Abstract
It is unclear whether differentiating live and dead diatoms would enhance the accuracy and precision of diatom-based stream bioassessment. We collected benthic diatom samples from 25 stream sites in the Northern Oregon Coast ecoregion. We counted live diatoms (cells with visible chloroplasts) and then compared the counts with those generated using the conventional method (clean counts). Non-metric multidimensional scaling (NMDS) showed that the diatom assemblages generated from the two counts were overall similar. The relationships between the two diatom assemblages (summarized as NMDS ordination axes) and the environmental variables were also similar. Both assemblages correlated well with in-stream physical habitat conditions (e.g., channel dimensions, substrate types, and canopy cover). The conventional diatom method provides taxonomic confidence while the live diatom count offers ecological reliability. Both methods can be used in bioassessment based on specific assessment objectives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Accuracy and precision in diatom-based bioassessment are continually being refined to meet the increasing demands of water resource management. Recently, researchers have examined whether sampling design (Potapova & Charles, 2005; Weilhoefer & Pan, 2007), lab analytical procedures (Alverson et al., 2003), and taxonomic consistency (Kelly, 2001; Prygiel et al., 2002) enhance the performance of diatoms in stream bioassessment. In general, bioassessment uses living organisms to evaluate water quality, with diatoms being an exception. A diatom sample comprises a mixture of live (containing chloroplast) and dead (empty) individuals which are not distinguished from each other in the traditional diatom analysis. It uses acid digestion to remove the organic material and to allow diatom identification to the lowest possible taxonomic level using the fine morphological features of their cell walls, which are otherwise obscured by the cell contents. This lack of distinction between live and dead diatoms in the traditional method is resolved by the assumption that the abundance of live diatoms is proportional to the abundance of dead diatoms (e.g., the most abundant live species is also the most abundant dead species), although, dead diatoms might include resident species, euplankton, and dislodged individuals from upstream and impoundments. A distinction between live and dead diatoms is usually made in assessments of growth rate, biovolume, and cell density (Peterson, 1996; Potapova & Charles, 2005) where the abundance of live diatoms is estimated from undigested samples.
Our knowledge on the abundance of live and dead diatoms in aquatic systems is limited. A few studies were conducted from natural substrates, e.g., estuarine sediments (Wilson & Holmes, 1981; Oppenheim, 1987; Sawai, 2001; Hassan et al., 2008), plants (Thomas, 1979), stream rocks (Pryfogle, 1975; Owen et al., 1979; Pryfogle & Lowe, 1979), and artificial substrates (Patrick et al., 1954). However, they found contradictory information about the abundance of dead diatoms in an assemblage. Proportions of dead diatoms in depositional habitats were variable and high (Wilson & Holmes, 1981; Oppenheim, 1987). Sawai (2001) recognized two trends between live and dead diatoms in tidal wetlands: species numbers were higher for dead diatoms, and many dead diatom frustules/valves were transported widely. Patrick et al. (1954) concluded that dead diatoms were rare on artificial substrates in streams, whereas others found that >50% of the diatoms in a sample were dead (Owen et al., 1979; Pryfogle & Lowe, 1979; Wilson & Holmes, 1981).
It is unclear whether differentiating live and dead diatoms in an assemblage would contribute significantly to the accuracy and precision of diatom-based bioassessment. The potential errors in diatom-based bioassessment when analyzing the entire diatom community have been discussed by a number of scientists (Cox, 1998; Round, 1998), but without the support of empirical data. Several researchers have suggested that assemblages generated by the traditional method may better integrate spatial and temporal variability of the sampled stream and its watershed, because the accumulation of live and dead diatoms over space and time may provide broad environmental information (Cox, 1998; Stevenson & Pan, 1999). However, Pryfogle & Lowe (1979) concluded that the traditional counting technique might mask the specific response of the resident taxa to the local environmental conditions and introduce more noise to the data by its inability to distinguish dead diatoms. The quantitative interpretation of the information embedded in dead diatoms in an assemblage is still unclear. It has been discussed that the contamination with dead diatoms from adjacent habitats might be a significant source of error in estimations of diatom communities and their signals about the local environment (Round, 1998). The advantages of live diatoms for bioassessment have been emphasized by Cox (1998), but without evidence for their data quality. She argued that live diatom identification will speed up and ease the process of bioassessment, decision making, and management. However, there are no comprehensive floras based on live diatom cell morphology (except Cox, 1996) which hinders their identification and use in bioassessment. Therefore, a study that would compare the response of the whole diatom assemblage and its live component to the measured environmental variables would benefit diatom-based stream bioassessment.
In this article, we compare the diatom assemblages generated by counting the traditional acid-cleaned samples, which do not allow separation of live from dead diatoms, to the live diatoms from the undigested samples. Specifically, we want to quantify the proportion of live individuals in an assemblage and assess if there is a correspondence in terms of species abundance with the diatoms of the whole assemblage. We anticipate that we will see mostly live diatoms on the sampled rock surfaces, and that their species composition will be very similar to the one of the whole assemblage. Next, we want to examine if the assemblages from the two counts differ in their responses to the measured environmental variables. Finally, we would like to see which environmental variables may account for the differences, if there are any.
Material and methods
Study area and site selection
Stream sites were located in the Northern Oregon Coast which is part of the Coast Range ecoregion (Level III ecoregions; Omernik, 1987). It is underlain by marine tuffaceous sandstones and shales with basaltic volcanic rocks (Jackson & Kimerling, 1993). The Northern Oregon Coast is characterized by high precipitation and maritime climate with cool, dry summers, and warm, wet winters (Naiman & Bilby, 1998). The Coastal Mountains receive most of their precipitation, which averages between 203 and 254 cm a year, between October and March or April (Jackson & Kimerling, 1993). Peak stream flows occur in late fall and winter. It is a forested region dominated by western hemlock (Tsuga heterophylla (Raf.) Sarg.), Sitka spruce (Picea sitchensis (Bong.) Carr.), Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco), and red alder (Alnus rubra Bong.) (Clarke et al., 1991). Land use in the area is mainly ungrazed forest and woodland (Jackson & Kimerling, 1993). Streams of the Coast Range ecoregion are naturally cool, clear, typically shaded, and oligotrophic in nutrient status. The sampling sites were selected based on geology, land cover and land ownership, and stream order (first- and second-order streams). From the initial pool of prospective sites, only watersheds dominated by volcanic bedrock lithology and well forested were retained. All sites were on public lands and allowed access for sampling.
Watershed characterization
Watershed boundaries above each sampling reach were delineated with the Spatial Analyst extension of ArcInfo using 10-m digital elevation models from USGS (CLAMS, 1996). Bedrock lithology categories within each watershed were quantified from the geological map of Oregon (USGS 1:500,000, http://www.gis.oregon.gov; Walker & MacLeod, 1991). Geologic types were grouped in two major classes (volcanic and sedimentary). Vegetation within each watershed was quantified from the Gradient Nearest Neighbor Vegetation Class developed by CLAMS (1996). Vegetation cover within the watersheds was grouped in three major categories: broadleaf, conifer, and mixed forest (broadleaf and conifer). Ownership data were also obtained from CLAMS (1996). ArcGIS (version 9, ESRI Inc. 2006) was used for all spatial analysis and calculations.
Field sampling
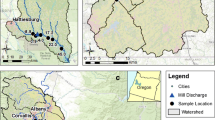
Diatom samples were collected from 25 stream sites in the Coast Range of Oregon in September 2006 (Fig. 1). The sampling procedures followed a modified US EPA’s Environmental Monitoring and Assessment Program (EMAP) protocol for wadeable rivers and streams (Lazorchak et al., 1998). The stream reach, 100 m in length, was divided into nine equally spaced transects. Along each transect, one diatom sample was collected at ¼ (left), ½ (center), or ¾ (right) of the distance from the stream bank after a random start. One rock from each of the nine sampling transects was removed from the stream, an 8-cm2 surface area was scraped with a toothbrush, and washed into a collection bottle with stream water. The scrapings from the nine rocks were combined into one composite sample which was frozen until the time of processing.
The water quality variables (e.g., pH, temperature, salinity, dissolved oxygen, and specific conductivity) were measured in the middle of the stream reach with YSI 556 MPS. Unfiltered water samples were taken from the middle of the stream for total Kjeldahl nitrogen and total phosphorus analysis. The physical habitat conditions were characterized at five points along each transect (two banks and 25%, 50%, 75% wetted width). At each point, stream depth, substrate size, and type were recorded. Substrate size and type for the reach were expressed as percent coverage according to Kaufmann et al. (1999). Stream width, canopy cover, and riparian vegetation along each transect were also measured. The stream depth and width were represented by their mean values for the reach. The canopy cover was measured with a spherical densiometer in the middle of the stream facing upstream, downstream, and the left and the right banks. The riparian vegetation, estimated as vegetation density and type for three layers (canopy, understory, and ground cover), was evaluated for each stream bank stretching 5 m upstream and 5 m downstream from the transect, and 10 m inland from the bank, and was expressed as percent coverage (Kaufmann et al., 1999).
Lab analysis
Water nutrients (total Kjeldahl nitrogen and total phosphorus) were analyzed according to EPA methods 351.2 and 365.4 (US EPA, 1983). Each composite algal sample was split into two. The first split was used for chlorophyll a analysis following standard methods (Clesceri et al., 1998). Chlorophyll a concentrations in 90% aqueous acetone before and after acidification with 0.1 N HCl were determined on Beckman DU-640 spectrophotometer. The second sample split was preserved with 40% formalin and used for composition analysis. Ten milliliters of this split were digested with HNO3 using a MARS™ 5 microwave for conventional diatom analysis (Charles et al., 2002). After repeated rinsing with distilled water, clean frustules were dried on a glass coverslip and mounted on a glass slide using Z-RAX® mounting medium. A minimum of 600 diatom valves were identified from this slide, and because no distinction between live and dead diatoms could be made it was termed LD count (Live + Dead). The remaining volume of the sample split was used for the making of a wet mount. It was prepared in the following way, a few drops of the undigested sample were placed on a glass coverslip which was air dried, inverted, and mounted in a drop of water on a glass slide. The coverslip was sealed with clear nail polish to prevent evaporation and allow the use of oil immersion. The wet mounts were used for the identification and enumeration of live and dead diatoms. The live diatoms were defined as the ones with visible cell contents while the dead diatoms were the ones with empty frustules. To be consistent with the LD count, 300 live diatom cells (L count) were identified and counted. During the analysis of live diatoms, dead diatoms (D count) were also identified and enumerated. The L and D counts from the wet mount were used to calculate the percentages of live and dead diatoms. All diatoms were identified to the lowest possible taxonomic level (mainly species) at 1000× magnification, using Leica DM LB2 light microscope with differential interference contrast (DIC). The diatom taxonomy followed predominantly Krammer & Lange-Bertalot (1997a, b, 2000, 2004) and Patrick & Reimer (1966, 1975).
Data analysis
Environmental variables which did not approximate normal distributions were log10 or square root transformed. Diatom counts were represented as relative abundances. To compare the three assemblages (LD, L, and D) we looked at: species richness, relative abundance of dominant species, similarity between samples, and site ordinations. Species richness between the samples and the relative abundances of the dominant species between the three different assemblages (LD, L, and D) were evaluated with paired t-tests because of their dependent nature. The similarity among the three assemblages for each stream site was assessed using Bray-Curtis similarity coefficient, after square-root transformation of the data to down weight the effect of dominant species. Bray-Curtis similarity values range between 0 (complete difference) and 100 (complete similarity). This coefficient takes into account both species presence and abundance, and is commonly used in the analysis of ecological communities (Clarke, 1993). To evaluate whether the similarity between LD and L counts is related to the species richness or percent live diatoms, a simple linear regression was performed. The inter-site similarities were used in non-metric multidimensional scaling (NMDS) ordination to project their relationships into a low-dimensional space and to best preserve the ranked distances between them. NMDS does not require any assumptions about the species distribution, and allows for user-specified distance measure. To assess if the inter-site relationships defined by their similarity coefficients were well projected into the NMDS plot, the stress value was calculated. It shows how good the correspondence between the calculated (plotted) and the actual distances (from the similarity matrix) between the sites is, where a lower value indicates a better ordination. NMDS was run separately for LD, L, and D assemblages. The resulting three NMDS plots were then compared using procrustes analysis with permutation test (Gower, 1971; Peres-Neto & Jackson, 2001). Procrustes analysis rotates two ordinations to maximize the similarities between them by minimizing the sum of their squared distances (m2 statistic, Gower, 1971). Higher values of the m2 statistic indicate better correspondence between two ordinations. Unlike Mantel test, procrustes analysis allows for a site-by-site comparison by returning pairwise residuals (Peres-Neto & Jackson, 2001). The significance of overall similarity between any two NMDSs was assessed using protest, a permutation test (Jackson, 1995). It evaluates whether the degree of resemblance between two ordinations is greater than random. To explore the relationship of each assemblage with its environment, a linear fitting function was then used. This function finds vector averages of the environmental variables, and fits them in the ordination space defined by the species data. The significance of each vector was assessed with a goodness-of-fit statistic (squared correlation coefficient, r 2) using 1,000 permutations. To further examine which environmental variables might be responsible for the differences in the comparisons between the three ordinations (LD, L, and D), we calculated the Pearson correlation coefficients between the residuals from the procrustes analysis and the environmental variables (Peres-Neto & Jackson, 2001). All data analyses were performed using the ‘vegan’ (Oksanen et al., 2008) and ‘MASS’ (Venables & Ripley, 2002) packages in R (R Development Core Team, 2008).
Results
The sampled stream reaches were generally small and well-shaded (Table 1). Most streams were narrower than 7.0 m, but their stream width ranged from 1.9 to 10.1 m. The streams were shallow, with a mean depth from 4.0 to 14.8 cm. Mid-channel canopy cover ranged between 50 and 100% with an average of 90%. The water temperature ranged from 9.5 to 13.0°C, and pH was slightly acidic to neutral (range 5.5–7.5). All streams had very low nutrient levels. The mean value of chlorophyll a was 0.025 mg/m2. Watershed area ranged between 2.4 and 23.7 km2.
A total of 135 species were recorded in the LD counts, with a mean species richness of 26 (Table 2). Fewer taxa (90) were identified in the L counts, with a mean species richness of 19. The D counts were slightly more diverse than the L ones with a total of 96 species, and a mean species richness of 20. The species richness of LD was significantly different from the other two assemblages (L and D, Fig. 2A, B). There were 47, 7, and 12 species unique to the LD, L, and D counts, respectively, occurring with less than 1% mean relative abundance. Most species encountered were periphytic in their habitat preferences. There were only a few euplanktonic species occurring with relative abundances below 1%. The paired t-tests revealed some interesting patterns in the differences in relative abundances of the dominant species between the LD, L, and D assemblages. Some species had significantly different relative abundances between LD and L, where they were either over- or underestimated (Fig. 3). A slightly smaller number of dominant species were found to differ significantly in their relative abundances between D and L (Fig. 4).
Scatterplots for different community characteristics: (A) species richness of LD and L, (B) species richness of D and L, (C) species richness of LD and percent similarity between LD and L, (D) percent live diatoms and percent similarity between LD and L. The lines in (A) and (B) are 1:1 ratio. * Significance at P < 0.05
Scatterplots for the percent relative abundances of the dominant species in LD and L assemblages with a 1:1 ratio line. Refer to Table 2 for complete species names. * Significance at P < 0.05
Scatterplots for the percent relative abundances of the dominant species in D and L assemblages with a 1:1 ratio line. Refer to Table 2 for complete species names. * Significance at P < 0.05
The mean percentage of live diatoms in a sample was 63.4% (range 50.5–79.4). The assemblages were overall similar. Their mean similarity values were 72.4%, 73.4%, and 75.4% for LD and L, L and D, and LD and D, respectively. The percentage of live diatoms did not exhibit a statistically significant relationship with the similarity among samples (Fig. 2D). The sites with the lowest similarity had the highest species richness in LD assemblages, while the sites with the highest similarity coefficients had some of the lowest species richness (Fig. 2C). The low similarity values were primarily due to many rare species (species with low abundances), while the high similarity coefficients were due to many common species. The two-dimensional NMDS plots for the LD, L, and D assemblages had stress values of 16.0, 16.7, and 17.0, respectively. The site with the highest similarity coefficient was dominated by Achnanthidium minutissimum, which reached its highest relative abundance here (85%, Fig. 5A). Only a few of the dominant species showed pronounced distributional patterns in their relative abundances among the sites, e.g., A. minutissimum had a similar pattern of increasing relative abundance along the first NMDS axis for both LD and L ordinations (Fig. 5).
The comparisons between the site ordinations were summarized in plots of the residuals for the procrustes rotations. The plot for the LD and L ordinations is presented in Fig. 6. Deviation in the species composition between LD and L count for a site is indicated by an arrow connecting the two counts. The longer the arrow, the greater the difference between the two counts. Some sites had larger differences in their species composition than others. The configuration for the procrustes rotation of the NMDS plots for LD and L counts, as well as the one for LD and D counts, matched well (m2 = 0.85, P < 0.001, 1,000 permutations). The configuration for the L and D ordinations was even better (m2 = 0.95, P < 0.001, 1,000 permutations). Seven environmental variables co-varied significantly with the species data (P ≤ 0.05, Table 3). These variables defined a gradient of primary production and physical habitat conditions (e.g., channel dimensions, substrate types, and canopy cover). There was a change from bigger streams with deciduous riparian vegetation and higher primary production to smaller less productive shaded streams with finer substrates (Fig. 7). Four variables (chlorophyll a, canopy cover, stream width, and fine sediments) were significant for all the three assemblages. The other three variables (smooth bedrock, percent sand, and riparian deciduous canopy) had varying influence on one or more of the assemblages. The Pearson correlation coefficients between the residuals from the procrustes analysis and the environmental variables showed which variables were responsible for the differences in the comparisons of the three ordinations (LD, L, and D). The only significant correlation was between chlorophyll a and the residuals from the procrustes rotation of LD and L.
Plot of the residuals from procrustes analysis for the comparison between the ordinations of LD and L. Each arrow connects the LD and the L count from one site. The arrows point in the direction of the L ordination. The arrow length corresponds to the difference in site location between the two ordinations
Non-metric multidimensional scaling plots for the LD (A) and L (B) assemblages, and significant environmental variables (P < 0.05) from the linear fitting function. Abbreviations: CanC—canopy cover, Chl—chlorophyll a, DecC—percent riparian deciduous canopy, Fines—percent fine substrates (silt/clay/muck), Sand—percent sandy substrates, SB—percent substrates as smooth bedrock, and Width—mean wetted width
Discussion
The traditional acid-cleaned diatom assemblage and the unprocessed one were overall similar. Pair-wise comparisons showed that the species composition generated by the two methods was overall similar (mean Bray-Curtis similarity = 72.4%). NMDS plots suggested that the overall relationships among stream sites based on their ranked Bray-Curtis similarity coefficients were similar. A more quantitative evaluation of the match between the two ordinations was achieved with the procrustes rotation which showed that there was a good agreement between the two ordination configurations (m2 = 0.85, P < 0.001, 1,000 permutations). In addition, both assemblages responded to a similar set of environmental conditions. In this sense, our results are consistent with several other studies that found similar species composition between the two assemblages in creeks, tidal wetlands, and estuaries (Pryfogle, 1975; Sawai, 2001; Hassan et al., 2008). Hassan et al. (2008) also concluded that the living diatom assemblage at the estuarine sediment surface was very similar to the subsurface one, and that their ecological preferences could be used to infer past changes in their environment. Our study reinforces the statement that the similarity between the two assemblages (LD and L) and the similarity in their relationships with the local environment justify their individual use for bioassessment purposes, because they will both provide similar results. We do not necessarily need to simultaneously identify live diatoms and acid-cleaned ones, but we can use each one of them independently.
Diatom analysis using the traditional counting method provides assemblage data with high taxonomic resolution and taxa richness. Both of them are of key importance in bioassessment. For instance, the multimetric (MMI) and multivariate (e.g., RIVPACS) indices commonly used in stream bioassessment, are based on species composition (Chessman et al., 1999; Hawkins, 2006; Cao et al., 2007). The finer taxonomic resolution facilitates bioassessment where species level identification improves the accuracy of prediction models because species have narrower environmental requirements than genera (Chessman et al., 1999; Hawkins, 2006). Our study shows that the LD had 33% higher total species richness and 27% higher average species richness than those generated in L (Table 2). Similar results have been reported by Wilson & Holmes (1981) who counted 300 frustules of live and dead diatoms to approximate the diversity of a cleaned assemblage and to calculate L/D ratios. They found that measurements of community richness and diversity increased when dead diatoms were included in their calculation. However, it is difficult to attribute the difference in species richness between the two assemblages to our inabilities to identify properly all live diatom taxa or to recognize if a large proportion of rare taxa in a benthic diatom assemblage were dead. It is a common practice in diatom analysis to identify and enumerate species from the LD count and then estimate total live diatom density by counting but not identifying all live diatoms from the wet mount or from a Palmer-Maloney counting cell (Charles et al., 2002). To estimate the density of each diatom taxon, its relative abundance from the LD count is multiplied by the total number of live diatoms from the wet mount or from the Palmer-Maloney counting cell. Potapova & Charles (2005) suggested that this extrapolation might overestimate live diatom species richness. This is in agreement with our results that L counts overly underestimated species richness (Fig. 2).
Technical difficulties may hinder the utilization of live diatoms in benthic bioassessment, despite the potential valuable information embedded in them. They are more difficult to identify because the protoplast obscures the cell wall morphology. Analysts still need to examine cleaned slides using the traditional method to aid identification of live diatoms. In addition, the lack of floras or keys for their identification requires every analyst to make their own and confirm questionable live taxa with cleaned material (Cox, 1998). As a result of their ambiguous identification, analysts might unintentionally lump similar species together and reduce taxa richness which is crucial for many bioassessment methods (e.g., many live Navicula’s are difficult to recognize, Thomas, 1979). Distinguishing live diatoms might be important in ecological studies where cell density, biovolume, and growth rate need to be estimated (e.g., experimental studies, Barnese & Lowe, 1992; Peterson, 1996) or to encourage non-specialists to recognize dominant or ecologically informative genera (Cox, 1998).
The assumption of proportional abundance for live and dead diatoms in a benthic diatom assemblage may not be valid for all species (Figs. 3 and 4). The discrepancy will affect bioassessment methods which heavily rely on good estimation of species abundance, e.g., weighted averaging, when the living component of the assemblages better reflects environmental conditions such as nutrients. The high numbers of dead diatoms in our samples contributed to the differences in the species composition between LD and L. Owen et al. (1979) reported that only a few of their samples from natural and artificial substrates contained less than 10% empty frustules. In our samples, the lowest abundance of dead diatoms was 20.6%. Such high numbers of dead diatoms were reported in other studies which found that half of the diatoms collected in the spring and summer were dead while for the rest of the year these same numbers were somewhat lower (Pryfogle & Lowe, 1979). Owen et al. (1979) observed high numbers of dead diatoms on the glass slides regardless of the period of their immersion. Only 12.5% of the dead species in our study had relative abundance above 1%. Owen et al. (1979) noticed that less abundant diatoms were often represented by empty frustules which have been observed for months after the species abundance have declined. The inclusion of dead diatoms might introduce some error to the data and its subsequent analysis by incorporating non-resident species. Our research revealed that the sampled erosional habitats, e.g., riffles with epilithic communities, accumulated high numbers of dead diatoms, but they were mainly resident to it.
In summary, our results showed that the benthic diatom assemblages generated by the two different counting methods were overall similar in the sampled small mountain streams. The LD count provides taxonomic confidence while the L count offers ecological reliability. Both can be used in bioassessment based on specific assessment objectives. We would recommend a visual examination of the undigested sample to check if a large fraction of it consists of live diatoms. It would be especially necessary for regions with large amount of depositional areas or low gradient rivers where the accumulation of dead diatoms might potentially be high.
References
Alverson, A. J., K. M. Manoylov & R. J. Stevenson, 2003. Laboratory sources of error for algal community attributes during sample preparation and counting. Journal of Applied Phycology 15: 357–369.
Barnese, L. E. & R. L. Lowe, 1992. Effects of substrate, light, and benthic invertebrates on algal drift in small streams. Journal of the North American Benthological Society 11: 49–59.
Cao, Y., C. P. Hawkins, J. Olson & M. A. Kosterman, 2007. Modeling natural environmental gradients improves the accuracy and precision of diatom-based indicators. Journal of the North American Benthological Society 26: 566–585.
Charles, D. F., C. Knowles & R. S. Davis, 2002. Protocols for the analysis of algal samples collected as part of the U.S. Geological Survey National Water-Quality Assessment Program. Report No. 02-06, Patrick Center for Environmental Research, The Academy of Natural Sciences, Philadelphia, PA. http://diatom.acnatsci.org/nawqa/.
Chessman, B., I. Growns, J. Curray & N. Plunkett-Cole, 1999. Predicting diatom communities at the genus level for the rapid biological assessment of rivers. Freshwater Biology 41: 317–331.
CLAMS (Coastal landscape analysis and modeling study), 1996. http://www.fsl.orst.edu/clams/data_index.html.
Clarke, K. R., 1993. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology 18: 117–143.
Clarke, S. E., D. White & A. L. Schaedel, 1991. Oregon, USA, Ecological regions and subregions for water quality management. Environmental Management 15: 847–856.
Clesceri, L. S., A. E. Greenberg & A. D. Eaton, 1998. Standard Methods for the Examination of Water and Wastewater, 20th edn. American Public Health Association, Washington, D.C.
Cox, E. J., 1996. Identification of Freshwater Diatoms from Live Material. Chapman & Hall, London, UK.
Cox, E. J., 1998. A rationale and some suggestions for developing rapid biomonitoring techniques using identification of live diatoms. In Proceedings of the 15th International Diatom Symposium: 43–50.
Gower, J. C., 1971. Statistical methods of comparing different multivariate analyses of the same data. In Hodson, F. R., D. G. Kendall & P. Tautu (eds), Mathematics in the Archaeological and Historical Sciences. Edinburgh University Press, Edinburgh: 138–149.
Hassan, G. S., M. A. Espinosa & F. I. Isla, 2008. Fidelity of dead diatom assemblages in estuarine sediments: how much environmental information is preserved? Palaios 23: 112–120.
Hawkins, C. P., 2006. Quantifying biological integrity by taxonomic completeness: its utility in regional and global assessments. Ecological Applications 16: 1277–1294.
Jackson, D. A., 1995. PROTEST: a PROcrustean Randomization TEST of community environment concordance. Ecoscience 2: 297–303.
Jackson, P. L. & A. J. Kimerling, 1993. Atlas of the Pacific Northwest. OSU Press, Corvallis, OR.
Kaufmann, P. R, P. Levine, E. G. Robison, C. Seeliger & D. V. Peck, 1999. Quantifying Physical Habitat in Wadeable Streams. EPA/620/R-99/003. U.S. Environmental Protection Agency, Washington, D.C.
Kelly, M., 2001. Use of similarity measures for quality control of benthic diatom samples. Water Resources 35: 2784–2788.
Krammer, K. & H. Lange-Bertalot, 1997a. Bacillariophyceae. Naviculaceae. In Ettl, J., J. Gerloff, H. Heynig & D. Mollenhauer (eds), Süßwasserflora von Mitteleuropa, 2/1. Spectrum Akademischer Verlag, Heidelberg-Berlin: 1–876.
Krammer, K. & H. Lange-Bertalot, 1997b. Bacillariophyceae. Bacillariaceae, Epithemiaceae, Surirellaceae. In Ettl, J., J. Gerloff, H. Heynig & D. Mollenhauer (eds), Süßwasserflora von Mitteleuropa, 2/2. Spectrum Akademischer Verlag, Heidelberg-Berlin: 1–610.
Krammer, K. & H. Lange-Bertalot, 2000. Bacillariophyceae. Centrales, Fragilariaceae, Eunotiaceae. In Ettl, J., J. Gerloff, H. Heynig & D. Mollenhauer (eds), Süßwasserflora von Mitteleuropa, 2/3. Spectrum Akademischer Verlag, Heidelberg-Berlin: 1–598.
Krammer, K. & H. Lange-Bertalot, 2004. Bacillariophyceae. Achnanthaceae. Kritische Ergänzungen zu Achnanthes s. l., Navicula s. str., Gomphonema. In Ettl, J., J. Gerloff, H. Heynig & D. Mollenhauer (eds), Süßwasserflora von Mitteleuropa, 2/4. Spectrum Akademischer Verlag, Heidelberg-Berlin: 1–468.
Lazorchak, J. M., D. J. Klemm & D. V. Peck, 1998. Environmental Monitoring and Assessment Program – Surface Waters: Field Operations and Methods for Measuring the Ecological Condition of Wadeable Streams. EPA/620/R-94/004F. U.S. Environmental Protection Agency, Washington, D.C.
Naiman, R. J. & R. E. Bilby, 1998. River ecology and management in the Pacific Coastal ecoregion. In Naiman, R. J. & R. E. Bilby (eds), River Ecology and Management: Lessons from the Pacific Coastal ecoregion. Springer-Verlag, New York: 1–10.
Oksanen, J., R. Kindt, P. Legendre, B. O’Hara, G. L. Simpson & M. H. H. Stevens 2008. Vegan: Community Ecology Package. R Package Version 1.11-2. http://cran.r-project.org/, http://vegan.r-forge.r-project.org/.
Omernik, J. M., 1987. Map supplement: ecoregions of the conterminous United States. Annals of the Association of American Geographers 77: 118–125.
Oppenheim, D. R., 1987. Frequency distribution studies of epipelic diatoms along an intertidal shore. Helgoland Marine Research 41: 139–148.
Owen, B. B., M. Afzal & W. R. Cody, 1979. Distinguishing between live and dead diatoms in periphyton communities. In Wetzel, R. L. (ed.), Methods and Measurements of Periphyton Communities: A Review. American Society for Testing and Materials, STP 690: 70–76.
Patrick, R. & C. W. Reimer, 1966. The Diatoms of the United States, Vol. 1. Monograph #13 of the Academy of Natural Sciences of Philadelphia.
Patrick, R. & C. W. Reimer, 1975. The Diatoms of the United States, Vol. 2. Monograph #13 of the Academy of Natural Sciences of Philadelphia.
Patrick, R., M. H. Hohn & J. H. Wallace, 1954. A new method for determining the pattern of the diatom flora. Notulae Naturae of the Academy of Natural Sciences of Philadelphia 259: 1–12.
Peres-Neto, P. R. & D. A. Jackson, 2001. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129: 169–178.
Peterson, C. G., 1996. Mechanisms of lotic microalgal colonization following space-clearing disturbances acting at different spatial scales. Oikos 77: 417–435.
Potapova, M. G. & D. F. Charles, 2005. Choice of substrate in algae-based water quality assessment. Journal of the North American Benthological Society 24: 415–427.
Pryfogle, P. A., 1975. Seasonal distribution of periphytic diatom communities of Tymochtee Creek. In Proceedings of the Sandusky River Basin Symposium. International Joint Commission Canada-U.S.A. Great Lakes Water Quality Agreement 1972: 153–173.
Pryfogle, P. A. & R. L. Lowe, 1979. Sampling and interpretation of epilithic lotic diatom communities. In Wetzel, R. L. (ed.), Methods and Measurements of Periphyton Communities: A Review. American Society for Testing and Materials, STP 690: 77–89.
Prygiel, J., P. Carpentier, S. Almeida, M. Coste, J.-C. Druart, L. Ector, D. Guillard, M.-A. Honoré, R. Iserentant, P. Ledeganck, C. Lalanne-Cassou, C. Lesniak, I. Mercier, P. Moncaut, M. Nazart, N. Nouchet, F. Peres, V. Peeters, F. Rimet, A. Rumeau, S. Sabater, F. Straub, M. Torrisi, L. Tudesque, B. V. de Vijver, H. Vidal, J. Vizinet & N. Zydek, 2002. Determination of the biological diatom index (IBD NF T 90-354): results of an intercomparison exercise. Journal of Applied Phycology 14: 27–39.
R Development Core Team, 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org.
Round, F. E., 1998. A problem in algal ecology: contamination of habitats from adjacent communities. Cryptogamie Algologie 19: 49–55.
Sawai, Y., 2001. Distribution of living and dead diatoms in tidal wetlands of northern Japan: relations to taphonomy. Palaeogeography, Palaeoclimatology, Palaeoecology 173: 125–141.
Stevenson, R. J. & Y. Pan, 1999. Assessing environmental conditions in rivers and streams with diatoms. In Stoermer, E. F. & J. P. Smol (eds), The Diatoms Applications for the Environmental and Earth Sciences. Cambridge University Press, New York: 11–40.
Thomas, D. P., 1979. Some problems connected with the use of cleared cells and relative proportions of species in diatom ecology. Beiheft für Nova Hedwigia 64: 503–511.
US EPA, 1983. Methods for Chemical Analysis of Water and Wastes. EPA-600/4-79-020. U.S. Environmental Protection Agency, Cincinnati, OH.
Venables, W. N. & B. D. Ripley, 2002. Modern Applied Statistics with S, 4th ed. Springer, New York.
Walker, G. W. & N. S. MacLeod, 1991. Geologic map of Oregon. US Geological Survey, Scale 1:500,000. http://geopubs.wr.usgs.gov/docs/geologic/or/oregon.html.
Weilhoefer, C. L. & Y. Pan, 2007. A comparison of periphyton assemblages generated by two sampling protocols. Journal of the North American Benthological Society 26: 308–318.
Wilson, C. J. & R. W. Holmes, 1981. The ecological importance of distinguishing between living and dead diatoms in estuarine sediments. European Journal of Phycology 16: 345–349.
Acknowledgments
We are grateful to Patrick Edwards, Miguel Estrada, Hsiao-Hsuan Lin, Jeff Meacham, Kalina Manoylov, and two anonymous reviewers who provided valuable feedback and critical review of earlier versions of the manuscript. This project was supported by the EPA-PSU cooperative agreement for WEMAP periphyton analysis (EPA R-82902601-0).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: J. Saros
Rights and permissions
About this article
Cite this article
Gillett, N., Pan, Y. & Parker, C. Should only live diatoms be used in the bioassessment of small mountain streams?. Hydrobiologia 620, 135–147 (2009). https://doi.org/10.1007/s10750-008-9624-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-008-9624-5