Abstract
Autologous bone transplantation which is a common treatment method for bone defects needs a large quantity of bone cells. In order to develop new treatments to regenerating bone tissues, this research aimed at identifying the key genes and finding their mechanism in human adipose-derived stem cells (hADSCs) osteogenesis. GSE63754, GSE89330 and GSE72429 were downloaded to perform GO functional and KEGG pathway analyses, construct a competing endogenous RNA (ceRNA) network, construct a PPI network and identify hub genes. The expression level of LMO3 during the osteogenesis of hADSCs was examined by quantitative reverse transcription polymerase chain reaction and western blot. Lentivirus transfection was used to knock down or overexpress LMO3, which enabled us to investigate the effect of LMO3 on osteogenic differentiation of hADSCs. Wortmannin were used to identify the mechanism of the LMO3/PI3K/Akt axis in regulating osteogenic differentiation of hADSCs. Moreover, ectopic bone formation in nude mice was used to investigate the effect of LMO3 on osteogenesis in vivo. In this study, we found the expression of LMO3 was significantly upregulated during the osteogenic differentiation of hADSCs. LMO3 knockdown remarkably suppressed osteogenic differentiation of hADSCs, while LMO3 overexpression promoted osteogenic differentiation of hADSCs both in vitro and in vivo. Moreover, we discovered that the enhancing effect of LMO3 overexpression on osteogenic differentiation was related to the activation of PI3K/Akt signaling pathway. Inhibition of PI3K/Akt signaling pathway with wortmannin effectively blocked the stimulation of osteogenic differentiation induced by LMO3 overexpression. In conclusion, based on transcriptomic analysis, we identified key genes involved in regulating the osteogenic differentiation of hADSCs. In addition, we found that LMO3 might act as a positive modulator of hADSC osteogenic differentiation by mediating PI3K/Akt signaling pathway. Manipulating the expression of LMO3 and its associated pathways might contribute to advances in bone regeneration and tissue engineering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone defects caused by tumors, infections and trauma affect tens of millions of patients around the world, and its repair and treatment is one of the major clinical problems. There are more than 10 million bone transplant operations worldwide each year, and it is still growing at a rate of 10% per year (Vos et al. 2020). At present, the treatment of bone defects and nonunion mainly includes two surgical methods: autologous bone transplantation and allogeneic bone transplantation (Si et al. 2015). Autologous bone transplantation is a usual method of treatment for bone defects, but the application is limited by its lack of source, poor plasticity, and causing damage to the donor site (Liu et al. 2021). The incidence of postoperative complications in patients with allogeneic bone transplantation is significantly higher, which has exceeded 30%, including fractures, insufficiency, infections, etc. (Wu et al. 2020). Therefore, a new treatment strategy with limited secondary trauma, low immunogenicity and ideal functional recovery is urgently needed.
Bone tissue engineering aims to develop bone graft substitutes which overcome the shortcomings of natural bone grafts (Duttenhoefer et al. 2013). Meantime, it should have the characteristics of being able to be shaped on demand and can be prepared in large quantities. These constructed bone graft substitutes are usually composed of good-performing scaffolds, abundant seed cells and osteoinductive factors. Tissue engineering has undergone substantial development, and human adipose-derived stem cells (hADSCs) have the capacity for multilineage differentiation and can perform paracrine functions (Chun et al. 2019; Hu et al. 2019; Park et al. 2020). hADSCs are an ideal source of seed cells for regenerative applications and tissue engineering, because of their convenience of access, large reserves in the body, absence of ethical issues and other characteristics. hADSCs transplantation have been recently studied as a prospective treatment due to their multiple sources, ease of collection and multipotency (Cheng et al. 2020; Paspaliaris and Kolios 2019). More importantly, hADSCs isolated from aged or osteoporotic humans or animals maintain their capability for osteogenic differentiation and are thus relatively promising for being used in autologous cell-based therapy to repair bone trauma. The osteogenic differentiation of hADSCs is regulated at the transcriptional, posttranscriptional and epigenetic levels (Kang et al. 2020). In the process of bone tissue repair, in addition to osteogenesis-promoting scaffolds and seed cells, high-efficiency osteoinductive factors are also required. Therefore, the development of efficient osteoinductive factors has become our research focus. The precise molecular mechanisms related to osteogenesis thus require further investigation.
In this study, three original microarray datasets, GSE63754, GSE89330 and GSE72429, which were selected from Gene Expression Omnibus (GEO) database were used to identify differentially expressed genes (DEGs). A total of 63 upregulated mRNAs common to these datasets were selected. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, and protein–protein interaction (PPI), lncRNA–miRNA–mRNA networks were constructed to evaluate the molecular mechanisms underlying osteogenic differentiation. In addition, the roles of the central node gene LMO3 in the regulation of osteogenic differentiation by hADSCs were evaluated, and the role of LMO3 in the regulation of hADSC osteogenic differentiation was further investigated in vivo and in vitro. Our study might provide a theoretical basis for the application of effective osteoinductive factors in bone tissue engineering.
Materials and methods
Data collection
In the present study, we sifted datasets from the publicly available Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Filters of publication date was set to be before April 22, 2020, as well as the organisms were characterized as ‘homo sapiens’. All selected datasets were collected for the fact that all the ADSCs were from human tissues and these cells underwent osteogenic induction. Finally, the following three independent datasets were screened: the mRNA dataset GSE63754 contained three undifferentiated ADSCs and three osteogenic differentiated ADSCs (Daniunaite et al. 2015); the miRNA dataset GSE72429 contained four undifferentiated ADSCs and four osteogenic differentiated ADSCs (Daniunaite et al. 2015); the LncRNA and mRNA dataset GSE89330 included four undifferentiated ADSCs and four osteogenic differentiated ADSCs (Huang et al. 2017). The series matrix files of GSE63754, GSE89330, and GSE72429 were based on the platforms GPL17077 (Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381), GPL16956 (Agilent-045997 Arraystar human lncRNA microarray V3) and GPL16770 (Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray), respectively.
Identification of DEGs
The differentially expressed genes (DEGs) between differentiated hADSCs and undifferentiated hADSCs in the GEO datasets were identified using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). P < 0.01 and log2 fold change (log2FC) > 2 or < − 1 were the thresholds for DEGs to be considered statistically significant. The volcano plot and heatmap were constructed to present the expression difference of DEGs in different samples. A Venn diagram tool was used to identify the overlapping DEGs between GSE63754 and GSE89330 (including 63-overlapping upregulated DEGs and 46-overlapping downregulated DEGs).
GO and KEGG enrichment analyses
To further analyze the potential biological processes (BPs), cellular components (CCs), molecular functions (MFs), and pathways of the 63-overlapping upregulated DEGs, GO analysis and KEGG pathway enrichment analysis were performed with the online tools Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/; Chen et al. 2020.) and Metascape (http://metascape.org; Li et al. 2019.). P < 0.05 and count > 2 were set as the threshold values. R software (version 3.5.3, https://www.r-project.org/) was used to construct a diagram.
PPI network construction and module analysis
PPI network analysis of the 63-overlapping upregulated DEGs was conducted via the STRING online database (version 11.0; https://string-db.org/). The network was visualized using Cytoscape software, a broadly used tool for the visual exploration of interaction networks among numerous biomolecules, including proteins and genes. The Molecular Complex Detection (MCODE) plugin was used to identify the most significant module in the PPI networks.
Construction of a competing endogenous RNA (ceRNA) network
DIANA-LncBase (http://carolina.imis.athena innovation.gr/diana_tools/web/index.php?r = lncbasev2/index-predicted) was used to predict lncRNA–miRNA interactions. mRNAs were retrieved with TargetScan version 7.2 (http://www.targetscan.org/vert_72/). A lncRNA–miRNA–mRNA ceRNA network was constructed. The interactions and visualizations were conducted in Cytoscape software (https://cytoscape.org/).
Cell culture
Human subcutaneous adipose tissue was collected from healthy people (average age: 25 years old) by liposuction aspiration and processed as previously described (Bunnell et al. 2008; Gaur et al. 2019). In brief, tissue samples were cut into 1–1.5 mm3 pieces. Adipose tissue was incubated with 0.2% collagenase (Solarbio, Beijing, China). After digestion, tissue was centrifuged at 200×g for 4 min, which separated it into three layers: the upper layer (lipids and undigested adipose tissue), the middle layer (the supernatant), and the lower layer (a precipitate of mixed cells, such as hADSCs and red blood cells). The upper and middle layers were discarded, and the cell precipitate was resuspended in 2 ml of basal medium (low-glucose Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) supplemented with 20% fetal bovine serum (Gibco, New York, NY, USA) and 100 U/ml penicillin–streptomycin). The culture medium was changed 36 h after resuspension and every 3 days thereafter. Cells were passaged at a 1:3 ratio at 80% confluence. Passage 3 cells were used for all experiments in this study. All protocols for human tissue collection and handling were approved by the Research Ethics Committee of Cancer Hospital of China Medical University (approval no: (2020G0328)). All patients included our study signed the informed consents.
Identification of hADSCs
Phenotypic characterization of hADSCs was performed as previously described (Bourin et al. 2013; Dominici et al. 2006). Briefly, hADSCs were harvested and washed three times in PBS. The cells were suspended in PBS and incubated with PerCP-conjugated anti-CD45, PE-conjugated anti-CD105, FITC-conjugated anti-CD44, and FITC-conjugated anti-CD73 antibodies (BD Biosciences, CA, USA) in the dark for 30 min at 37 °C. Cells were then washed and resuspended in PBS before detection via flow cytometry (BD Biosciences). The results were analyzed in FACSDiva Version 6.2 software (BD Biosciences).
Cell differentiation and staining
hADSCs were seeded in basal medium at a density of 2 × 105 cells/well in six-well plates. After cells reaching 75–85% confluence, the basal medium was replaced with osteogenic, adipogenic, or chondrogenic induction medium (Cyagen, Guangzhou, Guangdong, China). For osteogenesis induction, the medium was replaced with complete osteogenic medium when the cells reached 80% confluence, and was changed every 2 days for 14 days. Cells were harvested for further experiments at days 14. For adipogenic induction, the medium was replaced with complete adipogenic medium when the cells reached 75% confluence and was changed every 2 days for 14 days. For chondrogenesis induction, the cells were placed in a 15 mL centrifuge tube and cultured in chondrogenesis induction medium for 21 days. The cell pellets were taken to make paraffin sections. Cells were stained with Oil Red O (Sigma, MO, USA) for adipocyte detection, with Alizarin Red S (Santa Cruz Biotechnology, CA, USA) for osteoblast detection, and with Alcian Blue Solution (Cyagen) for chondrocyte detection.
Lentiviral transfection and cell treatment
In order to overexpress LMO3, the lentivirus expressing LMO3 and the scramble negative control (NC) were purchased from GenePharma Co. (Shanghai, China). For LMO3 knockdown, recombinant lentiviruses targeting LMO3 (shLMO3) and the non-targeting negative control (shNC) were also purchased from GenePharma Co. For virus transfection, cells were exposed to the lentiviral supernatant with the addition of polybrene (5 μg/mL, Sigma-Aldrich) for 24 h. After 72 h, antibiotic selection was conducted by adding puromycin (5 μg/mL, Sigma-Aldrich) to transfected cells. To inhibit PI3K signaling, the hADSCs in the Lv-NC group or Lv-LMO3 group cultured in complete osteogenic medium were treated with 0.5 μmol/L wortmannin (phosphatidyl inositol-3 kinase (PI3K) inhibitor; Solarbio; Beijing, China), as indicated in each experimental design. The hADSCs were divided into the following groups: the Control group,
LV-NC group, LV-LMO3 group, wortmannin group, LV-NC + wortmannin group, and LV-LMO3 + wortmannin group.
qRT-PCR
TRIzol® reagent (TaKaRa, Tokyo, Japan) was used to extract total RNA. First-strand cDNA was synthesized with a PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa). Real-time PCR was carried out using a SYBR® Green PCR kit (TaKaRa). Gapdh RNA levels were used for normalization as an endogenous reference. The primer sequences are shown in Table 1.
Western Blot
Total protein extraction kit (KeyGen, Nanjing, China) and BCA protein determination kit (KeyGen) were used to extract and quantify total protein. The protein samples were mixed with loading buffer, boiled for 5 min, separated by SDS-PAGE, and transferred to polyvinylidene fluoride membranes (PVDF, Millipore, Billerica, MA, USA) in turn. The PVDF membranes were then incubated with 5% bovine serum albumin in Tris-buffered saline-0.5% Tween-20. Using primary antibodies against Runx2 (1:1000, Cell Signaling Technology), ALP (1:5000, Abcam), OSX (1:10,000, Abcam), LMO3 (1;2000, Abcam), phosphorylated-PI3K (1:1000, Cell Signaling Technology) or phosphorylated-Akt (1:1000, Cell Signaling Technology), the membranes were incubated at 4℃ overnight. After incubating the membrane with the horseradish peroxidase-conjugated secondary antibody, the ECL kit (Pierce, Rockford, IL, USA) was used to observe the antigen–antibody complex. An enhanced chemiluminescence detection system (Bio-Rad, Hercules, USA) was used for protein quantification. Protein levels are presented as the fold change normalized to an endogenous reference (β-actin protein).
Heterotopic osteogenesis animal model
Eighteen male BALB/c nude mice (5 weeks) obtained from the Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) were housed under the Specific Pathogen Free conditions at the Department of Laboratory Animal Science of China medical University. All animal studies were approved by the Animal Care and Use Committee of China Medical University (IACUC-2020012). The nude mice were divided into three groups (n = 6); Control (the hADSCs without transfection but osteogenic differentiation in vitro for 3 days), LV-NC, and LV-LMO3. After transfection, hADSCs were induced osteogenic differentiation in vitro for 3 days. Bio OSS collagen scaffold was cut into 5 mm × 5 mm × 5 mm pieces and incubated with 5 × 105 hADSCs at 37 ℃ for 1 h. Then, the scaffold and hADSCs were transplanted into the armpit region. Eight weeks after injection, mice were sacrificed and the transplants were extracted and fixed in 4% paraformaldehyde in PBS. Specimens were fixed, decalcified, dehydrated and processed for paraffin embedding, and cut into 4 μm thick slices. The pathological sections of each group were evaluated by Hematoxylin and eosin staining (H&E, Beyotime, China), Masson’s trichrome staining, and images were captured with a light optical microscope (Olympus, Japan). Computer-assisted histomorphometric measurements of the newly formed bone were obtained using an image analysis system (IBAS, Contron, Erching, Germany). The percentages of the newly formed bone within the specimen outline were then calculated. For the histomorphometric analysis, the significant differences for the amount of new bone formed were identified by ANOVA (analysis of variance). Data were considered significant with a P < 0.05, and the statistical analysis was performed using the statistical software package, SPSS 16.0 software (SPSS Inc., Chicago, IL, USA).
Statistical analyses
Statistical differences between groups were analyzed using Student’s t-test or one-way ANOVA by using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The results were presented as the mean ± SD of three separate experiments. Values of P less than 0.05 were considered statistically significant.
Results
Identification of DEGs expressed during osteogenic differentiation
The GEO2R online analysis tool was used to investigate the DEGs in the selected GEO datasets. Volcano plots and heatmaps were constructed to visualize the results of the differential gene expression analysis (Fig. 1A–D). The 63-overlapping upregulated DEGs between the GSE63754 and GSE89330 datasets were shown in Fig. 1E. The 46-overlapping downregulated DEGs between the GSE63754 and GSE89330 datasets were shown in Fig. 1F. These genes were selected and presented in a Venn diagram. The DEGs are listed in Table 2.
The commonly and differentially expressed genes between GSE63754 and GSE89330. Volcano plot and heatmap of significant differential gene expression in GSE63754 (A and B) and GSE89330 (C and D). For the volcano plot, the x-axis shows the gene expression differences as log-transformed fold changes, while the y-axis shows significance as –log10-transformed p-values. A gene was considered to be significantly differentially expressed if its |log (FC)|> 1 and P-value < 0.05. Red dots represent upregulated genes, while green dots represent downregulated genes. E and F Results for overlapping upregulated DEGs (E) and downregulated DEGs (F) in the indicated GEO datasets
GO enrichment and KEGG pathway analyses
To investigate the biological classification of the 63-overlapping upregulated DEGs, all genes in the two datasets were identified using DAVID and Metascape software. The cutoff criterion was established as P < 0.05. The 63-overlapping upregulated DEGs were significantly enriched in the BP terms organism process, response to stimulus, cellular component organization or biogenesis and cell differentiation (Fig. 2A). The 63-overlapping upregulated DEGs were significantly enriched in the CC terms membrane and extracellular region (Fig. 2B). The 63-overlapping upregulated DEGs were significantly enriched in the MF terms protein binding, receptor binding and oxidoreductase activity (Fig. 2C). The KEGG pathway enrichment analysis results were visualized using a bubble chart; the overlapping upregulated DEGs were enriched in metabolism-associated pathways, including tyrosine metabolism, phenylalanine metabolism, histidine metabolism and drug metabolism–cytochrome P450 (Fig. 2D). Metascape software was used to evaluate the biological classification of the overlapping downregulated DEGs. As shown in Fig. 2E, the overlapping downregulated DEGs were enriched in muscle structure development, extracellular structure organization and muscle tissue development.
GO and pathway enrichment analysis. A–C Significant biological processes of overlapping upregulated genes, including biological processes (A), cell components (B), and molecular functions (C). D Significant biological pathways of the overlapping upregulated genes. E Significantly biological process of overlapping upregulated genes
Construction of a PPI network and module analysis
To further investigate the interactions among the 63 upregulated DEGs, the PPI network of the 63 upregulated genes was constructed with the STRING and Cytoscape (v3.1.2) tools (Fig. 3A). We identified two significant modules via cluster analysis of the PPI network with the Cytoscape MCODE plugin based on the degree of importance (Fig. 3B). Specifically, the central node genes in significant module 1 were LOM3, SAA1, SAA2, HP, APOB, and the central node genes in significant module 2 were ADH1C, ADH1A, MAOB, MAOA. The central node genes might potentially play an important role in regulating ADSC osteogenic differentiation. We then performed GO and pathway enrichment analyses of the genes in these modules. The genes in these two modules were mainly enriched in tyrosine metabolism, retinoid metabolic process, binding and uptake of ligands by scavenger receptors, drug catabolic process and G alpha (i) signaling events (Fig. 3C).
Construction of lncRNA–miRNA–mRNA network
The GSE89330 and GSE72429 datasets were used to identify upregulated lncRNAs and downregulated miRNAs (Fig. 4A, B). A total of 240 upregulated lncRNAs and 11 downregulated miRNAs were identified. The DIANA-LncBase (www.microrna.gr/LncBase) and TargetScan version 7.2 (http://www.targetscan.org/vert_72/) databases were used to predict the binding between lncRNAs and the mRNA transcripts of miRNAs. The intersection of the predicted lncRNAs and mRNAs with the upregulated lncRNAs and mRNAs mentioned above was determined to identify the lncRNAs in the ceRNA network. Among the genes in the network, we found LMO3 are upregulated in osteogenic differentiation (Fig. 4C).
Construction of lncRNA–miRNA–mRNA network based on GSE89330 and GSE72429. A and B Volcano plot and heatmap of significance gene expression difference in GSE72429. C Construction of lncRNA–miRNA–mRNA regulatory network. The lncRNA–miRNA–mRNA regulatory network included 10 lncRNAs, 10 miRNAs and 40 mRNAs
Identification and osteogenic differentiation of hADSCs
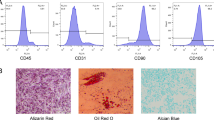
Using flow cytometry analysis, we found CD73, CD44, and CD105 but not CD45 expressing in hADSCs (Fig. 5A). hADSCs could be induced to differentiate into adipocytes, osteoblasts, or chondrocytes in the appropriate induction culture medium (Fig. 5B–E). Cells were stained with Oil Red O for adipocyte detection, Alizarin Red S for osteoblast detection, and Alcian Blue Solution for chondrocyte detection.
Human ADSC characteristics. A Flow cytometry analysis showing positive and negative expression of hADSC surface markers. B Representative photomicrographs of hADSC morphological characteristics. Scale bar, 120 μm. C Adipogenesis shown as red lipid droplets in the cytoplasm after 14 days of adipogenic induction (oil red O staining). Scale bar, 120 μm. D Osteogenesis shown as orange-red mineralized nodule formation after 21 days of osteogenic induction (Alizarin Red staining). Scale bar, 120 μm. E Chondrogenesis shown as round, multilayered cells with blue nuclei and cytoplasm after 21 days of chondrogenic induction (Alcian Blue solution staining). Scale bar, 20 μm
LMO3 knockdown inhibited osteogenic differentiation of hADSCs
Among those key genes, we found that LMO3 was one of the central node genes in significant module 1 generated by the PPI network. In addition, in the competing endogenous RNA (ceRNA) network, we found LMO3 is a key upregulated node in osteogenic differentiation. To further verify our analysis results, we successfully induced the differentiation of hADSCs. We demonstrated that the expression of LMO3 was upregulated during the osteogenic differentiation of hADSCs by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 6A). After knocking down LMO3 expression, the expression of ALP and RUNX2 was decreased (Fig. 6B-D). In addition, Alizarin Red S staining and ALP staining after osteogenic induction for two weeks revealed that the number of calcium nodules was significantly lower in the LMO3 shRNA group than in the shNC group and control group (Fig. 6E). Besides, it was detected by Western blot that the endogenous RUNX2, ALP and OSX levels showed a clear decrement in hADSCs which were transfected with shLMO3 on 14 days of differentiation (Fig. 6F).
LMO3 knockdown inhibited osteogenic differentiation of hADSCs. A The expression level of LMO3 was significantly upregulated in hADSCs undergoing osteogenic differentiation. B Dramatic downregulation of mRNA expression of LMO3 following shLMO3 transfection. C and D Inhibition of the expression of osteogenic differentiation markers by LMO3 downregulation. qRT-PCR demonstrated that the mRNA levels of ALP and RUNX2 were suppressed in shLMO3 hADSCs undergoing osteogenic differentiation. Bars indicate SD. NS refers to no statistical significance between groups. *Refers to statistical significance between groups (P < 0.05). **Refers to statistical significance between groups (P < 0.01). E Alkaline phosphatase (ALP) staining and Alizarin Red S staining in control, shNC and shLMO3 groups. Scale bar, 100 μm. F and G Inhibition of the expression of osteogenic differentiation markers by LMO3 downregulation. Western blot demonstrated that the protein levels of LMO3, ALP, Runx2 and OSX were suppressed in shLMO3 hADSCs undergoing osteogenic differentiation. Bars indicate SD. NS refers to no statistical significance between groups. *Refers to statistical significance between groups (P < 0.05). **Refers to statistical significance between groups (P < 0.01)
LMO3 overexpression enhanced osteogenic differentiation of hADSCs
The effect of LMO3 on osteogenesis was further evaluated by overexpressed LMO3 in hADSCs. Western blot showed the expression level of LMO3 increased by more than 2.5 folds in the LV-LMO3 group in comparison to the control or LV-NC group (Fig. 7A, B). After culturing the infected hADSCs in osteogenic medium for 14 days, Western blot was performed to detect the protein expression levels of osteogenic markers, which showed that LMO3 overexpression promoted the expression of RUNX2, ALP, and OSX(Fig. 7A, C–E). ALP staining and ARS staining further supported the trend found in Western blot analysis (Fig. 7F).
LMO3 overexpression enhanced osteogenic differentiation of hADSCs. A and B Dramatic upregulation of LMO3 expression following LMO3 overexpression. C–E Western blot demonstrated that upregulation of the expression of osteogenic differentiation markers by LMO3 overexpression. (*P < 0.05, **P < 0.01, compared with the control group). F Alkaline phosphatase (ALP) staining and Alizarin Red S staining in the control, LV-NC and LV-LMO3 groups. Scale bar, 100 μm
Effect of LMO3 on hADSCs osteogenic differentiation in nude mice
To validate our findings in vitro, we examined whether LMO3 could affect the bone formation ability of hADSCs in vivo. There was trabecular bone as well as an amorphous calcified matrix in all the samples. The trabeculae included many osteocytes and were regularly lined with many osteoblasts, indicating bone-forming activity. There was no evidence of inflammation or foreign-body reaction in the host tissue adjacent to the new bone, nor was there any evidence of cartilage generation. As shown in Fig. 8A, HE staining and Masson staining revealed nude mice implanted with LV-LMO3 hADSCs showed much more bone-like tissue. Furthermore, quantitative measurements confirmed that the volume of newly formed bone was increased in LV-LMO3 hADSCs (Fig. 8B). The percentages of newly formed bone were 11.1% ± 0.6% in the control group, 10.7% ± 0.81% in the LV-NC group, and 68.2% ± 1.7% in the LV-LMO3 group. These results indicated that LMO3 might promote osteogenic differentiation of hADSCs in vivo.
Effect of LMO3 on hADSCs osteogenic differentiation in nude mice. A At 56 days post-transplantation, Heterotopic osteogenesis animal model demonstrated that LMO3 overexpression promoted bone formation capacity of hADSCs., as determined by HE staining (HE) and Masson staining (Masson). Scale bar, 100 μm. B Quantitative measurements of bone-like tissue showed the area of bone-like tissue was significantly increased in LV-LMO3 group. Scale bar = 20 μm. Bars indicate SD. NS refers to no statistical significance between groups. **Refers to statistical significance between groups (P < 0.01)
LMO3 overexpression promoted osteogenesis by mediating PI3K/Akt signaling pathway
To further explore the mechanism by which LMO3 regulates osteogenic differentiation, we examined several osteogenesis-related signaling pathways and found the expression of phosphorylated Akt (p-Akt) and phosphorylated PI3K (p-PI3K) were upregulated after LMO3 overexpression (Fig. 9A–C). We hypothesized that PI3K/Akt signaling might be involved in LMO3- mediated osteogenic differentiation. To further clarify the specific mechanism by which LMO3 regulated osteogenic differentiation of hADSCs, the PI3K inhibitor (wortmannin) were added in. The hADSCs were divided into the following groups: the Control group, LV-NC group, LV-LMO3 group, wortmannin group, LV-NC + wortmannin group, and LV-LMO3 + wortmannin group. Western blots were performed on Day 7 after treatment (Fig. 9D). The level of OSX were significantly upregulated in the LV-LMO3 group, and significantly downregulated in the wortmannin group (Fig. 9E). The level of p-PI3K and p-Akt were upregulated in the LV-LMO3 group compared with the control group. The level of p-PI3K and p-Akt in the wortmannin group and LV-NC + wortmannin group was significantly decreased by treating with wortmannin (Fig. 9F–G). The wortmannin-induced inhibition of p-PI3K and p-Akt expression was attenuated by LMO3 overexpression (Fig. 9F–G). These data suggested that LMO3 overexpression might activate PI3K/Akt signaling pathway.
LMO3 overexpression promoted osteogenesis by mediating PI3K/Akt signaling pathway. A–C key proteins of PI3K/Akt signaling pathway analysed by Western blot after LMO3 overexpression (*P < 0.05, **P < 0.01, compared with NC). (D-G) Western blots were performed on Day 7 after the PI3K inhibitor (wortmannin) were added in. Bars indicate SD. NS refers to no statistical significance between groups. *Refers to statistical significance between groups (P < 0.05). **Refers to statistical significance between groups (P < 0.01)
Discussion
That hADSCs have self-renewal ability, high proliferation ability and pluripotency, makes them an attractive cell resource for repairing bone trauma. It is important to find the key molecules that promote the osteogenic differentiation of hADSCs. In this study, we identified 63 significantly upregulated mRNAs common to the three selected datasets. Among these genes, LMO3 was selected for further study. Based on GO and pathway enrichment analyses of the significantly upregulated genes common to GSE63754 and GSE89330, the BP terms organism process, response to stimulus, cellular component organization or biogenesis and cell differentiation had the highest enrichment scores, indicating that these processes play active roles in the osteogenic differentiation of hADSCs. Among the CC terms, membrane and extracellular region had the highest enrichment scores. Interestingly, the top eleven CCs most strongly associated with hADSC osteogenic differentiation were located in either the cell membrane or the extracellular space, indicating that cell‐to‐cell signaling is critical for the osteogenic differentiation of hADSCs. The most highly affected MF terms were protein binding, receptor binding and oxidoreductase activity.
In the KEGG analysis, the top significantly altered pathways were related to metabolism-associated pathways, including tyrosine metabolism, phenylalanine metabolism, histidine metabolism and drug metabolism (Tang et al. 2017)-cytochrome P450. The tyrosine metabolism-related molecules Src homology-2 (SH2) domain-containing phosphatase 1 (SHP-1) and tyrosine-rich amelogenin peptide (TRAP) have been reported to affect the process of osteogenic differentiation (Amin and Ethier 2016), indicating that tyrosine metabolism has an important influence on the osteogenic differentiation of hADSCs.
PPI network analysis is considered a powerful tool for developing a mechanistic understanding of the functions of complex biological systems in health and of their dysfunctions in disease. We constructed 2 subnetworks containing 9 nodes via the Cytoscape MCODE plugin. In subnetwork 1, the central node genes were SAA2, LMO3, APOB, SAA1, and HP, and those in subnetwork 2 were ADH1C, ADH1A, MAOA, and MAOB.
lncRNAs, miRNAs and mRNAs form large-scale ceRNA crosstalk networks, which have interesting implications on posttranscriptional gene regulation during multiple physiological and pathophysiological processes (Ala et al. 2013; Gu et al. 2017; Salmena et al. 2011; Wang et al. 2015). The expression of some lncRNAs is altered during osteogenic differentiation in different types of MSCs (Dong et al. 2014). For example, the lncPCAT1/miR-106a-5p/E2F5 axis can regulate the osteogenic differentiation of periodontal ligament stem cells (Jia et al. 2019). In our study, we found 240 lncRNAs upregulated during osteogenesis in hADSCs. Through ceRNA network analysis, we identified 10 lncRNAs that are upregulated during osteogenic differentiation in hADSCs and may regulate the expression of 40 mRNAs, thus promote osteogenic differentiation.
In our study, we constructed a lncRNA–miRNA–mRNA network which is composed of the associated genes. In this network, LMO3 was upregulated during osteogenic differentiation. LIM-only proteins (LMO), which consist of LMO1, LMO2, LMO3, and LMO4, are involved in cell fate determination and differentiation during embryonic development (Aoyama et al. 2005). Sun et al. reported that LMO3 promoted human preadipocyte differentiation by enhancing peroxisome proliferator-activated receptor γ (PPARγ) transcriptional activity, which is the master regulator of adipogenesis. Wagner et al. reported that LMO3 was a new key player in the development of human adipose tissue, acting as a new partner in GC-dependent signaling to modulate the key adipogenic master switch PPARγ in human, but not mouse, visceral adipose progenitors (Wagner et al. 2021). In this study, we first showed that LMO3 overexpression enhanced hADSC osteogenesis through PI3K/Akt signaling. Combining with the previous researches, we speculated that LMO3 might play important role in the hADSC stemness. However, we didn’t explore in this study.
The PI3K/Akt pathway is an intracellular signaling pathway of great importance in the cell cycle process (Xie et al. 2019). In recent years, many researches have reported that the PI3K/Akt signaling pathway was associated with cell proliferation and differentiation (Dong et al. 2016; Meng et al. 2006; Peltier et al. 2007). This axis has also been proven to be a key regulator in the differentiation of mesenchymal stem cells (Gao et al. 2021). However, it is not clear whether LMO3 could promote osteogenesis through the PI3K/Akt axis. Accordingly, the experiment explored whether LMO3 regulated the osteogenic differentiation of human adipose derived stem cells via PI3K/Akt signaling pathway. We overexpressed LMO3 in hADSCs, and found that the protein expression of phosphorylated Akt and phosphorylated PI3K were both upregulation. Wortmannin is an effective inhibitor of PI3K/Akt (Yang et al. 2021), and it can reduce the expression of phosphorylated Akt (Ahmed et al. 2021). Then our study proved that wortmannin successfully blocked the PI3K/Akt signaling pathway. We detected the related indicators and found that the activating of the PI3K/Akt signaling pathway due to overexpression LMO3 was significantly inhibited with wortmannin pretreatment. These results suggested that LMO3 could promote osteogenesis through the PI3K/Akt axis.
With the development of tissue engineering to this day, more and more biocompatible scaffolds are used in the repair of bone defects. Thus, it can be seen that the development of effective osteoinductive factors is essential. These studies prove the reliability of our analysis results. In addition, we demonstrated that the expression of LMO3 was upregulated during the osteogenic differentiation of hADSCs. LMO3 expression affected the osteogenic potential of hADSCs in vivo. We provided the first evidence that LMO3 can regulate the osteogenic differentiation of hADSCs via the PI3K/Akt axis. Our research provides a new therapeutic target for bone defect repair.
Conclusions
In conclusion, based on transcriptomic analysis, we identified key genes involved in regulating the osteogenic differentiation of hADSCs. In addition, we found that LMO3 might act as a positive modulator of hADSC osteogenic differentiation by mediating PI3K/Akt signaling pathway. Manipulating the expression of LMO3 and its associated pathways might contribute to advances in bone regeneration and tissue engineering.
References
Ahmed LA et al (2021) Boosting Akt pathway by rupatadine modulates Th17/Tregs balance for attenuation of isoproterenol-induced heart failure in rats. Front Pharmacol 12:651150
Ala U et al (2013) Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA 110(18):7154–7159
Amin HD, Ethier CR (2016) Differential effects of tyrosine-rich amelogenin peptide on chondrogenic and osteogenic differentiation of adult chondrocytes. Cell Tissue Res 364(1):219–224
Aoyama M et al (2005) LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Can Res 65(11):4587–4597
Bourin P et al (2013) Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy 15(6):641–648
Bunnell BA et al (2008) Adipose-derived stem cells: isolation, expansion and differentiation. Methods 45(2):115–120
Chen S et al (2020) Identification of the key genes and microRNAs in adult acute myeloid leukemia with FLT3 mutation by bioinformatics analysis. Int J Med Sci 17(9):1269–1280
Cheng C, Wentworth K, Shoback DM (2020) New frontiers in osteoporosis therapy. Annu Rev Med 71:277–288
Chun SY et al (2019) Preparation and characterization of human adipose tissue-derived extracellular matrix, growth factors, and stem cells: a concise review. Tissue Eng Regen Med 16(4):385–393
Daniunaite K et al (2015) Epigenetic regulation of human adipose-derived stem cells differentiation. Mol Cell Biochem 410(1–2):111–120
Dominici M et al (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8(4):315–7
Dong R et al (2014) Comparison of long noncoding RNA and mRNA expression profiles in mesenchymal stem cells derived from human periodontal ligament and bone marrow. Biomed Res Int 2014:317853
Dong X et al (2016) GPR39 activates proliferation and differentiation of porcine intramuscular preadipocytes through targeting the PI3K/AKT cell signaling pathway. J Recept Signal Transduct Res 36(2):130–138
Duttenhoefer F et al (2013) 3D scaffolds co-seeded with human endothelial progenitor and mesenchymal stem cells: evidence of prevascularisation within 7 days. Eur Cells Mater 26(4):49–65
Gao F et al (2021) Cornuside I promoted osteogenic differentiation of bone mesenchymal stem cells through PI3K/Akt signaling pathway. J Orthop Surg Res 16(1):397
Gaur M, Dobke M, Lunyak VV (2019) Methods and strategies for procurement, isolation, characterization, and assessment of senescence of human mesenchymal stem cells from adipose tissue. Methods Mol Biol 2045:37–92
Gu X et al (2017) Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet 18(1):100
Hu C, Zhao L, Li L (2019) Current understanding of adipose-derived mesenchymal stem cell-based therapies in liver diseases. Stem Cell Res Ther 10(1):199
Huang G et al (2017) Identification and characterization of long non-coding RNAs in osteogenic differentiation of human adipose-derived stem cells. Cell Physiol Biochem 42(3):1037–1050
Jia B et al (2019) A feed-forward regulatory network lncPCAT1/miR-106a-5p/E2F5 regulates the osteogenic differentiation of periodontal ligament stem cells. J Cell Physiol 234(11):19523–19538
Kang Y et al (2020) Differential circular RNA expression profiling during osteogenic differentiation in human adipose-derived stem cells. Epigenomics 12(4):289–302
Kanis JA et al (2019) European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int 30(1):3–44
Li W et al (2019) Comprehensive bioinformatics analysis of acquired progesterone resistance in endometrial cancer cell line. J Transl Med 17(1):58
Liu T et al (2021) Advances of adipose-derived mesenchymal stem cells-based biomaterial scaffolds for oral and maxillofacial tissue engineering. Bioact Mater 6(8):2467–2478
Meng Q et al (2006) Role of PI3K and AKT specific isoforms in ovarian cancer cell migration, invasion and proliferation through the p70S6K1 pathway. Cell Signal 18:2262–2271
Park JS et al (2020) Osteoporotic conditions influence the activity of adipose-derived stem cells. Tissue Eng Regen Med 17(6):875–885
Paspaliaris V, Kolios G (2019) Stem cells in osteoporosis: from biology to new therapeutic approaches. Stem Cells Int 2019:1730978
Peltier J et al (2007) PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol 67:1348–1361
Salmena L et al (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Si L et al (2015) Projection of osteoporosis-related fractures and costs in China: 2010–2050. Osteoporos Int 26(7):1929–1937
Sorger JI et al (2001) Allograft fractures revisited. Clin Orthop Relat Res 382:66–74
Sun W et al (2019) Resveratrol inhibits human visceral preadipocyte proliferation and differentiation in vitro. Lipids 54(11–12):679–686
Tang XL et al (2017) Protein tyrosine phosphatase SHP-1 modulates osteoblast differentiation through direct association with and dephosphorylation of GSK3β. Mol Cell Endocrinol 439:203–212
Vos T et al (2020) Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396(10258):1204–1222
Wagner G et al (2021) LMO3 reprograms visceral adipocyte metabolism during obesity. J Mol Med (berl) 99(8):1151–1171
Wang L et al (2015) Differential expression of long noncoding ribonucleic acids during osteogenic differentiation of human bone marrow mesenchymal stem cells. Int Orthop 39(5):1013–1019
Wu Z et al (2020) Immunomodulatory effects of mesenchymal stem cells for the treatment of cardiac allograft rejection. Exp Biol Med (Maywood). https://doi.org/10.1177/1535370220978650
Xie Y et al (2019) PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia. Mol Med Rep 19(2):783–791
Yang B et al (2021) Selenium attenuates ischemia/reperfusion injury-induced damage to the blood-brain barrier in hyperglycemia through PI3K/AKT/mTOR pathway-mediated autophagy inhibition. Int J Mol Med 48(3):178
Funding
This study was supported by the Joint Funds of the Natural science Foundation of Liaoning Province, Grant Number 2020-ZLLH-40.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experimental procedures involving animals were done in accordance with the Guide for the Care and Use of Laboratory Animals and the institutional ethical guidelines for animal experiments.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kang, Y., Pei, W. Transcriptomic analysis and biological evaluation reveals that LMO3 regulates the osteogenic differentiation of human adipose derived stem cells via PI3K/Akt signaling pathway. J Mol Histol 53, 379–394 (2022). https://doi.org/10.1007/s10735-021-10047-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-021-10047-5