Abstract
Ancient lakes represent one of the most stable freshwater environments on Earth, with a species richness clearly exceeding that of younger and more short-lived limnic habitats. In most cases, the biological colonization of old lake systems must have occurred via the surrounding rivers. Two ancient lake systems of Sulawesi (Malili lake system and Lake Poso) have been studied in terms of the taxonomy and phylogeny of freshwater crabs (Decapoda: Brachyura: Gecarcinucidae: Parathelphusinae). Both systems have been colonized twice independently, and in both systems we can find three trophic niches which are always occupied by different crab species: molluskivores, omnivores, and detritivores. In the present study, we reconstruct phylogenies of freshwater crabs from more than 20 river systems of Sulawesi. We thereby confirm two independent colonization events for both ancient lake systems, with subsequent radiations. The phylogenies imply that the lineages which evolved into the molluskivore forms were the first ones to colonize, whereas omnivores and detritivores are derived from later colonization events and, based on their monophyletic relationship, resulted from minor lacustrine radiations. Most of the diversity of freshwater crabs from Sulawesi remains taxonomically undescribed. This study uncovers several undocumented phylogenetic units, with long independent evolutionary histories according to patristic distances.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Southeast Asia is a well-known hotspot of biodiversity (Myers et al., 2000), with its insular part being subdivided into three regions: Sundaland, the Philippines, and Wallacea. Each of these regions shows a remarkably high degree of biodiversity (Sodhi et al., 2004). Within these areas, the freshwater systems have a high share of biodiversity and endemism (e.g., amphibians in Sodhi et al., 2004; Dudgeon et al., 2006). Therefore, animals that are restricted to freshwater habitats can be considered as good indicators for overall biodiversity and, conversely, we can assume to find high diversity of freshwater organisms within the hotspots of biodiversity in Southeast Asia. However, most of the studies on the biodiversity of Southeast Asia so far focused on terrestrial animals (Inger & Voris, 2001; Gorog et al., 2004; Steiper, 2006; Outlaw & Voelker, 2008; Meijaard, 2009; Blackburn et al., 2010; den Tex et al., 2010; Lim et al., 2010; Sheldon et al., 2010), and the aquatic diversity often still needs to be estimated and documented.
Freshwater crabs can be considered good zoogeographic indicators (Ng & Rodriguez, 1995). They are completely independent from the marine habitat in terms of their ontogeny (direct development), and they are probably restricted to their respective drainage system (e.g., Daniels et al., 2002b; Daniels, 2003), which leads to a high rate of endemism and, therefore, to high overall diversity. The freshwater crab fauna of Southeast Asia is of special interest, because of the insular habitats and past connections during lowered sea levels at the ice ages. Recently, Klaus et al. (2013) reconstructed the phylogenetic history and geographic origin of the crab genus Parathelphusa and related genera, postulating a center of origin on the island of Borneo and revealing high undocumented biodiversity. It also became evident that all representatives of Sulawesi are monophyletic, with inclusion of one ancestral lineage from southeast Borneo. But despite of these new insights and earlier studies by Schubart et al. (2008) and Schubart & Ng (2008), the phylogenetic history, phylogeographic structure, and genetic diversity of the freshwater crabs from Sulawesi remains mostly unknown.
The freshwater crabs of Sulawesi had evoked interest, due to the presence of different ecotypes and consequent sympatric diversity in the ancient lakes of the Central Plateau (Poso Lake and Malili lake system). These lake representatives belong to the genera Parathelphusa, Nautilothelphusa, Migmathelphusa, Syntripsa, and Sundathelphusa (see Chia & Ng, 2006), showing a high rate of lacustrine diversity within the relatively small area of these lake systems. This is paralleled by high diversity levels in freshwater shrimps (von Rintelen et al., 2010a, 2012), snails (von Rintelen et al., 2004, 2010b, 2012), and fishes (Herder et al., 2012a, b; von Rintelen et al., 2012), which have been shown to be the outcome of adaptive radiations. Previous studies on Asian freshwater crabs had shown that Sulawesian representatives of the genus Sundathelphusa belong to different phylogenetic lineages than the genus Parathelphusa (see Klaus et al., 2009, 2010), whereas Nautilothelphusa, Migmathelphusa, and Syntripsa are found to be phylogenetically embedded within the genus Parathelphusa (see Schubart & Ng, 2008; Schubart et al., 2008). The latter authors reconstructed phylogenetic relationships of Sulawesi freshwater crabs, described a new species, and showed that colonization took place at least twice independently in both lake systems, with only minor subsequent radiations. However, only a small number of the overall freshwater drainage systems of Sulawesi and tributaries to the lakes were included in these studies. Thus, the real dimension of biodiversity that is to be expected within the freshwater crabs on Sulawesi and the potential colonization pathways must be considered in a larger framework. In a recent effort, we were able to obtain mtDNA sequences from freshwater crabs inhabiting more than 20 river systems of Sulawesi. This leads to a better insight into the real biodiversity of Sulawesi river systems. Although we are far from providing a complete picture of rivers from Sulawesi and small neighboring islands, due to the lack of available material, our aim is to show that the biodiversity in terms of evolutionary lineages is much higher than that currently recognized in the scientific literature (Bott, 1970). Currently, 13 species of Parathelphusa are considered valid for Sulawesi (Chia & Ng, 2006). Here, we examine and quantify the biodiversity in a number of freshwater systems on Sulawesi to clarify the evolutionary history and to estimate potential numbers of species of freshwater crabs on Sulawesi.
Materials and methods
Sampling and sequencing
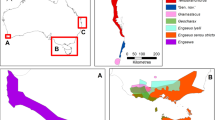
Freshwater crabs were collected in Sulawesi during several expeditions between 2000 and 2008. They were preserved in ethanol and often walking legs were punctured with sterile needles for better alcohol penetration into the tissue. Many crabs were made available by Thomas and Kristina von Rintelen, Matthias Glaubrecht, and Fabian Herder. At the laboratories of the University of Regensburg, genomic DNA was isolated using a modified Puregene method (Gentra Systems, Minneapolis). In order to amplify the so-called “Folmer region” of the mitochondrial gene cytochrome oxidase subunit 1 (Cox1), polymerase chain reactions (PCR) were carried out with the primers LCO 1490 (GGT CAA CAA ATC ATA AGA TAT TGG) (Folmer et al., 1994), COL6b (ACA AAT CAT AAA GAT ATY GG) and COH6 (TAD ACT TCD GGR TGD CCA AAR AAY CA) (Schubart & Huber, 2006; Schubart, 2009) and the following PCR profile: 40 cycles with 45 s 94°C, 1 min 48–50°C, and 1 min 72°C for denaturing, annealing, and extension, respectively. PCR products were purified with the Quick-Clean kit (Bioline) or Microcon filters (Millipore), cycle sequenced with the Big Dye Terminator v1.1 (AB Applied Biosystems) and analyzed with the automated capillary DNA sequencer ABI PRISM® 310 Genetic Analyzer (Applied Biosystems) (see also Schubart et al., 2008). In addition, 71 Cox1 sequences were obtained after sending tissue of samples to the Barcode of Life Database (BOLD) in Guelph (Canada) (see Ratnasingham & Hebert, 2007). All available Cox1 sequences of Sulawesi freshwater crabs originating from the previous studies on the relationship of the Gecarcinucidae or the colonization of the Sulawesi lakes were retrieved from GenBank (Schubart et al., 2008; Klaus et al., 2013: AB265250, AB601861, FM177599, FM177607–609, FM177611, FM177615–616, FM177618, FM177622, FM177625, FM177628–629, FM177635–643, GQ289661, GQ982588). Overall, this allowed aligning a total of 116 Cox1 sequences with a length of 658 base pairs using the software BioEdit (v.7.0.5.3) (Hall, 1999). Cox1 sequences from Oziothelphusa hippocastanum (GQ982588, Beenaerts et al., 2010), Sayamia cf. germaini (AB265250, Shih et al., 2007), and Siamthelphusa improvisa (AB601861, Shih et al., 2011) were included as outgroups or reference for intergeneric genetic distances. The sampled geographic area of the ingroups covers more than 20 river systems from central and southern Sulawesi (Fig. 1). However, many river systems could not be included in this sampling and, therefore, the overall analysis remains incomplete.
Phylogenetic analyses
Two alignments, one for our complete set of sequences and one for all unique haplotypes from Sulawesi freshwater crabs of the genera Parathelphusa, Nautilothelphusa, Migmathelphusa, and Syntripsa, were converted with FaBox (Villesen, 2007) to Nexus files as input for Mr.Bayes (v. 3.2.1) (Huelsenbeck and Ronquist, 2001). The best evolutionary model was determined with MrModeltest (v.2.3) (Nylander, 2004) and MrMTgui (v.1.0) (Nuin, 2005) in PAUP (v.4.0b10) (Swofford, 2003). The evolutionary model that best described our data was selected with the Akaike information criterion (Posada & Buckley, 2004). For the Mr.Bayes run, we used 3 million generations with 4 chains (one heated) and a sample frequency of one tree per 1,000 generations.
Patristic distances
Based on the smaller alignment (restricted to sequences of Parathelphusa and related genera from Sulawesi, i.e., Parathelphusa lineage), of 116 sequences, we used R (R Core Team, 2012) with the Ape package (v.3.0-6) (Paradis et al., 2004) to calculate patristic distances for each taxon pair along the final tree (Fig. 3). For determining an estimate of the number of potentially to be expected operational taxonomic units (OTUs), we initially applied a threshold of 0.16 as suggested in Lefébure et al. (2005).
Results
The alignment contained no indels or stop codons, making the inclusion of pseudogenes less likely. Analyzing the dataset with MrModeltest (Nylander, 2004) revealed GTR+G+I (Rodriguez et al., 1990) as the best fitting evolutionary model for the alignment containing only the Parathelphusa lineage and HKY+I+G (Hasegawa et al., 1985) as the best fitting evolutionary model for the alignment including Sundathelphusa and other outgroups. Therefore, these models were chosen for subsequent Bayesian inference analyses.
The resulting phylogenetic tree based on the larger alignment (Fig. 3) shows a deep early split between two clades, corresponding to Sundathelphusa on one hand, and Parathelphusa and allied genera on the other. As stated by Schubart & Ng (2008) and Klaus et al. (2013), the genera Syntripsa, Migmathelphusa, and Nautilothelphusa are deeply nested within the genus Parathelphusa, which currently renders the genus paraphyletic (Figs. 3, 4). Hereafter, we will consider all these genera from Sulawesi as a monophyletic lineage and refer to it as the Parathelphusa lineage (see above).
The overall tree (Fig. 3) suggests an early colonization of Lake Poso by the genus Sundathelphusa, while the following colonization events of Lake Poso and the Malili lake system took place from within the Parathelphusa lineage more recently (Fig. 3, see also Schubart et al., 2008).
Even though there are only 14 valid species of Parathelphusa currently recognized for Sulawesi (see Chia & Ng, 2006), the clusters reflected in the topology of the Parathelphusa lineage phylogeny from Sulawesi (Fig. 4) and patristic distances along this tree suggest a total potential number of up to 35 OTUs, if a threshold of 0.16 is applied as suggested by Lefébure et al. (2005).
The base of the tree for the Parathelphusa lineage (Fig. 4) is polytomous and formed by four big lineages. The first of these lineages consists of up to eight potential OTUs (Table 1, 1–6). Only two of them could be identified and connected to a valid species name, Parathelphusa linduensis Roux, 1904 (4b) and P. lokaensis De Man, 1892 (6). The second lineage contains three potential OTUs (Table 1, 7–9), and none of them could be assigned a valid species name.
The third lineage is the largest and can be subdivided into two clades and several groups. The first clade (10a–14) includes seven potential OTUs (Table 1, 10a–14). One of them could be identified as Parathelphusa pareparensis De Man, 1892 (13) whereas the others could not be assigned to one of the existing scientific names. The second clade within the third lineage contains three distinct groups. The first one (15a–24b) is formed by five OTUs (Table 1, 15a–24b) that are currently recognized as Parathelphusa pallida Schenkel, 1902 sensu lato (15a–17 and 23), a clade uniting Parathelphusa ferruginea Chia & Ng, 2006 and Nautilothelphusa zimmeri Balss, 1933 from lakes Towuti and Mahalona (19), a clade of Nautilothelphusa zimmeri from Lake Matano (20), an unnamed species of Parathelphusa from Lake Masapi (21), a clade of Parathelphusa pantherina Schenkel, 1902 (22), and two clades that could not be identified (24a and 24b) to species level. The second group consists of two clades (Table 1, 25–26) that could not be assigned to a scientific valid name (25 and 26). The third group is formed by four distinct clades (Table 1, 27–30), Parathelphusa possoensis Roux, 1904 (27), Parathelphusa sarasinorum Schenkel, 1902 (28), Migmathelphusa olivacea Chia & Ng, 2006 (29), Syntripsa matannensis Chia & Ng, 2006, and S. flavichela Chia & Ng, 2006 (30). The fourth of the basal lineages of the Parathelphusa lineage on Sulawesi consists of three potential OTUs (Table 1, 31–33), of which only one could be linked to a species name, i.e., Parathelphusa sorella Chia & Ng, 2006 (33).
Unfortunately, for some samples (FM177635; R619-A06; R619-A07; R619-A10; R619-A11; R619-C01; R619-C02; R619-C03; R619-C04; R619-C07; R619-C08; R619-H10; R619-H11; Seb; Seb-1; Seb-3; ZRC2000.1681; ZRC2000.1684; ZRC2000.1695) we were not able to achieve the exact coordinates and, therefore, they are not shown in Figs. 1 and 2.
Discussion
The overall phylogenetic tree (Fig. 3) shows that both the ancient lake systems (Malili lake system and Lake Poso) have been colonized twice independently with subsequent radiations. A similar pattern of multiple independent colonizations and a subsequent radiation can also be found in other groups like gastropods (von Rintelen et al., 2004) and shrimps (von Rintelen et al., 2010a) from the ancient lakes in Sulawesi. In both systems, we can find three ecological niches which are always occupied by different crab species: molluskivores, omnivores, and detritivores. This confirms the results based on 16S mtDNA and a much smaller dataset by Schubart & Ng (2008). Furthermore, the trees in Figs. 3 and 4 imply that the lineages which today are represented by molluskivore species were the first ones to colonize the lake, whereas omnivores and detritivores are derived from later colonization events and the outcome of local lacustrine radiations. This may be explained by the fact that mollusks represent the richest and most reliable source of proteins within the lake system and, therefore, the best available food source for the freshwater crabs. Von Rintelen et al. (2004) also described a long arms race between crabs and snails that may have driven diversification and adaptive radiation in the mollusks. The oldest lineage of the lake dwellers appears to be Sundathelphusa molluscivora Schubart & Ng, 2008, which occupies the molluskivore niche in Lake Poso. The other niches in this lake are filled by species from the Parathelphusa lineage: Parathelphusa sarasinorum is the generalist in Lake Poso, but based on its chelar morphology also includes mollusks in its diet, whereas Migmathelphusa olivacea with its broadened last pair of legs occupies the niche of the soft sediment dwelling detritivore within this lake. In the Malili lake system, Syntripsa flavichela and Syntripsa matannensis are obviously the molluskivore forms, with notoriously large molariform armed chelae. Parathelphusa ferruginea and Parathelphusa pantherina appear to be the generalists, and Nautilothelphusa zimmeri, with the smallest forceps-shaped pincers, the detritivores.
Phylogenetic tree based on 658 base pairs from the mitochondrial Cox1 gene created with MrBayes (v.3.2.1) with 3 million generations, a sample frequency of 1,000, HKY+I+G as evolutionary model, and Oziothelphusa hippocastanum as outgroup. The standard deviation of split frequencies by the end of the analysis was 0.006649. A more detailed version of the tree is available as electronic supplementary material
Phylogenetic tree based on 658 base pairs from the mitochondrial Cox1 gene created with Mr.Bayes (v.3.2.1) with 3 million generations, a sample frequency of 1,000, GTR+G+I as evolutionary model, and a Parathelphusa sp. (34) from Borneo as outgroup. The standard deviation of split frequencies by the end of the analysis was 0.006903. A more detailed version of the tree is available as electronic supplementary material
The phylogenetic tree restricted to the Parathelphusa lineage as defined above (Fig. 4) reveals a huge proportion of cryptic diversity within the freshwater crabs of Sulawesi. The number of well-supported clades in the tree (Fig. 4) drastically exceeds the number of available scientific names for the genus Parathelphusa, which currently consists of 13 (see Chia & Ng, 2006). Even with our alignment lacking sequences from the type locality for P. ceophallus Ng, 1993; P. tenuipes Schenkel, 1902; and P. crocea Schenkel, 1902; the number of well-supported clades is clearly exceeding the number of available scientific names. The patristic distances, with the threshold of 0.16 (as applied by Lefébure et al., 2005) suggest up to 35 different potential OTUs within the sampled freshwater crabs of the Parathelphusa lineage from Sulawesi. Patristic estimates of species numbers are only rough estimates of taxonomic diversity and remain highly subjective and dependent on the threshold selected. Also relying on a single mitochondrial marker like Cox1 can lead to a wrong estimation of biodiversity. Our clade 19 consists of Parathelphusa ferruginea and Nautilothelphusa zimmeri which cannot be separated from each other using Cox1 as mitochondrial marker due to possible mitochondrial introgression (see also Schubart et al., 2008). This could lead to an underestimation of biodiversity. Relying solely on Cox1 sequences, or other single-locus datasets, bares a risk of including pseudogenes or other paralogs, which would lead to an overestimation of biodiversity. We partly addressed this issue by controlling our sequence electropherograms for double peaks and testing for indels and stop codons to avoid inclusion of pseudogenes in the analysis. In any case, our aim was not to give exact species numbers as a measure for diversity within the Parathelphusa lineage. Nevertheless, the bottomline message remains that the phylogeny reveals a huge potential for cryptic diversity within this group and on a long term will lead to a drastic increase of recognized species.
With a threshold for the patristic distance of 0.063, derived from our own data and corresponding to the distance between Nautilothelphusa zimmeri and a morphologically clearly distinct undescribed species of Parathelphusa from Lake Masapi (21), the number of potentially distinguishable OTUs rises to 43; Even with a more conservative estimate of threshold of 0.24 (subjective increase by 50% compared to the one used by Lefébure et al., 2005), still 23 OTUs could be postulated and many new species possibly described. Due to the fact that genetic distances must not be directly correlated to species boundaries and the use of only one single gene in the analysis, the patristic distances probably do not reflect the real number of valid species within our dataset, but they are a good indicator to show that there may be indeed a high number of unrecognized species hidden within the freshwater systems of Sulawesi. The real number of cryptic or unrecognized freshwater crab species in Sulawesi is probably even higher, because large parts of Sulawesi are still completely unsampled and leave a high potential for additional species being present in different drainage systems and on several neighboring islands. The Sulawesi Parathelphusa lineage has its next relatives on southeastern Borneo, as suggested by Klaus et al. (2013). Here we show that the monophyletic Parathelphusa lineage rapidly splits up into several distinct clusters. This indicates a very quick radiation into the different available niches after the arrival of their common ancestor from Borneo.
Also the biodiversity within the genus Sundathelphusa must be considered highly underrated until now, as crabs of this genus also show a high potential for endemism. Our study already reveals at least two undescribed species of Sundathelphusa (both here determined as “Sundathelphusa Mayoa,” see Fig. 3). To address this problem, further studies must be carried out to morphologically examine the different clades of freshwater crabs that are distinguished here.
A number of published studies already revealed a large proportion of cryptic diversity in freshwater crabs in South Africa (Daniels et al., 2001, 2002a, 2003, 2006; Daniels & Bayliss, 2012) belonging to the family Potamonautidae, in those along the Aegean region (Jesse et al., 2010, 2011), the Middle East (Keikhosravi & Schubart, unpublished data) and East Asia (Shih et al., 2006; Shih & Ng, 2011) belonging to the family Potamidae, and in freshwater crabs from Borneo (Klaus et al., 2013) belonging to the Gecarcinucidae. In contrast, studies on neotropical freshwater crabs of the family Pseudothelphusidae showed a relatively low rate of endemism and diversity (Cook et al., 2008; Poettinger et al., 2011; Schubart et al., 2011; Rivera et al., unpublished data). This is probably due to the fact that the different families of freshwater crabs may not be monophyletic and colonized freshwater systems independently (see Tsang et al. unpublished data). Thus, they may differ in their ability to migrate over land. The neotropical Pseudothelphusidae seem to have a greater potential for dispersal over land and crossing hydrographic barriers. This may be due to adaptations of their branchial chamber to air breathing (see Díaz & Rodríguez, 1977). Therefore, they may be less geographically restricted and show less potential for endemism than Gecarcinucidae, Potamidae, and Potamonautidae, which constitute a monophyletic assemblage (Klaus et al., 2006, 2009, 2010; Marijnissen et al., 2006; Tsang et al., submitted). In contrast to the neotropical freshwater crabs, the Parathelphusa lineage from Sulawesi (this study) and representatives of Parathelphusa from Borneo (see Klaus et al., 2013) are genetically highly structured and have a strong potential for endemism.
Overall, it remains to be said that the species richness of Wallacea is remarkably high (Sodhi et al., 2004), but in the studied freshwater systems it is evidently far from being satisfactorily explored and documented. In order to obtain a complete picture of the real diversity of the freshwater crabs from Sulawesi and the colonization history of the island, far more comprehensive sampling and analyses must be carried out in the future.
Conclusion
Two large ancient lake systems on Sulawesi have been colonized twice independently by freshwater crabs. In both systems, the molluskivore niche was occupied by the lineages that colonized the corresponding lake system first and the following species evolved into detritivores and generalists within the lakes. In addition, our trees show that a huge part of the biodiversity within the freshwater crabs of Sulawesi remains undescribed. Even without sampling all of the drainage systems and neighboring islands, the number of clades in the tree (Fig. 3) and potential species is clearly exceeding the number of available scientific names.
References
Beenaerts, N., R. Pethiyagoda, P. K. L. Ng, D. C. Yeo, G. J. Bex, M. M. Bahir & T. Artois, 2010. Phylogenetic diversity of Sri Lankan freshwater crabs and its implications for conservation. Molecular Ecology 19: 183–196.
Blackburn, D. C., D. P. Bickford, A. C. Diesmos, D. T. Iskandar & R. M. Brown, 2010. An ancient origin for the enigmatic flat-headed frogs (Bombinatoridae: Barbourula) from the islands of Southeast Asia. PLoS One 5(8): e12090.
Bott, R., 1970. Die Süßwasserkrabben von Europa, Asien, Australien und ihre Stammesgeschichte. Eine Revision der Potamoidea und der Parathelphusoidea (Crustacea, Decapoda). Abhandlungen der Senckenbergischen Naturforschenden Gesellschaft 526: 1–338, pls. 1–58.
Chia, O. K. S., P. K. L. Ng, 2006. The freshwater crabs of Sulawesi, with descriptions of two new genera and four new species (Crustacea: Decapoda: Brachyura: Parathelphusidae). The Raffles Bulletin of Zoology 54: 381–428.
Cook, B. D., C. M. Pringle, J. M. Hughes, 2008. Phylogeography of an island endemic, the Puerto Rican freshwater crab (Epilobocera sinuatifrons). Journal of Heredity. doi:10.1093/jhered/esm126.
Daniels, S. R., 2003. Examining the genetic structure among populations of the common Cape river crap Potamonautes perlatus from river systems in South Africa reveals hydrographic boundaries. Journal of Crustacean Biology 23: 936–950.
Daniels, S. R. & J. Bayliss, 2012. Neglected refugia of biodiversity: mountainous regions in Mozambique and Malawi yield two novel fresh water crab species (Potamonautidae: Potamonautes). Zoological Journal of the Linnean Society 164: 498–509.
Daniels, S. R., B. A. Stewart & L. Burmeister, 2001. Geographic patterns of genetic and morphological divergence amongst populations of a river crab (Decapoda: Potamonautidae) with the description of a new species from mountain streams in the Western Cape. Zoologica Scripta 30: 181–197.
Daniels, S. R., B. A. Stewart & P. A. Cook, 2002a. Congruent patterns of genetic variation in a burrowing freshwater crab revealed by allozymes and mtDNA sequence analysis. Hydrobiologia 468: 171–179.
Daniels, S. R., B. A. Stewart, G. Gouws, M. Cunningham & C. A. Matthee, 2002b. Phylogenetic relationships of the southern African freshwater crab fauna (Decapoda: Potamonautidae: Potamonautes) derived from multiple sets reveal biogeographic patterning. Molecular Phylogenetics and Evolution 25: 511–523.
Daniels, S. R., G. Gouws, B. A. Stewart & M. Coke, 2003. Molecular and morphological data demonstrate the presence of cryptic lineages among freshwater crabs (Decapoda: Potamonautidae: Potamonautes) from the Drakensberg Mountains, South Africa. Biological Journal of the Linnean Society 78: 129–147.
Daniels, S. R., N. Cumberlidge, M. Pérez-Losada, S. A. Marijnissen & K. A. Crandall, 2006. Evolution of Afrotropical freshwater crab lineages obscured by morphological convergence. Molecular Phylogenetics and Evolution 40: 227–235.
den Tex, R. J., R. Thorington, J. E. Maldonado & J. A. Leonard, 2010. Speciation dynamics in the SE Asian tropics: putting a time perspective on the phylogeny and biogeography of Sundaland tree squirrels, Sundasciurus. Molecular Phylogenetics and Evolution 55: 711–720.
Díaz, H. & G. Rodríguez, 1977. The branchial chamber in terrestrial crabs: a comparative study. The Biological Bulletin 153: 485–504.
Dudgeon, D., A. H. Arthington, M. O. Gessner, Z. Kawabata, D. J. Knowler, C. Lévêque, R. J. Naiman, A. Prieur-Richard, D. Soto, M. L. Stiassny & C. A. Sullivan, 2006. Freshwater biodiversity: importance, threats, status and conservation challenges. Biological Reviews 81: 163–182.
Folmer, O., M. Black, W. Hoeh, R. Lutz, R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular marine Biology and Biotechnology 3: 294–299.
Gorog, A. J., M. H. Sinaga & M. D. Engstrom, 2004. Vicariance or dispersal? Historical biogeography of three Sunda shelf murine rodents (maxomys surifer, Leopoldamys sabanus and Maxomys whiteheadi). Biological Journal of the Linnean Society 81: 91–109.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Hasegawa, M., H. Kishino & T. Yano, 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 21: 160–174.
Herder, F., R. K. Hadiaty & A. W. Nolte, 2012a. Pelvic-fin brooding in a new species of riverine ricefish (Antherinomorpha: Beloniformes: Adrianichthyidae) from Tana Toraja, Central Sulawesi, Indonesia. The Raffles Bulletin of Zoology 60: 467–476.
Herder, F., U. K. Schliewen, F. M. Geiger, R. K. Hadiaty, S. M. Gray, S. J. McKinnon, R. P. Walter & J. Pfaender, 2012b. Alien invasion in Wallace’s Dreamponds: records of the hybridogenic “flowerhorn” cichlid in Lake Matano, with an annotated checklist of fish species introduced to the Malili Lakes system in Sulawesi. Aquatic Invasions 7: 521–535.
Huelsenbeck, J. P. & F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Inger, R. F. & H. K. Voris, 2001. The biogeographical relations of the frogs and snakes of Sundaland. Journal of Biogeography 28: 863–891.
Jesse, R., C. D. Schubart & S. Klaus, 2010. Identification of a cryptic lineage within Potamon fluviatile (Herbst) (Crustacea: Brachyura: Potamidae). Invertebrate Systematics 24(4): 348–356.
Jesse, R., M. Grudinski, S. Klaus, B. Streit & M. Pfenninger, 2011. Evolution of freshwater crab diversity in the Aegean region (Crustacea: Brachyura: Potamidae). Molecular Phylogenetics and Evolution 59: 23–33.
Klaus, S., C. D. Schubart & D. Brandis, 2006. Phylogeny, biogeography and a new taxonomy for the Gecarcinucoidea Rathbun, 1904 (Decapoda: Brachyura). Organisms, Diversity and Evolution 6(3): 199–217.
Klaus, S., D. Brandis, P. K. L. Ng, D. C. J. Yeo & C. D. Schubart, 2009. Phylogeny and biogeography of Asian freshwater crabs of the family Gecarcinucidae (Brachyura: Potamoidea). In Martin, J. W., K. A. Crandall & D. L. Felder (eds), Crustacean Issues 18: Decapod Crustacean Phylogenetics. Taylor & Francis/CRC Press, Boca Raton, FL: 509–531.
Klaus, S., C. D. Schubart, B. Streit & M. Pfenninger, 2010. When Indian crabs were not yet Asian – biogeographic evidence for Eocene proximity of India and Southeast Asia. BMC Evolutionary Biology 10: 287.
Klaus, S., S. Selvandran, J. G. Goh, D. Wowor, D. Brandis, P. Koller, C. D. Schubart, B. Streit, R. Meier, P. K. L. Ng & D. C. J. Yeo, 2013. Out of Borneo: Neogene diversification of Sundaic freshwater crabs (Crustacea: Brachyura: Gevarcinucidae: Parathelphusa). Journal of Biogeography 40: 63–74.
Lefébure, T., C. J. Douady, M. Gouy & J. Gibert, 2005. Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Molecular Phylogenetics and Evolution 40: 435–447.
Lim, H. C., F. Zou, S. S. Taylor, B. D. Marks, R. G. Moyle, G. Voelker & F. H. Sheldon, 2010. Phylogeny of magpie-robins and shamas (Aves: Turdidae: Copsychus and Trichixos): implications for island biogeography in Southeast Asia. Journal of Biogeography 37: 1894–1906.
Marijnissen, S. A. E., E. Michel, S. R. Daniels, D. Erpenbeck, S. B. J. Menken & F. R. Schram, 2006. Molecular evidence for recent divergence of Lake Tanganyika endemic crabs (Decapoda: Platythelphusidae). Molecular Phylogenetics and Evolution 40: 628–634.
Meijaard, E., 2009. Solving mammalian riddles along the Indochinese–Sundaic zoogeographic transition: new insights from mammalian biogeography. Journal of Biogeography 36: 801–802.
Myers, N., R. A. Mittermeier, C. G. Mittermeier, G. A. B. da Fonseca, J. Kent, 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858.
Ng, P. & G. Rodriguez, 1995. Freshwater crabs as poor zoogeographical indicators: a critique of Banarescu 1990. Crustaceana 68: 636–645.
Nuin, P. A. S., 2005. MTgui – A Simple Interface to Model Test. Program distributed by the author. University of Toronto. Available from: http://www.genedrift.org/mtgui.php.
Nylander, J. A. A., 2004. MrModeltest v2.2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
Outlaw, D. C. & G. Voelker, 2008. Pliocene climatic change in insular Southeast Asia as an engine of diversification in Ficedula flycatchers. Journal of Biogeography 35: 739–752.
Paradis, E., J. Claude & K. Strimmer, 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Poettinger, T., I. S. Wehrtmann & C. D. Schubart, 2011. Genfluss in Flussoberläufen des zentralen Hochlands von Costa Rica? Vorläufige Ergebnisse bei Süßwasserkrabben (Pseudothelphusidae). In Erweiterte Zusammenfassung der Jahrestagung 2010. Deutsche Gesellschaft für Limnologie, Hardegsen: 242–246. ISBN 978-3-9813095-0-8.
Posada, D. & T. R. Buckley, 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793–808.
R Core Team, 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. Available from http://www.R-project.org/.
Ratnasingham, S. & P. D. N. Hebert, 2007. BOLD: the Barcode of Life Data System (www.barcodinglife.org). Molecular Ecology Notes 7: 355–364.
Rodriguez, F. J., J. L. Oliver, A. Marin & J. R. Medina, 1990. The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142: 485–501.
Schubart, C. D., 2009. Mitochondrial DNA and decapod phylogenies; the importance of pseudogenes and primer optimization. In Martin, J. W., K. A. Crandall & D. L. Felder (eds), Crustacean Issues 18: Decapod Crustacean Phylogenetics. Taylor & Francis/CRC Press, Boca Raton, FL: 47–65.
Schubart, C. D. & M. G. J. Huber, 2006. Genetic comparisons of German populations of the stone crayfish, Austropotamobius torrentium (Crustacea: Astacidae). Bulletin Français de la Pêche et de la Pisciculture 380–381: 1019–1028.
Schubart, C. D. & P. K. L. Ng, 2008. A new molluscivore crab from Lake Poso confirms multiple colonisation of ancient lakes in Sulawesi by freshwater crabs (Decapoda: Brachyura). Zoological Journal of the Linnean Society 154: 211–221.
Schubart, C. D., T. Santl & P. Koller, 2008. Mitochondrial patterns of intra- and interspecific differentiation among endemic freshwater crabs of ancient lakes in Sulawesi. Contributions to Zoology 77: 83–90.
Schubart, C. D., N. T. Rivera, K. A. Crandall & T. Santl, 2011. Shallow phylogeographic structure of Puerto Rico freshwater crabs: an evolutionary explanation for low species diversity compared to Jamaica. In Held, C., S. Koenemann & C. D. Schubart (eds), Crustacean Issues 19: Phylogeography and Population Genetics in Crustacea. Taylor & Francis/CRC Press, Boca Raton, FL: 352–365, 378–381.
Sheldon, F. H., A. Styring & P. A. Hosner, 2010. Bird species richness in a Bornean exotic tree plantation: a long-term perspective. Biological conservation 143: 399–407.
Shih, H. T. & P. K. L. Ng, 2011. Diversity and biogeography of freshwater crabs (Crustacea: Brachyura: Potamidae, Gecarcinucidae) from East Asia. Systematics and Biodiversity 9(1): 1–16.
Shih, H. T., H. C. Hung, C. D. Schubart, C. A. Chen & H. W. Chang, 2006. Intraspecific diversity of the endemic freshwater crab Candidiopotamon rathbunae (Crustacea: Decapoda, Brachyura, Potamidae) reflects five million years of geological history of Taiwan. Journal of Biogeography 33: 980–989.
Shih, H. T., S. H. Fang & P. K. L. Ng, 2007. Phylogeny of the freshwater crabs genus Somanniathelphusa Bott (Decapoda: Parathelphusidae) from Taiwan and the coastal regions of China, with notes on their biogeography. Invertebrate Systematics 21: 29–37.
Shih, H. T., J. Y. Shy, T. Naruse, H. T. Hung, D. C. J. Yeo & P. K. L. Ng, 2011. Introduction of an Indochinese freshwater crab Sayamia germaini (Crustacea: Brachyura: Gecarcinucidae) to Taiwan: morphological and molecular evidence. Raffles Bulletin of Zoology 59: 83–90.
Sodhi, N.s., L. P. Koh, B. W. Brook & P. K. L. Ng, 2004. Southeast Asian biodiversity: an impending disaster. Trends in Ecology and Evolution 19: 654–660.
Steiper, M. E., 2006. Population history, biogeography, and taxonomy of orangutans (Genus: Pongo) based on a population genetic meta-analysis of multiple loci. Journal of Human Evolution 50(5): 509–522.
Swofford, D. L., 2003. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0b10.
Villesen, P., 2007. FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes 7(6): 965–968.
von Rintelen, T., A. B. Wilson, A. Meyer & M. Glaubrecht, 2004. Escalation and trophic specialization drive adaptive radiation of freshwater gastropods in ancient lakes on Sulawesi, Indonesia. Proceedings of the Royal Society 271: 2541–2549.
von Rintelen, K., M. Glaubrecht, C. D. Schubart, A. Wessel & T. von Rintelen, 2010a. Adaptive radiation and ecological diversification of Sulawesi’s ancient lake shrimps. Evolution 64(11): 3287–3299.
von Rintelen, T., K. von Rintelen & M. Glaubrecht, 2010b. The species flocks of the viviparous freshwater gastropod Tylomelania (Mollusca: Cerithioidea: Pachychilidae) in the ancient lakes of Sulawesi, Indonesia: the role of geography, trophic morphology and color as driving forces in adaptive radiation. In Glaubrecht, M. (ed.), Evolution in Action. Case Studies in Adaptive Radiation, Speciation and the Origin of Biodiversity. Springer-Verlag, Berlin: 485–512.
von Rintelen, T., K. von Rintelen, M. Glaubrecht, C. D. Schubart & F. Herder, 2012. Aquatic biodiversity hotspots in Wallacea: the species flocks in the ancient lakes of Sulawesi, Indonesia. In Goer, D. J., K. G. Johnson, J. E. Richardson, B. R. Rosen, L. R. Williams & S. T. Williams (eds), Biotic Evolution and Environmental Change in Southeast Asia. Cambridge University Press, Cambridge: 290–315.
Acknowledgments
Our thanks go to Matthias Glaubrecht, Thomas and Kristina von Rintelen (all Naturkundemuseum Berlin), Fabian Herder (Alexander-Koenig Museum Bonn), Daisy Wowor (LIPI, Bogor), Tobias Santl, Peter Koller, and Florian Kolbinger (Universität Regensburg) for helping us to gather a large collection of freshwater crabs from Sulawesi. Sebastian Klaus shared with us important sequences. He and one anonymous reviewer gave valuable feedback on a first version. We also want to thank Dirk Steinke and BOLD systems for their help to produce a large dataset of Cox1 sequences. Funding was provided through the Deutsche Forschungsgemeinschaft (DFG, project Schu1460/8) and part of the laboratory supplies from the budget of Biologie 1, Universität Regensburg (thanks to Jürgen Heinze).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: T. von Rintelen, R. M. Marwoto, G. D. Haffner & F. Herder / Speciation in Ancient Lakes – Classic Concepts and New Approaches
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Poettinger, T., Schubart, C.D. Molecular diversity of freshwater crabs from Sulawesi and the sequential colonization of ancient lakes. Hydrobiologia 739, 73–84 (2014). https://doi.org/10.1007/s10750-013-1643-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-013-1643-1