciliates (Protozoa, Ciliophora) from continental Antarctica, with notes on the morphogenesis of Sterkiella histriomuscorum. Polar Records 33: 307–326.
Petz, W., W. Song & N. Wilbert, 1995. Taxonomy and ecology of the ciliate fauna (Protozoa, Ciliophora) in the endopagial and pelagial of the Weddel Sea, Antarctica. Stapfia 40: 1–223.
Petz, W., A. Valbonesi, U. Schiftner, A. Quesada & C. Ellis-Evans, 2007. Ciliate biogeography in Antarctic and Arctic freshwater ecosystems: endemism or global distribution of species? FEMS Microbial Ecology 59: 396–408.
Posada, D. & K. A. Crandall, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818.
Ronquist, F. & J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Schmidt, H. A., K. Strimmer, M. Vingron & A. von Haeseler, 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504.
Scott, F. J. & H. J. Marchant, 2005. Antarctic Marine Protists. Australian Biological Resources Study, Camberra.
Sims, P. A., D. G. Mann & L. K. Medlin, 2006. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45: 361–402.
Sorokin, Y. I., 1999. Aquatic Marine Ecology. Backhuys, Leiden.
Souffreau, C., H. Verbruggen, A. P. Wolfe, P. Vanormelingen, P. A. Siver, E. J. Cox, D. G. Mann, B. Van de Vijver, K. Sabbe & W. Vyverman, 2011. A time-calibrated multi-gene phylogeny of the diatom genus Pinnularia. Molecular Phylogenetics and Evolution 61: 866–879.
Swofford, D. L., 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, Massachusetts.
Taylor, F. J. R., M. Hoppenrath & J. F. Soldiarrage, 2008. Dinoflagellate diversity and distribution. Biodiversity Conservation 17: 407–418.
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin & D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 4: 4876–4882.
Valbonesi, A. & P. Luporini, 1990a. Description of two new species of Euplotes (Ciliophora, Hypotrichida) from Antarctica. Polar Biology 11: 47–53.
Valbonesi, A. & P. Luporini, 1990b. A new marine species of Euplotes (Ciliophora, Hypotrichida) from Antarctica. Bulletin of the British Museum of Natural History (Zoology) 56: 57–61.
Valbonesi, A. & P. Luporini, 1993. Biology of Euplotes focardii, an Antarctic ciliate. Polar Biology 13: 489–493.
Valbonesi, A., C. Ortenzi & P. Luporini, 1992. The species problem in a ciliate with a high multiple mating type system, Euplotes crassus. Journal of Protozoology 39: 45–54.
Vallesi, A., C. Alimenti, G. Di Giuseppe, F. Dini & P. Luporini, 2012. Coding genes and molecular structures of the diffusible signaling proteins (pheromones) of the polar ciliate, Euplotes nobilii. Marine Genomics 8: 9–13.
Vallesi, A., C. Alimenti, G. Di Giuseppe, F. Dini, B. Pedrini, K. Wüthrich & P. Luporini, 2010. The water-born protein pheromones of the polar protozoan ciliate, Euplotes nobilii: coding genes and molecular structures. Polar Science 4: 237–244.
Vincent, W. F., 1988. Microbial Ecosystems of Antarctica. Cambridge University Press, Cambridge.
Wilbert, N. & W. Song, 2005. New contributions to the marine benthic ciliates from the Antarctic area, including description of seven new species (Protozoa, Ciliophora). Journal of Natural History 39: 935–973.
Wilbert, N. & W. Song, 2008. A further study on littoral ciliates (Protozoa, Ciliophora) near King George Island, Antarctica, with description of a new genus and seven new species. Journal of Natural History 42: 979–1012.
Acknowledgments
The authors warmly thank anonymous reviewers for insightful and constructive criticisms, and Dr. Gill Philip for editing the English usage. The research was financially supported by the “Programma Nazionale di Ricerca in Antartide” (PNRA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: Diego Fontaneto & Stefano Schiaparelli / Biology of the Ross Sea and Surrounding Areas in Antarctica
Rights and permissions
About this article
Cite this article
Di Giuseppe, G., Dini, F., Vallesi, A. et al. Genetic relationships in bipolar species of the protist ciliate, Euplotes . Hydrobiologia 761, 71–83 (2015). https://doi.org/10.1007/s10750-015-2274-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-015-2274-5