Abstract
Microbial eukaryote diversity was assessed in Arctic Kongsfjorden (Svalbard), by constructing SSU rDNA clone libraries. Samples were collected from different depths at the outer basin in summer (2006), plus an additional one glacial and one sediment sample. The libraries displayed diversity based on 284 full-length sequences. Four main phyla, namely, Alveolates, Stramenopiles, Cercozoans, and Metazoans were often screened in this fjord. Alveolate occupied the highest percentage of taxa in the library of surface sea water, besides the Metazoan-related clones. Moreover, dinoflagellates, diatoms, and pico-Prasinophytes were detected as prevalent phytoplankton through the analysis of libraries. Questions related to the quantity of these phytoplankton and their roles in the microbial food loop arose from an ecological viewpoint.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Recent environmental studies based on molecular data have revealed a high diversity of eukaryotic lineages in the marine environment, such as “pico-sized” fraction of deep-sea (López-García et al., 2001), the ocean surface (Díez et al., 2001; Fuller et al., 2006; Moon-van der Staay et al., 2001; Zeidner et al., 2003), and the coasts (Massana et al., 2004). Meanwhile, cloning of environmental rDNA has been applied into surveys of some extreme environments as permanently anoxic deep-sea waters (Stoeck & Epstein, 2003). These surveys indicate a contrasting distribution of organisms (Countway et al., 2005; Edgcomb et al., 2002; Not et al., 2007), which is particularly evident for the picoeukaryotic community (Guillou et al., 2004; Romari & Vaulot, 2004; Vaulot et al., 2004). Clear differences in the picoeukaryotic communities could be observed in shallow versus deep sea samples (Not et al., 2007). Because marine eukaryotes belonged to very different phylogenetic groups, the extent of diversity and distribution are changing the views in ecology and evolutionary biology (Not et al., 2005).
The North polar region is a highly vulnerable environment, predicted to warm rapidly as a result of global climate change and where significant impacts have already been observed. Polar coastal ecosystems are particularly sensitive to factors such as surrounding afflux and increased anthropogenic activities, and have attracted scientific interest concerning diversity and the ecological role of marine microbes in these extreme regions. However, marine microbial eukaryotes are still poorly described in these highly influenced regions of Arctic latitudes. Recently, Lovejoy (2006) reported a pan-Arctic survey of the microbial eukaryote from three Canadian sites and two European Arctic sites, and discovered important radiolarians and the presence of novel lineages in diverse protists from such extreme cold environments.
Kongsfjorden is one of the largest fjords of the Svalbard Archipelago. It is a part of the Kongsfjorden–Krossfjorden twin fjord system located on the northwest coast of Spitsbergen Island (Svendsen et al., 2002). Marine microbial eukaryotes have been poorly described in this fjord yet. As one of the northern-most coast ecosystems, the microbial eukaryotic diversity and distribution deserved further investigation, made more feasible due to the increased reliability of the nucleic acid-based approaches as rDNA, psbA etc. Therefore, surveys of eukaryotic microbes inhabiting natural environments have enhanced, and will continue to enhance, our observation of their different ecological roles (Takishita et al., 2005), especially in these extremely harsh and rapidly changing environments.
This article reports the molecular diversity of microbial eukaryotes over a range of one polar coastal site, based on phylogenetic information. With the collection of different environmental samples from glacial, sea water along a vertical profile of the outer basin, and sediment from the high Arctic fjord, we were aiming to fill the gap in our knowledge of microbial eukaryotes, and helping to understand the microbial ecosystem in Kongsfjorden. As a vulnerable component of global genetic diversity, refinement of the phylogenetic data would improve our predictions of fast changing ecosystem.
Materials and methods
Oceanographic sampling and environmental data
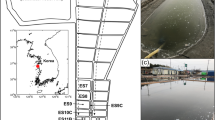
Fresh glacial melt water sample of 500 ml was collected from Austre Brøggerbreen (Site I). Sea water samples (Site II) were collected in Kongsfjorden, Spitzbergen (N 78°59.29′, E 11°39.60′) Svalbard (Fig. 1) on August 22, 2006. This site was one of the annual oceanographic and ecology observatories of the Polar Research Institute of China. Temperature, pressure, and conductivity measurements were collected using a (CTD) profiler mounted on a General Oceanic rosette carousel equipped with 2.5 l Niskin bottles. Chlorophyll a concentrations were determined with a TD-800 laboratory fluorometer, after 24-h extraction in 90% acetone at 5°C (Table 1). Among sea water samples, 100 ml aliquots were filtered through 0.45-μm pore-size GF/F filters and fixed with HgCl solution at each depth and site, which were stored at −20°C. The concentrations of nitrate–nitrogen, phosphate–phosphorus, and silica were measured by a continuous flow nutrient analyzer (Skalar San++, Skalar UK (Ltd.), York, UK).
Map showing sites sampled in Kongfjorden (Svalbard), and glaciers around the fjord. Sampling sites are marked by asterisks; site II was one of the annual sea ecology monitoring sites of PRIC. Glacial sample (Site I) was collected from the Austre Brøggerbreen. The sediment sample (Site III) was collected from 40-m depth adjacent to Kongsbreen
Sea water samples for DNA analysis were collected directly into the Niskin bottles, and into clean bottles that had been rinsed with acid and then with MilliQ water, followed by three rinses of sample water prior to filtering. Here, we collected four sea water samples from the outer basin site II (Fig. 1) at depths of 2, 20, 30 and 200 m, operated with the winch. The microbial samples were collected by filtering 500–1000 ml of seawater under <5 mmHg pressure. The microbial biomass was successively trapped onto 47-mm diameter, 0.2-μm pore size nucleopore membrane filters (Whatman) after 50-μm mesh prefilter. Filters were frozen at −80°C in lysis buffer (40 mM EDTA, 50 mM Tris–HCl, 0.75 M sucrose) until nucleic acid was extracted.
Owing to the limited sampling capability and stony geology of site II, the sediment sample (Site III) was taken by coring the sediments to approximately 37-cm depth using a gravity core sampler from the sea bed at 40-m depth in the vicinity of Kongsbreen (Fig. 1). A subsample was separated from the bottom 2 cm of the core (at a depth of 35–37 cm), and stored at −80°C until molecular processing.
DNA extraction
Sample filters were thawed on ice. The microbial organisms were rinsed off from the filters, and then digested using lysozyme (final concentration, 1 mg ml−1) and proteinase K (0.2 mg ml−1). Lysates were recovered, and nucleic acids extracted with phenol–chloroform–isoamyl alcohol (25:24:1), followed by chloroform–isoamyl alcohol (24:1) protocol. DNA in the sediment sample was extracted by using the FastDNA spin Kit for Soil (Qbiogene).
DNA amplification, cloning, and sequencing
A total of six clone libraries were constructed. Eukaryotic 18S rDNA genes were amplified by PCR with eukaryote-specific primers EukA and EukB (Medlin et al., 1988). Amplified rDNA gene products from several individual PCRs were pooled (Luo et al., 2006). The polymerase chain reaction was performed with an initial “hot start” for 10 min at 95°C, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min 30 s, and by a final extension at 72°C for 15 min. A total of 100 μl PCR products were cleaned using a QIAGEN purification kit, and then cloned with the Cloning kit (pGEM-T, Promega) according to the manufacturer’s directions. Libraries were screened for the whole 18S rDNA inserted by PCR with M13 primers.
Full-length sequencing was done by ABI 3730 Sequencer with four conserved primers: two internal to the PCR products (570 F: 5′-CCA GCA GCC GCG GTA ATT C-3′; 905 F: 5′-GTC AGA GGT GAA ATT CTT GG-3′), and two targeted to the plasmid (M13F and M13R).
Phylogenetic analysis
The closest match to each sequence was obtained from NCBI blast. Poor-quality sequences and suspected chimeras were checked by using BLAST with sequence segments separately, and then using the Chimera check program at Ribosomal Data Project II. The sequences that passed Chimeric screening were phylogenetically grouped and aligned using Clustal X v.1.83; alignments were manually checked by using the “multicolor sequence alignment editor” of Hepperle (2003). Some ambiguously aligned positions have been removed manually. Phylogenetic analyses were conducted with various modules from the Phylip 3.62 package, by using neighbour joining (NEIGHBOR) with the Kimura 2-parameter correction algorithm in MEGA version 4. Support for trees was obtained by bootstrapping 1,000 datasets. Meanwhile, maximum likelihood (ML) by bootstrapping 100 data sets was involved into four major phyla analysis.
Molecular phylogenies inferred from the data set of SSU rDNA are summarized in Figs. 2, 3, and 4. The tree topology shown was obtained by neighbor-joining analysis (Figs. 3, 4); and maximum likelihood (Fig. 3) using the full length data set of 18S rDNA. The bootstrap values >50% are indicated in the trees.
Diagram of taxonomic affiliation of sequences for the complete vertical depth profile, and the number of rDNA sequences that contributed to first rank (Adl et al., 2005) taxonomic groups
Phylogenetic relationship of 18S rDNA sequences within four main phyla of total libraries: a Alveolate; b Stramenopile; c Cercozoan; and d Metazoan. The numbers in parentheses are the number of closely related sequences with >97% similarity in the same library. The phylogenetic trees shown were inferred by maximum likelihood method. Bootstrap support values (>50%) of maximum likelihood (100 replicates) and neighbor-joining analysis (1,000 replicates) are marked in the tree. The clones of different libraries are marked by specified squares. The scale bar indicates the estimated number of base changes per nucleotide sequence position
Phylogeny tree of SSU rDNA gene sequences for the glacial sample (NPK97) and 2-m surface seawater (NPK2) from other taxonomic clades by neighbor-joining analysis. Labels in bold type are from this study. Palmata palmate is the red algal group. Bootstrap support values (>50%) neighbor-joining analysis (1,000 replicates) are marked in the tree. The scale bars represent nucleotide substitutions per site; the actual value depends on the branch lengths in the tree. A total of 1837 positions were considered from an alignment of 48 sequences
OTU richness estimation
In order to conduct richness estimation and to rigorously compare the diversity among the communities, sequences were placed into operational taxonomic units (OTUs) at a level of sequence similarity of ≥97%. All the OTU richness and sample coverage calculations were performed with the program EstimateS (version 8.0). For the purposes of inputting the data into the program, each cloned sequence was treated as a separate sample, and 100 randomizations were conducted for all the tests. Further randomizations did not change the results. The OTU richness was calculated for each of the sediment samples using the nonparametric estimator Chao 1 (Chao, 1987). Extrapolation using best-fit regression analysis was performed (where necessary) to calculate the point at which 95% confidence intervals (CIs) did not overlap (Hughes et al., 2001).
Nucleotide sequence accession numbers
Gene sequences reported in this study have been deposited in the GenBank database under accession number EU371117-EU371397. The alignments are available from the authors on request.
Results
Sample sites
The Kongsfjorden site varies in depth from 40 m to over 400 m, and is surrounded by many glaciers (Fig. 1). Library NPK97 was constructed from the glacial melt water sample from Austre Brøggerbreen (site I). Four 18S rDNA gene libraries NPK2, NPK55, NPK57, and NPK60 of sea water were constructed from samples at 2, 20, 30, and 200 m from site II (Fig. 1) individually. NPKS2 were retrieved from the sediment sample of site III.
The vertical sampling of sea water covered a range of temperature, salinity, and Chl a according to the CTD at sample site II of the fjord (Table 1). The sea surface temperature was 8°C, decreasing to nearly 5°C at 200 m depth, which was caused by summer surface warming and influenced by the North Atlantic boundary currents. Summer glacial melt inputs a large volume of fresh water into the sea, and, therefore, salinity was the lowest in the surface water at the sampling site (31.96 psu), and higher in deeper waters below the halocline. At 200-m depth, the salinity increased into 35 psu. The overall profile showed a similar trend to our five annual monitoring observatories (not shown). Chl a levels increased from 0.89 μg l−1 (2 m) to the highest point of 2.90 μg l−1 (20 m), then began to decrease. Beyond 84 m, it became nondetectable.
Analysis of 18S rDNA clone libraries
Investigations of microbial eukaryote diversity covered samples from a range of different environments (Table 1). Each library yielded between 47 and 93 positive clones, except for the sediment sample where only 18 positive clones were sequenced (Table 1). Overall, full-length 18S rDNA sequences were aligned and taxonomically assigned to known groups from GenBank. The percentage of taxonomic affiliation to known taxanomic groups is shown in Fig. 2.
The glacial sample revealed a rich diversity of seven taxon groups: Alveolates, Stramenopiles, Cercozoans, Metazoans, Telonema, Chlorophyta, and Chytridiaceae. The tree topology corresponded well with recently proposed revisions of the structuring and classification of eukaryotes (Adl et al., 2005). The former five phyla were screened as sea water samples, while the other two represented as glacial in particular. Entering into the euphotic zone of the fjord, four taxa (Alveolates, Stramenopiles, Cercozoans, and Metazoans) were recovered in the clone library NPK2 from the sea surface at 2-m depth, as well as four additional taxa (Prasinophyceae, Cryptophyta, Haptophyceae, and Choanoflagellida) (Fig. 2). However, the library NPK55 from sea water at 20-m depth showed surprisingly poor diversity. Only one Thalassiosira-related clade (98% of total 51 sequences), and one clone NPK55.54 linked to cercozoan were traced. The library NPK57 for 30-m depth also retrieved poor genotype diversity, with one Thalassiosira clade (42% of all the 26 sequences) and one Calanus clade (Metazoan). Meanwhile, at the clone library NPK60 of 200-m depth, the eukaryotic diversity clearly increased, integrating related clones of Metazoans, Alveolates, Telonema, and Stramenopiles. The sediment sample was collected from the sea bed near the biggest glacier (Kongsbreen), determined by sampling boat accessibility and the stony geology of the outer basin. Only two cercozoan sequences and sixteen Strombidium-related sequences were screened in NPKS2. None of the other eukaryotes in the sediment library was unveiled in this study.
Analysis of main detected phyla
Clone libraries are not quantitative, but still provide a crude approximation of the potential prevalence of a phylotype in a natural sample. Taking all the libraries together, the diagram of phylogenetic diversity showed us clearly that four main phyla (Alveolates, Stramenopiles, Cercozoans, and Metazoans) were detected in the fjord. Owing to insufficient prefiltered sample volume, Metazoans are not involved while calculating the percentage of phylogenetic groups compared with Antarctic environment (Table 2). The relative distribution of the major phylogenetic groups within each library is shown in Fig. 3. Selective clones sequences of these total libraries were integrated into the major phyla analysis. Meanwhile, selected clones represented by each of the different clades are integrated into Table 3.
According to this analysis, 20% of all the sequences belonged to Alveolates, separated into three lineages (Strombidiidae, NAI (Novel Alveolate I), and Dinophyceae) (Fig. 3a). Alveolates were not recovered in the NPK55 of 20-m and NPK57 of 30-m samples. Dinophyceae-related clones were found in the NPK60 of 200 m, while most sequences of the sediment were blasted similar to Strombidium. Two different genotypes were differentiated into the Strombidiidae clade, and two NPK2 clones were 97% similar to Laboea strobila. One glacial clone NPK97.252 and four NPK60 clones were clustered into NAI. Nearly, 30 dinoflagellate sequences were recovered from our libraries. Most of them were closely related to characterized dinoflagellates Gymnodinium sp. The dinoflagellate phylotype clones were prevalent from NPK97 (eight clones) and NPK2 (19 clones). Two NPK60 clones from 200 m showed 98.9% similarity to the culture Dinophyceae sp. from the Antarctic, which was defined as novel dinoflagellates.
Within the phototrophic zone, stramenopiles were sequenced closest to five sub-groups (Corethraceae, Hemidiscaceae, Thalassiosiraceae, Skeletonemataceae and Chaetocerotaceae) over the vertical profile (Fig. 3b). Skeletonema (Skeletonemataceae), Thalassiosira (Thalassiosiraceae) and Corethron (Corethraceae) were constructed from the glacial water library. In addition, Skeletonema and Thalassiosira, along with Chaetoceros (Chaetocerotaceae), were retrieved at NPK2. These were confirmed from direct light microscopic observation of the surface sample. In addition, only one clone NPK60.71 from the 200-m library was 99% similar to Actynocyclus (Hemidiscaceae) (Table 3). Clone NPK97.67 of the glacial sample was 98.7% similar to Skeletonema costatum, and six clones from NPK2 were 98% similar to Skeletonema grevillea. NPK 97.24 was retrieved at 99.6% similarity to Corethron criophilum. At depths of 20 and 30 m, most sequences were grouped into the Thalassiosira clade, which was predicted as one of the most prevalent phytoplankton in this fjord (20.7% of the total 284 sequences) at the time of sampling time, as confirmed by an in situ microscopy and DGGE analysis as well.
Cercozoans were well represented in four clone libraries. The phylogeny tree yielded different phylotypes that grouped into Cercozoa (Fig. 3c). Clone NPK2.123 was 98% closest to an uncultured clone defined as unknown clade I (Table 3). Two NPK2 clones were clustered with an uncultured freshwater cercozoa clone PCFIAU2004, defined as unknown cercozoan clade II. Clone NPK97.40 from the glacial sample was 97.4% closest to Spongomonas. Two NPKS2 sequences from the sediment were defined as unknown clade III. Seven clone sequences of NPK2 were 98% similarity to Cryothecomonas and defined as unknown cercozoan IV. Altogether twelve sequences from the libraries were constructed into the phylum Cercozoan of the algal predator Cryothecomonas.
The metazoans were recovered from four clone libraries. These sequences fell within three major groups: Chordata, Anthropoda, and one unknown metazoan clade. NPK60.38 from 200 m depth was grouped with Oikopleura dioica (Larvacean) (95% similarity). Fifteen NPK57 sequences were over 99% similar to Calanus (Copepoda), separated into three different genotypes (I, II, III). The unknown clade IV included most metazoan related sequences (99 clones), and was separated into three different sub-genotypes (Fig. 3d).
The most diversed phyla were detected in the libraries of NPK97 and NPK2. The other phyla in both libraries were shown in Fig. 4 except the above four major phyla. The sea sample library of NPK2 from this glacially influenced fjord corresponded well with the microbial eukaryotic diversity from the European Arctic Ocean, with the exception of two Cryptophyceae clones linked to Teleaulax amphioxeia. Ten clones relating to Prasinophyceae were detected. Two Chlorophyta related clones as NPK97.402 and NPK97.11 were picked out in the library of “fresh water” environment. Clone NPK97.11 was traced 98% closest to Characium saccatum, whereas clone NPK97.402 and two NPK2 clones were grouped with Pyramimonas (99% similarity). The other five clones in NPK2 were clustered with Micromonas isolation CCMP2099 from Arctic.
One choanoflagellate phylotype was recovered here, grouped with Diaphanoeca grandis (96% similarity). Only one fungus clone NPK97.82, was distantly related to Chytriomyces (93% similarity). Five clones from NPK97, one clone of NPK2, and two clones of NPK60 were clustered with a common marine phagotrophic flagellate Telonema antarcticum. Clone NPK2.66 grouped with the unicellular and photosynthetic flagellate Chrysochromulina (Haptophyceae) (99% similarity).
As the clone libraries were constructed at the same time, the diversity of the Kongsfjorden samples was subjected to comparative analysis to extrapolate species richness. The relative richness among three libraries (NPK97, NPK2, and NPK 60) is shown in Fig. 5, while the other libraries represented low diversity. Plotting the cumulative number of OTUs estimated against the sampling effort gives species-richness curves. The highest estimated number of species presented in the surface sea water sample NPK 2 with 99 OTUs, subsequently the NPK 97 and NPK 60 were predicted as 40 OTUs, and 12 OTUs. As the species-richness curve did nearly complete an asymptote, it was suggested that the species richness has been estimated.
Discussion
Kongsfjorden is influenced by both Atlantic and Arctic water masses, and additionally influenced by the inputs from large tidal glaciers that create steep environmental gradients in sedimentation and salinity along the length of this fjord in summer (Hop et al., 2002). The biodiversity and animal populations in this fjord are strongly structured by the different physical factors that influence the fjord (Svendsen et al., 2002). The sea water sample site is an outer basin that opens directly to the ocean, containing tide-water calving glaciers and receiving considerable fresh water runoff. Thus, we selected sampling sites including one glacial, four sea water of different depths at the outer basin, and one sediment, in aiming to provide characterization of eukaryotes in the glacial influencing coastal area at Arctic high latitude.
Molecular biodiversity in Kongsfjorden
Owing to the particular sample site locations, there was marked diversity resemblance between the glacial sample and sea water samples, since the coastal level was strongly affected by the large glacial melt water input into this fjord ecosystem. Some genotypes were detected in both two different water environments: Pyramimonas, Thalassiosira, Skeletonema, Telonema, Gyrodinium, Gymnodinium, Dinophyceae, Pentapharsodinium, and even “big” Metazoan-related clones (Table 3). Somehow, the phytoplankton of the outer basin resembled the glacial “inputs.” Other genotypes as Micromonas, fungi, Actinocyclus, Corethron, Haptophyceae, Cryptophyceae, and Cercozoan related were distributed in either glacial or sea water environments. Meanwhile, fungi as Chytridiales related in glacial water have been recorded as parasites of freshwater algae (Gromov et al., 1999), which were reported primitively in high Arctic. Some “swimming” organisms as Choanoflagellates (Thomsen, 1982; Buck & Garrison, 1988), Chrysochromulina (Haptophyceae) (Edvardsen et al., 2000), andTelonema (Klaveness et al., 2005) were reported in polar environments.
It was reported that Alveolates, Stramenopiles, and Chlorophyta occurred very frequently in polar ecosystem (Vaulot et al., 2008). Alveolates with a highest percentage played an important role in the upper level of the study coastal system, while Stramenopiles covered the highest percentage from libraries of Antarctic (Díez et al., 2001). Except most often traced Dinophyceae-related clones, two lineages were clustered into the important group Strombidiidae of the planktonic food webs-oligotrich ciliates (Michaela et al., 2003). Laboea was obligate mixotrophic, previously reported in the Arctic Sea (Sime-Ngando et al., 1997). However, the species occasionally contributed significantly to the biomass of mixotrophic ciliates or oligotricha due to its large size (Agatha et al., 2004). As to Novel Alveolate I, which was reported recently as parasites related to Duboscquella or parasites of radioralian/phaeodarian protists (Harada et al., 2007).
Meanwhile, similar to the pan-Arctic Sea survey (Lovejoy et al., 2006), only cercozoa was found through all the libraries, which showed much less rhizaria diversity in this European Arctic sea region than in the Canadian Basin. NPK 2 clones of sea water were clustered with uncultured freshwater cercozoa from a dimictic and oligomesotrophic Lake Pavin (Lefèvre et al., 2007). The ubiquitous flagellates Cryothecomonas occur in polar and temperate waters, tolerating salinity from 0.0 to 34.0 psu (Ikävalko & Thomsen, 1997). In this study, they were recovered from all water depths except for at 200 m where the salinity exceeded 35.0 psu. Apart from being important heterotrophic nanoflagellates in marine pelagic food webs, Cryothecomonas are also major consumers of bacteria and picophytoplankton. They feed on either nanoplanktonic algae, or diatoms which are much larger than themselves by gradually phagocytizing host protoplasm (Kühn et al., 2000).
As regards Metazoans, five clades representing different genotypes were screened in this study. Their diversity suggests retention of either dissolved free DNA adhering to small particles or DNA—containing particles (Lovejoy et al., 2006; Vaulot et al., 2008). Kwasniewski et al. (2003) investigated the distribution of copepods in Kongsfjorden in the summers of 1996 and 1997, and found that Calanus finmarchicus and Calanus glacialis were the dominant species. Moreover, most sequences defined as the unknown clade IV were grouped with copepodes such as Eucyclops and Macrocyclops from the northern European region (Alekseev et al., 2006).
The glacial and five sea water samples were well represented by the snapshot of diversity in the fjord under estimates analysis, while the diversity of sediments probably needs further investigation. In order to address the temporal and spatial succession of the microbes in the highly glacial influenced fjord, more frequent horizontal and vertical sampling depending on DGGE would be suggested.
Ecological implications
Along the coastal vertical depth, as Chl a reached its maximum further deep into 20 m, eukaryotic algae diversity was poorly represented and mainly dominated by diatom as Thalassiosira, which was further confirmed by DGGE analysis. Lack of other photosynthetic eukaryotes in these depths at the sample site might be due to the decreasing light and salinity concentration gap caused by the huge sediment inputs from the surroundings along with the glacial melt water. However, the genotype of 200 m resolved into more diverse clades. The salinity at this depth was much closer to “real” sea water, and the stably stratified flow at this depth could be taken into consideration.
The dinoflagellates were suggested to be an ecologically significant organism in some Antarctic marine ecosystems (Gast et al., 2006). The frequently detected Dinophyceae sequences in the surface waters of Kongsfjorden were well traced by the libraries screen. Diatom sequences were another prevalent phyla throughout our results. In Canadian Arctic Basin, most sequences are related to pennate diatoms Fragilariopsis as one key genus in these waters (Lovejoy et al., 2006), whereas Thalassiosira in this European Arctic water. Skeletonema-related clones of the sea waters were highly clustered with isolations from the Arabian sea (Sarno et al., 2007). Pico-prasinophytes were shown to be both spatially and temporally prevalent in open ocean water, as well as in coastal ecosystems (Lovejoy et al., 2007; Not et al., 2004; Throndsen and Kristiansen, 1991). Micromonas pusilla was the most abundant phototrophic in Norwegian Arctic seas (Not et al., 2005), and was recently confirmed as the most widely distributed unique pan-Arctic ecotype (Lovejoy et al., 2007). The euphotic zone library (NPK2) suggested that the pico-Prasinophytes played an important role in this fjord microbial ecosystem as well. The nanoplankton (<5 μm) presented the most distinctive biomass in this fjord from our annual observation in summer (Dai., unpublished), so that monitoring the abundance of the most advantaged microbial (pico)eukaryotes, such as Prasinophyceae, would be beneficial in high Arctic fjord.
The DAPI analysis in this fjord confirmed that microbial trophic pathways were important in energy and material cycling in the high latitude shelf (Wang et al., 2009). In order to characterize the diverse eukaryotes distribution temporally and spatially in the polar fjord, more frequent samples through the use of DGGE were suggested to be executed, with emphasis understanding on ecological roles as dinoflagellates, diatoms and Prasinophytes.
References
Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson & J. R. Barta, 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protist. Journal of Eukaryotic Microbiology 52: 399–451.
Agatha, S., M. C. Strüder-Kypke & A. Beran, 2004. Morphologic and genetic variability in the marine planktonic ciliate Laboea strobila Lohmann, 1908 (Ciliophora, Oligotrichia), with notes on its ontogenesis. Journal of Eukaryotic Microbiology 51: 267–281.
Alekseev, V., H. J. Dumont, J. Pensaert, D. Baribwegure & J. R. Vanfleteren, 2006. A redescription of Eucyclops serrulatus (Fischer, 1851) (Crustaceae: Copepoda: Cyclopoida) and some related taxa, with a phylogeny of the E. serrulatus-group. Zoologica Scripta 35: 123–147.
Buck, K. & D. Garrison, 1988. Distribution and abundance of choanoflagellates (Acanthoecidae) across the ice-edge zone in the Weddell Sea, Antarctica. Marine Biology 98: 263–269.
Chao, A., 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43: 783–791.
Countway, P. D., R. J. Gast, P. Savai & D. A. Caron, 2005. Protistan diversity estimates based on 18S rDNA from seawater incubations in the Western North Atlantic. Journal Eukaryotic Microbiology 52: 95–106.
Díez, B., C. Pedrós-Alió & R. Masanna, 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Applied and Environmental Microbiology 67: 2932–2941.
Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez & M. L. Sogin, 2002. Benthic eukaryotic diversity in the Guaymas basin hydrothermal vent environment. Proceedings of the National Academy of Sciences United States of America 99: 7658–7662.
Edvardsen, B., W. Eikrem, J. C. Green, R. A. Andersen, S. Y. Moon-van der Staay & L. K. Medlin, 2000. Phylogenetic reconstructions of the Haptophyta inferred from 18S ribosomal DNA sequences and available morphological data. Phycologia 39: 19–35.
Fuller, N. J., C. Campbell, D. J. Allen, F. D. Pitt, K. Zwirglmaier, F. Le Gall, D. Vaulot & D. J. Scanlan, 2006. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquatic Microbial Ecology 43: 79–93.
Gast, R. J., D. M. Moran, D. J. Beaudoin, J. N. Blythe, M. R. Dennett & D. A. Caron, 2006. Abundance of a novel dinoflagellate phylotype in the Ross Sea, Antarctica. Journal of Phycology 42: 233–242.
Gromov, B. V., A. V. Plujusch & K. A. Marnkaeva, 1999. Morphology and possible host range of Rhizophydium algavorum sp. nov. (Chytridiales)—an obligate parasite of algae. Protistology 1: 62–65.
Guillou, L., W. Eikrem, M. J. Chrétiennot-Dinet, F. Le Gall, R. Massana, K. Romari, C. Pedrós-Alió & D. Vaulot, 2004. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155: 193–214.
Harada, A., S. Ohtsuka & T. Horiguchi, 2007. Species of the Parasitic Genus Duboscquella are members of the Enigmatic Marine Alveolate Group I. Protist 158: 337–347.
Hepperle, D., 2003. Align, Manual Sequence Alignment Editor for PCs. Distributed by the author. http://wwwuser.gwdg.de/~dhepper/.
Hop, H., T. Pearson, E. N. Hegseth, K. M. Kovacs, C. Wiencke, S. Kwasniewski, K. Eiane, F. Mehlum, B. Gulliksen, M. Wlodarska-Kowalezuk, C. Lydersen, J. M. Weslawski, S. Cochrane, G. W.Gabrielsen, R. J. G. Leakey, O. J. Lonne, M. Zajaczkowski, S. Falk-Petersen, M. Kendall, S. A. Wangberg, K. Bischof, A. Y. Voronkov, N. A. Kovaltchouk, J. Wiktor, M. Poltermann, G. di Prisco, C. Papucci & S. Gerland, 2002. The marine ecosystem of Kongsfjorden, Svalbard. Polar Research 21: 167–208.
Hughes, J. B., J. J. Hellmann, T. H. Ricketts & B. J. M. Bohannan, 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Applied and Environmental Microbiology 67: 4399–4406.
Ikävalko, J. & H. A. Thomsen, 1997. The Baltic Sea biota (March 1994): A study of the protistan community. European Journal of Protistology 33: 229–243.
Klaveness, D., K. Shalchian-Tibrizi, H. A. Thomsen, W. Eikrem & K. S. Jakobsen, 2005. Telonema antarcticum sp. nov., a common marine phagotrophic flagellate. International Journal of Systematic and Evolutionary Microbiology 55: 2595–2604.
Kühn, S., M. Lange & L. K. Medlin, 2000. Phylogenetic position of Cryothecomonas inferred from Nuclear-Encoded small subunit ribosomal RNA. Protist 151: 337–345.
Kwasniewski, S., H. Hop, S. Falk-Petersen & G. Pedersen, 2003. Distribution of Calanus species in Kongsfjorden, a glacial fjord in Svalbard. Journal of Plankton Research 25: 1–20.
Lefèvre, E., C. Bardot, C. Noēl, J. F. Carrias, E. Viscoglios, C. Amblard & T. Sime-Ngando, 2007. Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: evidence from a molecular diversity study in a deep meromictic lake. Environmental Microbiology 9: 61–71.
López-García, P., F. Rodriguez-Valera & C. Pedrós-Alió, 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409: 603–606.
Lovejoy, C., R. Massana & C. Pedrós-Alió, 2006. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and Adjacent seas. Applied and Environmental Microbiology 72: 3085–3095.
Lovejoy, C., W. F. Vincent, S. Bonilla & S. Roy, 2007. Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in Arctic seas. Journal of Phycology 43: 78–89.
Luo, W., S. Pflugmacher, T. Pröschold, N. Walz & L. Krienitz, 2006. Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 157: 315–333.
Massana, R., V. Balagué, L. Guillou & C. Pedrós-Alió, 2004. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiology Ecology 50: 231–243.
Medlin, L. K., H. J. Elwood, S. Stickel & M. L. Sogin, 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71: 491–499.
Michaela, C., M. C. Strüder-Kypke & D. H. Lynn, 2003. Sequence analyses of the small subunit rRNA gene confirm the paraphyly of oligotrich ciliate sensu lato and support the monophyly of the subclasses Oligotrichia and Choreotrichia (Ciliophora, Spirotrichea). Journal of Zoology 260: 87–97.
Moon-van der Staay, S. Y., R. De Wachter & D. Vaulot, 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409: 607–610.
Not, F., M. Latasa & D. Marie, 2004. A single species Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Applied Environmental Microbiology 70: 4064–4072.
Not, F., R. Masssana, M. Latasa, D. Marie, C. Colson, W. Eikrem, C. Pedrós-Alió, D. Vaulot & N. Simon, 2005. Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnology and Oceanography 50: 1677–1686.
Not, F., R. Gausling, F. Azam, J. F. Heidelberg & A. Z. Worden, 2007. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environmental Microbiology 9: 1233–1252.
Romari, K. & D. Vaulot, 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of English Channel from 18S rDNA sequence. Limnology and Oceanography 49: 784–798.
Sarno, D., W. H. C. F. Kooistra, S. Balzano, P. E. Hargraves & A. Zingone, 2007. Diversity in the genus Skeletonema (Bacillariophyceae): III. Phylogenetic position and morphological variability of Skeletonema costatum and Skeletonema grevillei, with the description of Skeletonema ardens sp. nov. Journal of Phycology 43: 156–170.
Sime-Ngando, T., M. Gosselin, S. K. Juniper & M. Lavasseur, 1997. Changes in sea-ice phagotrophic microprotists (20–200 μm) during the spring algal bloom, Canadian Arctic Archipelago. Journal of Marine Systems 11: 163–172.
Stoeck, T. & S. Epstein, 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Applied Environmental Microbiology 69: 2657–2663.
Svendsen, H., A. Beszczynska-Moller, J. O. Hagen, B. Lefauconnier, V. Tverberg, S. Gerland, J. B. Orbaek, K. Bischof, C. Papucci, M. Zajaczkowski, R. Azzolini, O. Bruland, C. Wiencke, J. G. Winther & W. Dallmann, 2002. The physical environment of Kongsfjorden-Krossfjorden, an Arctic fjord system in Svalbard. Polar Research 21: 133–166.
Takishita, K., H. Miyake, M. Kawato & T. Maruyama, 2005. Genetic diversity of microbial eukaryotes in anoxic sediment around fumaroles on a submarine caldera floor based on the small-subunit rDNA phylogeny. Extremophiles 9: 185–196.
Thomsen, H., 1982. Planktonic choanoflagellates from Disko Bugt, West Greenland, with a survey of the marine nanoplankton of the area Meddelelser om Gronland. Bioscience 8: 3–36.
Throndsen, J. & S. Kristiansen, 1991. Micromonas pusilla (Prasinophyceae) as part of picoplankton and nanoplankton communities of the Barents Sea. Polar Research 10: 201–207.
Vaulot, D., F. L. Gall & D. Marie, 2004. The Roscoff culture collection (RCC): A collection dedicated to marine picoplankton. Nova Hedwigia 79: 49–70.
Vaulot, D., W. Eikrem, M. Viprey & H. Moreau, 2008. The diversity of small eukaryotic phytoplankton (<3 μm) in marine ecosystems. FEMS Microbiology Review 32: 795–820.
Wang, G. Z., C. Y. Guo, W. Luo, M. H. Cai & J. F. He, 2009. The distribution of picoplankton and nanoplankton in Kongsfjorden during late summer 2006. Polar Biology 32: 1233–1238.
Zeidner, G., C. M. Preston & E. F. Delong, 2003. Molecular diversity among marine picophytoplankton as revealed by psbA analyses. Environmental Microbiology 5: 212–216.
Acknowledgments
This research is supported by National Natural Science Foundation of China (No. 40806073, 40876097); the Youth Innovation foundation of Polar research Institute of China (JDQ200701); Polar Strategic Research Foundation of China; Youth Marine Science Foundation (2008128), SOA; and Resource-sharing Platform of Polar Samples (2005DKA21406). We acknowledge the logistic support from Kingsbay AS, Svalbard. We also appreciate the technical assistance from Q. Luo, YY. Lou, CJ. Zhu, HB. Teng, and CY. Guo. We also thank the reviewers for their advices.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: J. Padisak
Rights and permissions
About this article
Cite this article
Luo, W., Li, H., Cai, M. et al. Diversity of microbial eukaryotes in Kongsfjorden, Svalbard. Hydrobiologia 636, 233–248 (2009). https://doi.org/10.1007/s10750-009-9953-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-009-9953-z