Abstract
Despite the efficacy of tamoxifen in preventing disease relapse, a large portion of breast cancer patients show intrinsic or acquired resistance to tamoxifen, leading to treatment failure and unfavorable clinical outcome. MYB proto-oncogene like 2 (MYBL2) is a transcription factor implicated in the initiation and progression of various human cancers. However, its role in tamoxifen resistance in breast cancer remained largely unknown. In the present study, by analyzing public transcriptome dataset, we found that MYBL2 is overexpressed in breast cancer and is associated with the poor prognosis of breast cancer patients. By establishing tamoxifen-resistant breast cancer cell lines, we also provided evidence that MYBL2 overexpression contributes to tamoxifen resistance by up-regulating its downstream transcriptional effectors involved in cell proliferation (PLK1, PRC1), survival (BIRC5) and metastasis (HMMR). In contrast, inhibiting those genes via MYBL2 depletion suppresses cancer progression, restores tamoxifen and eventually reduces the risk of disease recurrence. All these findings revealed a critical role of MYBL2 in promoting tamoxifen resistance and exacerbating the progression of breast cancer, which may serve as a novel therapeutic target to overcome drug resistance and improve the prognosis of breast cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common cancer in women and a leading cause of cancer-related mortality worldwide. As a large portion (~ 80%) of breast cancer patients show positive estrogen receptor (ER) expression, blocking the proliferative effects of estrogen with ER antagonists, such as tamoxifen, is most frequently used to treat breast cancer and prevent cancer recurrence (Jordan 2003). However, more than half of the patients with advanced disease stages show intrinsic resistance to tamoxifen treatment. Even in patients initially responding well to tamoxifen, up to 40% of them develop acquired drug resistance during the treatment process, which eventually leads to tumor recurrence and has been considered as a serious obstacle in the management of breast cancer (Ring and Dowsett 2004). In this context, a deeper understanding of the molecular mechanism underlying the pathophysiology of tamoxifen resistance is essential for developing novel therapeutic strategies to overcome drug resistance and improve the overall survival of breast cancer patients.
MYB Proto-Oncogene Like 2 (MYBL2) is a highly conserved member of the MYB (myeloblastosis) family of transcription factors critical for normal cell growth and survival, while deregulated MYBL2 function is implicated in the initiation and progression of various human cancers (Musa et al. 2017). As a transcription factor, MYBL2 is responsible for the expression of multiple cell cycle regulators involved in G2/M transition such as cyclin B1 and cyclin A2, and promote cell proliferation when overexpressed in cancer cells (Osterloh et al. 2007; Zhu et al. 2004). In addition, MYBL2 activity coordinates with the expression of stem cell markers Oct4, Nanog and Sox, which further sustains the self-renewal capacity and maintains cancer cells in an undifferentiated or stem cell-like status (Tarasov et al. 2008a, b; Zhan et al. 2012). Moreover, MYBL2 also show potential to promote cancer cell survival under stress conditions by enhancing the expression of apoptosis and autophagy suppressors such as Bcl-2, clusterin and VDAC2, and thus confers resistance to cancer treatments (Cervellera et al. 2000; Grassilli et al. 1999; Santilli et al. 2005; Yuan et al. 2015). Consistent with this, MYBL2 is upregulated in ovarian cancer cells with impaired response to carboplatin (Peters et al. 2005), and is also involved in the drug resistance against various DNA interactive agents such as doxorubicin in neuroblastoma and fibrosarcoma cells (Cervellera et al. 2000; Levenson et al. 2000). All these prior findings support the oncogenic potential of MYBL2.
In breast cancer, MYBL2 has been found to drive abnormal cell growth and is predictive of poor prognosis (Inoue and Fry 2016; Tao et al. 2014). There is also evidence suggesting a role of MYBL2 overexpression in breast cancer metastases via disrupting cell adhesion and promoting epithelial-to-mesenchymal transition, while MYBL2 depletion is able to restore cell junction and suppress cell invasion (Tao et al. 2015). However, up to now, we still have a lack of knowledge about the impact of MYBL2 on breast cancer treatment, particularly tamoxifen, the oldest and most widely used adjuvant therapy for breast cancer patients. Therefore, in the current study, we established tamoxifen-resistant cell line to evaluate the association of MYBL2 with tamoxifen efficacy and explore the underlying mechanism. The results from our study will not only lead to a better understanding of the function of MYBL2 in breast cancer progression, but also provide novel insights into cancer therapy.
Materials and methods
Breast cancer transcriptome analysis
Gene Expression Profiling Interactive Analysis (GEPIA) web server was used to study gene profiling data of 1085 breast tumor tissues and 291 normal tissues extract from The Cancer Genome Atlas (TCGA) database (Tang et al. 2017). Publicly available transcriptome data of breast cancer patients was also obtained from the Gene Expression Omnibus (GEO) database (GSE4922) (Ivshina et al. 2006). The association of gene expression with disease-free survival was evaluated using Kaplan-Meier survival analysis and log-rank test, in which patients with gene expression level higher than average were in the high group and the rest were in the low group. Chromatin Immunoprecipitation Sequencing (ChIP-seq) data was obtained from ChIP-Atlas database (Oki et al. 2018) and visualized by Integrative Genomics Viewer (IGV) (Robinson et al. 2011).
Tamoxifen-resistant breast cancer cell lines
Two breast cancer cell lines MCF7 and T47D obtained from the American Type Culture Collection (ATCC) were used in the current study. MCF7 cells were cultured in Eagle’s minimal essential medium (MEM) (Hyclone) and T47D cells were cultured in RPMI-1640 medium (Hyclone). Both cell culture media were supplemented with 10% fetal bovine serum (FBS) (Hyclone) and incubated with 5% CO2 at 37 °C. Tamoxifen resistant cell lines MCF7/TamR and T47D/TamR were produced by continuously exposing parental cells to tamoxifen (Sigma) for over 12 months. The treatment started with initial tamoxifen concentration of 0.05 µM, which was gradually increased to 1 µM. Tamoxifen-resistant cell lines were established when the cells could survival under 1 µM Tamoxifen treatment for more than 6 months (Li et al. 2018; Zhu et al. 2018).
MYBL2 knockdown by lentivirus-mediated RNA interference
shRNAa targeting MYBL2 sequence 5′-CCCAGATCAGAAGTACTCCAT-3′ (shMYBL2) as well as nonsliencing scramble sequence 5′-GAAACTACCAGATCTGACTCG-3′ (shCtrl) were cloned into lentivirus vector GV115 carrying green fluorescent protein (GFP). Recombinant lentiviruses expressing shMYBL2 and shCtrl were produced by co-transfecting 293T cells (ATCC) with shRNA-encoding plasmid, packaging plasmid and envelop plasmid using Lipofectamine2000 (Invitrogen). Lentiviral supernatant was harvested after 48 hrs and was used to transduce tamoxifen-resistant cells. Transduction efficiency can be monitored by GFP marker under fluorescence microscopy.
Cell growth assay
For dose-dependent cytotoxicity assays, 5⋅103 tamoxifen-resistant cells and parental cells were plated in 96-cell plate and were treated with different concentrations of tamoxifen or 2 days. Cell viability was measured using Cell Counting Kit 8 (CCK-8) (Abcam). Cells treated with 0.1% ethanol were used as control. For cell proliferation assays, 1⋅103 tamoxifen-resistant cells transfected with shMYBL2 and shCtrl were plated in 96-cell plate and were stained with CCK-8 solution at different time points. OD values were measured at 450 nm. Experiment was performed in triplicate.
Apoptosis assay
Tamoxifen-resistant cells transfected with shMYBL2 and shCtrl were treated with 1 µM tamoxifen for 48 hrs. The cells were then trypsinized, stained with Annexin V-FITC and propidium iodide (PI) using Apoptosis Staining/Detection Kit (Abcam), and analyzed by flow cytometry.
Quantitative real-time PCR
Total RNA was extracted from parental cells and tamoxifen-resistant cells using Trizol reagent (Invitrogen) and was reverse transcribed to cDNA using TaqMan Reverse Transcription Reagents (Thermo) in triplicate. Quantitative real-time PCR was performed using SYBR Premix Ex Taq (Takara) with primers specific to MYBL2 and GAPDH. Gene abundance was quantified using 2−ΔΔCt method normalized to internal standard GAPDH.
Wound healing assay
Tamoxifen-resistant cells transfected with shMYBL2 and shCtrl were grown in 6-well plates. At around 80 ~ 90% confluence, wounds were generated on cell monolayers using 1 ml pipette tips. Wound closure rate was determined as the change of wound widths after 24 hrs using a microscopy with camera.
Transwell invasion assay
Tamoxifen-resistant cells transfected with shMYBL2 and shCtrl were plated into the upper 24-well Transwell chamber coated with Matrigel (Corning) and cultured in FBS-free medium. The lower chamber was filled with the same culture medium but supplemented with 10% FBS. After incubation at 37 °C for 24 hrs, Matrigel-coated filters with invaded cells were fixed with methanol and stained with crystal violet. Stained cells were then visualized under a microscopy with camera.
Western blotting
Whole protein lysate was obtained from breast cancer cells with RIPA lysis buffer. After quantification with BCA Assay Kit (Thermo), 30 µg total protein from each sample was loaded onto SDS-PAGE and transferred onto PVDF membrane. The membrane was blocked with 5% skim milk and then incubated with primary antibodies against MYBL2, Tubulin (Abcam), PLK1, PRC1, BIRC5, Caspase 3 (Cell Signaling) and HMMR (Sigma), followed by secondary antibody. Protein signal was visualized using enhanced Chemiluminescent Detection Kit (Millipore).
Results
MYBL2 expression in breast cancer
We first used large-scale breast cancer transcriptome data to evaluate MYBL2 expression in breast cancer and its association with disease progression. As revealed by GEPIA analysis of the RNA sequencing data from 1085 breast tumor tissues and 291 normal tissues (Tang et al. 2017), breast cancer was associated with dramatic MYBL2 overexpression (p < 0.001) (Fig. 1a). In addition, according to TNM staging, more elevated MYBL2 expression was observed at more advanced disease stages (p < 0.001) (Fig. 1b), indicating a role of MYBL2 in breast cancer progression. This result was further confirmed by using other cancer grading system, Nottingham grading system, in different breast cancer datasets (GSE4922) (Ivshina et al. 2006), in which, MYBL2 level was again highly correlated with cancer severity (p < 0.001) (Fig. 1c). Moreover, we also found that, compared to those with relatively low MYBL2 levels, patients with high MYBL2 expression were at higher risk to develop recurrent disease (HR, 2.80; 95% CI, 1.78–4.41; p < 0.001) (Fig. 1d), suggesting the association of MYBL2 overexpression with poor prognosis of breast cancer.
MYBL2 expression in breast cancer. a MYBL2 expression in breast cancer tissue and normal tissue according to GEPIA analysis of RNA sequencing data obtained from TCGA project. b MYBL2 expression in different breast cancer stages classified by TNM staging system. c MYBL2 expression in different breast cancer grades classified by Nottingham grading system. Data was obtained from public dataset (GSE4922). d The association of MYBL2 expression with disease-free survival in breast cancer patients
MYBL2 overexpression in tamoxifen-resistant breast cancer cells
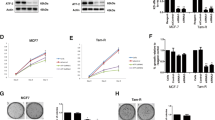
As cancer progression and recurrence are frequently accompanied by treatment failure, we thus hypothesized that MYBL2 may be associated with drug resistance in breast cancer treatment, and aimed to explore the influence of MYBL2 on the action of tamoxifen, the standard first-line treatment for a major part of breast cancer patients. To this end, we generated tamoxifen-resistant breast cancer cell lines MCF7/TamR and T47D/TamR by treating the parental cells MCF7 and T47D with gradually increased concentrations of tamoxifen. As shown in Fig. 2a, tamoxifen-resistant cells demonstrated increased viability on treatment, and the tamoxifen concentration required for 50% growth inhibition (IC50) was significantly higher for both MCF7/TamR and T47D/TamR cells compared to parental cells (p < 0.001) (Fig. 2b). More importantly, as revealed by RT-PCR and western blot analyses, we found that MYBL2 expression was largely enhanced in tamoxifen resistant cells at both transcription and translation levels (Fig. 2c, d), while knockdown of MYBL2 expression by small hairpin RNA (shRNA) showed potential to resensitize the cells to tamoxifen (Fig. 2d, e) and thus decrease the IC50 concentrations (p < 0.01) (Fig. 2f). All these results indicated the association of MYBL2 with tamoxifen resistance in breast cancer.
MYBL2 overexpression in tamoxifen-resistant breast cancer cells. a Tamoxifen cytotoxicity analysis in parental cells and tamoxifen-resistant breast cancer cells. b The IC50 concentration of tamoxifen for parental cells and tamoxifen-resistant breast cancer cells. c The expression level of MYBL2 mRNA. d Western blotting analysis of MYBL2 protein expression in parental cells and tamoxifen-resistant breast cancer cells as well as in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. e Tamoxifen cytotoxicity analysis in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. f The IC50 concentration of tamoxifen for tamoxifen-resistant cells transfected with shCtrl and shMYBL2. *p < 0.05; **p < 0.01; ***p < 0.001
MYBL2 overexpression enhances cell proliferation in tamoxifen-resistant cells
As a transcription factor, MYBL2 is responsible for the expression of a wide range of target genes, and investigating the downstream transcriptional network will be informative for understanding MYBL2 function in tamoxifen response. We obtained the target genes of MYBL2 identified by ChIP-seq from ChIP-Atlas database and ranked these genes based on their binding score with MYBL2 (Oki et al. 2018). As analyzed by Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005), MYBL2-regulated genes were significantly enriched in cell cycle, consistent with the regulatory role of MYBL2 in cell cycle progression (Fig. 3a). Of these cell cycle targets, polo-like kinase 1 (PLK1) and protein regulator of cytokinesis 1 (PRC1) showed high binding affinity to MYBL2 (Fig. 3b), and a strong MYBL2 peak was detected at the promoter region of both PLK1 and PRC1 (Fig. 3c). This was further validated in tamoxifen-resistant cells by ChIP-qPCR analysis. As shown in Fig. 3d, when DNA immunoprecipitated from MCF7/TamR and T47D/TamR cells by MYBL2 antibody was used as template, strong PCR amplification signal was only observed for the primers designed for the promoter region of PLK1 and PRC1, but not for other regions, which further confirmed a direct role of MYBL2 in transcriptional control of PLK1 and PRC1 in breast cancer cells, and indicated that tamoxifen resistance-related MYBL2 overexpression may enhance PLK1 and PRC1 production. Moreover, similar to MYBL2, the overexpression of PLK1 and PRC1 was also associated with poor prognosis of breast cancer and increased the risk of cancer recurrence (PLK1: HR, 1.64; 95% CI, 1.08–2.49; p = 0.019; PRC1: HR, 2.37; 95% CI, 1.54–3.66; p < 0.001) (Fig. 3e). And knockdown of MYBL2 was found to decrease the expression of PLK1 and PRC1 in both MCF7/TamR and T47D/TamR cells (Fig. 3f) and thus slow the growth of tumor cells (Fig. 3g). All these results suggested that MYBL2 may promote the rapid proliferation of tamoxifen-resistant breast cancer cells by enhancing the expression of cell cycle regulators.
MYBL2 expression and cell proliferation in tamoxifen-resistant breast cancer cells. a GSEA analysis of MYBL2 target genes identified a role of MYBL2 in regulating the expression of cell cycle regulators. b MYBL2 target genes involved in cell cycle are ranked according to the binding affinity to MYBL2. PLK1 and PRC1 show high MYBL2 binding affinity and thus are direct targets of MYBL2. c The binding of MYBL2 to the promoter region of PLK1 and PRC1 are visualized by IGV. d MYBL2 binding to the promoter region of PLK1 and PRC1 is validated by ChIP-qPCR in tamoxifen-resistant breast cancer cells. e Kaplan-Meier analysis of the association of PLK1 and PRC1 expression with disease-free survival in breast cancer patients. f Western blotting analysis of PLK1 and PRC1 tamoxifen-resistant cells transfected with shCtrl and shMYBL2. g Cell proliferation analysis of tamoxifen-resistant cells transfected with shCtrl and shMYBL2. *p < 0.05; **p < 0.01; ***p < 0.001
MYBL2 overexpression inhibits apoptosis in tamoxifen-resistant cells
In addition to modulating cell cycle progression, MYBL2 is also critical for maintaining cell viability. According to MYBL2 ChIP-seq data, MYBL2 was largely enriched at the promoter region of BIRC5 gene, an apoptosis inhibitor also known as survivin (Fig. 4a). ChIP-qPCR analysis further verified the binding of MYBL2 to the promoter region of BIRC5 in MCF7/TamR and T47D/TamR cells (Fig. 4b), which may subsequently activate BIRC5 production. Given that BIRC5 is an anti-apoptotic factor and breast cancer patients with high BIRC5 levels showed poor disease-free survival (HR, 2.06; 95% CI, 1.33–3.17; p < 0.001) (Fig. 4c), it is thus possible that MYBL2 facilitates breast cancer cell survival under stress condition by enhancing BIRC5 expression and finally leads to disease recurrence after treatment. In contrast, decreasing BIRC5 expression via MYBL2 silencing showed potential to restore drug sensitivity in tamoxifen-resistant cells (Fig. 4d). As indicated by the accumulation of activated Caspase 3 fragment, MYBL2 silencing is associated with apoptosis activation in MCF7/TamR and T47D/TamR cells (Fig. 4d) and more cancer cells were undergoing apoptosis on tamoxifen treatment (p < 0.001) (Fig. 4e, f). We thus concluded that MYBL2 may inhibit cell death and confer resistance to tamoxifen treatment in breast cancer cells through inducing BIRC5 overexpression.
MYBL2 expression and cell survival in tamoxifen-resistant breast cancer cells. a IGV visualization of MYBL2 intensity at the promoter region of BIRC5. b ChIP-qPCR validation of the binding of MYBL2 to the promoter region of BIRC5 but not to the other region. c The association of BIRC5 expression with disease-free survival in breast cancer patients. d Western blotting analysis of the expression of BIRC5 and other apoptosis markers in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. e Analysis of tamoxifen-induced apoptosis in tamoxifen-resistant cells. f The rate of tamoxifen-induced apoptosis in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. *p < 0.05; **p < 0.01; ***p < 0.001
MYBL2 overexpression promote the motility of tamoxifen-resistant cells
From MYBL2 targets, we also identified genes involved in cell migration, such as hyaluronan-mediated motility receptor (HMMR) and a MYBL2 peak was clearly detected at the promoter region of HMMR (Fig. 5a). The transcriptional interaction between MYBL2 and HMMR, as revealed by ChIP-qPCR, was also preserved in MCF7/TamR and T47D/TamR cells (Fig. 5b), indicating that MYBL2 may increase the motility of tamoxifen-resistant breast cancer cells via activating HMMR and lead to metastatic cancer recurrence. This result was further supported by the association of HMMR with breast cancer recurrence (HR, 2.00; 95% CI, 1.28–3.12; p = 0.002) (Fig. 5c). Moreover, we also found that MYBL2 silencing was able to decrease HMMR expression in MCF7/TamR and T47D/TamR cells (Fig. 5d) As a result, the migratory ability (Fig. 5e, f) and invasive ability (Fig. 5g h) of tamoxifen-resistant cells was largely suppressed. Taken together, these results suggested that MYBL2 overexpression in tamoxifen-resistant cells may promote cell migration, while MYBL2 was capable of suppressing cell motility and decrease the risk of distant metastasis.
MYBL2 expression and cell motility in tamoxifen-resistant breast cancer cells. a The enrichment of MYBL2 at the promoter region of HMMR as visualized by IGV. b ChIP-qPCR analysis of MYBL2 binding at different regions of HMMR gene. c Disease-free survival analysis in breast cancer patients with high and low HMMR expression. d Western blotting analysis of HMMR expression in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. e The influence of MYBL2 on the migratory ability of tamoxifen-resistant breast cancer cells was analyzed by wound healing assay. f Wound closure rate in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. g Transwell invasion analysis of the influence of MYBL2 on the invasive ability of tamoxifen-resistant breast cancer cells. h The number of invasive cells in tamoxifen-resistant cells transfected with shCtrl and shMYBL2. *p < 0.05; **p < 0.01; ***p < 0.001
Discussion
Despite the recent advances in hormonal therapy and immunotherapy for breast cancer, blocking the effects of hormones (estrogen) on cancer cells with tamoxifen remains as the mainstay of adjuvant treatment for breast cancer due to its effectiveness in reducing cancer recurrence and improving overall survival. However, a large part of the patients show intrinsic resistance against tamoxifen or develop acquired resistance during the treatment, which largely limits therapeutic efficacy and remains as a major challenge in breast cancer treatment (Jordan 2003; Ring and Dowsett 2004). To overcome therapeutic resistance and develop more effective and individualized therapeutic strategies, it is thus critical to identify the key alterations during cancer progression. It has been shown that aberrant expression of transcription factors may play a critical role in breast cancer development and drug resistance via regulating downstream transcriptional network (Kohno et al. 2005). In the current study, we reported that transcription factor MYBL2 is overexpressed in tamoxifen-resistant breast cancer cells and increases the risk of cancer recurrence. We also integrated ChIP-seq data to determine the target genes of MYBL2, and found that MYBL2 overexpression is directly linked to the upregulation of PLK1, PRC1, BIRC5 and HMMR, which are essential for activating cell proliferation, survival and migration and eventually exacerbate breast cancer malignancy. Moreover, we also provided evidence that knockdown of MYBL2 expression decreases the expression of downstream effectors and shows potential to resensitize cancer cell to tamoxifen treatment and inhibit disease progression, which may serve as a novel target for effective risk stratification and development of individualized medicine.
In line with our findings, several recent studies have provided evidence supporting the role of MYBL2 in driving the initiation and evolution of multiple human cancers. It has been shown that MYBL2 expression is significantly enhanced in esophageal squamous-cell carcinoma tumors compared to nontumor tissue and is responsible for promoting G2/M-transition as well as DNA synthesis and repair (Qin et al. 2019). MYBL2 overexpression has also been reported to facilitate the epithelial-to-mesenchymal transition in glioma cells (Zhang et al. 2017) and accelerate tumor growth in gallbladder cancer (Liang et al. 2017). Recently, several studies have suggested that MYBL2 functions are not limited to transcriptional control of cell growth and apoptosis. It has been reported that MYBL2 encodes an oncogenic circular RNA (circ-MYBL2), which serves as a sponge for miR-361-3p to promote cervical cancer cells proliferation and invasion (Wang et al. 2019). And MYBL2 was also found to regulate cell proliferation by directly interacting with serine–threonine kinase receptor-associated protein (STRAP) and inhibiting the following signaling cascades mediated by TGFβ (Seong et al. 2011). Notably, MYBL2 has also been identified with a role in the biosynthesis of brassinosteroids, which are a unique class of plant polyhydroxysteroids regulating cell proliferation and are structurally similar to the cholesterol-derived animal steroid hormones (Ye et al. 2012). Since tamoxifen exerts antitumor activity by targeting cholesterol biosynthesis, such as blocking cholesterol esterification and inhibiting cholesterol epoxide hydrolase (de Medina et al. 2004, 2009, 2010, 2011; Payre et al. 2008), it is thus very likely that cholesterol metabolism is involved in the oncogenic function of MYBL2 in breast cancer, particularly the breast cancer resistant to tamoxifen treatment. More importantly, it has been increasingly recognized that MYBL2 does not function alone but is part of a larger transcriptional complex network comprising multiple functionally distinct cofactors, such as Cyclin A/E-CDK2, p300, MuvB and FOXM1, which team up to control the level of expression of downstream effectors in multiple levels (Sadasivam and DeCaprio 2013; Sadasivam et al. 2012; Sala et al. 1997; Schubert et al. 2004). Given these, the function network of MYBL2 is far more complex than expected. And unraveling the complex regulatory network that initiates breast cancer and interferes with cancer treatment will not only inform the biology of disease, but also provide novel insight into more rationally intervene in disease therapy.
In conclusion, we study identified the overexpression of transcription factor MYBL2 associated with tamoxifen resistance in breast cancer, while knockdown of MYBL2 restored tamoxifen sensitivity and suppressed disease progression by inhibiting cell proliferation, survival and migration.
Data availability
All data that supports the findings of this study are included in this published article.
References
Cervellera M et al (2000) Direct transactivation of the anti-apoptotic gene apolipoprotein J (clusterin) by B-MYB. J Biol Chem 275:21055–21060. doi:https://doi.org/10.1074/jbc.M002055200
de Medina P et al (2004) Tamoxifen is a potent inhibitor of cholesterol esterification and prevents the formation of foam cells. J Pharmacol Exp Ther 308:1165–1173. doi:https://doi.org/10.1124/jpet.103.060426
de Medina P, Silvente-Poirot S, Poirot M (2009) Tamoxifen and AEBS ligands induced apoptosis and autophagy in breast cancer cells through the stimulation of sterol accumulation. Autophagy 5:1066–1067. https://doi.org/10.4161/auto.5.7.9820
de Medina P, Paillasse MR, Segala G, Poirot M, Silvente-Poirot S (2010) Identification and pharmacological characterization of cholesterol-5,6-epoxide hydrolase as a target for tamoxifen and AEBS ligands. Proc Natl Acad Sci U S A 107:13520–13525. doi:https://doi.org/10.1073/pnas.1002922107
de Medina P et al (2011) Importance of cholesterol and oxysterols metabolism in the pharmacology of tamoxifen and other AEBS ligands. Chem Phys Lipids 164:432–437. doi:https://doi.org/10.1016/j.chemphyslip.2011.05.005
Grassilli E, Salomoni P, Perrotti D, Franceschi C, Calabretta B (1999) Resistance to apoptosis in CTLL-2 cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2 induction. Cancer Res 59:2451–2456
Inoue K, Fry EA (2016) Novel molecular markers for breast cancer. Biomark Cancer 8:25–42. https://doi.org/10.4137/BIC.S38394
Ivshina AV et al (2006) Genetic reclassification of histologic grade delineates new clinical subtypes of. breast cancer Cancer Res 66:10292–10301. doi:https://doi.org/10.1158/0008-5472.CAN-05-4414
Jordan VC (2003) Tamoxifen: a most unlikely pioneering medicine. Nat Rev Drug Discov 2:205–213. doi:https://doi.org/10.1038/nrd1031
Kohno K et al (2005) Transcription factors and drug resistance. Eur J Cancer 41:2577–2586. doi:https://doi.org/10.1016/j.ejca.2005.08.007
Levenson VV, Davidovich IA, Roninson IB (2000) Pleiotropic resistance to DNA-interactive drugs is associated with increased expression of genes involved in DNA replication, repair, and stress response. Cancer Res 60:5027–5030
Li J, Lu M, Jin J, Lu X, Xu T, Jin S (2018) miR-449a Suppresses Tamoxifen Resistance in Human Breast Cancer Cells by Targeting ADAM22. Cell Physiol Biochem 50:136–149. doi:https://doi.org/10.1159/000493964
Liang HB et al (2017) MYBL2 is a Potential Prognostic Marker that Promotes Cell Proliferation in Gallbladder Cancer. Cell Physiol Biochem 41:2117–2131. doi:https://doi.org/10.1159/000475454
Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG (2017) MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 8:e2895. doi:https://doi.org/10.1038/cddis.2017.244
Oki S et al (2018) ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data EMBO Rep 19 doi:https://doi.org/10.15252/embr.201846255
Osterloh L et al (2007) The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26:144–157. doi:https://doi.org/10.1038/sj.emboj.7601478
Payre B et al (2008) Microsomal antiestrogen-binding site ligands induce growth control and differentiation of human breast cancer cells through the modulation of cholesterol metabolism. Mol Cancer Ther 7:3707–3718. doi:https://doi.org/10.1158/1535-7163.MCT-08-0507
Peters D, Freund J, Ochs RL (2005) Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells. Mol Cancer Ther 4:1605–1616. doi:https://doi.org/10.1158/1535-7163.MCT-04-0311
Qin H, Li Y, Zhang H, Wang F, He H, Bai X, Li S (2019) Prognostic implications and oncogenic roles of MYBL2 protein expression in esophageal squamous-cell carcinoma. Onco Targets Ther 12:1917–1927. doi:https://doi.org/10.2147/OTT.S190145
Ring A, Dowsett M (2004) Mechanisms of tamoxifen resistance. Endocr Relat Cancer 11:643–658. doi:https://doi.org/10.1677/erc.1.00776
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer Nat Biotechnol 29:24–26. doi:https://doi.org/10.1038/nbt.1754
Sadasivam S, Duan S, DeCaprio JA (2012) The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 26:474–489. doi:https://doi.org/10.1101/gad.181933.111
Sadasivam S, DeCaprio JA (2013) The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13:585–595. doi:https://doi.org/10.1038/nrc3556
Sala A et al (1997) Activation of human B-MYB by cyclins. Proc Natl Acad Sci U S A 94:532–536. doi:https://doi.org/10.1073/pnas.94.2.532
Santilli G, Schwab R, Watson R, Ebert C, Aronow BJ, Sala A (2005) Temperature-dependent modification and activation of B-MYB: implications for cell survival. J Biol Chem 280:15628–15634. doi:https://doi.org/10.1074/jbc.M411747200
Schubert S, Horstmann S, Bartusel T, Klempnauer KH (2004) The cooperation of B-Myb with the coactivator p300 is orchestrated by cyclins A and D1. Oncogene 23:1392–1404. doi:https://doi.org/10.1038/sj.onc.1207255
Seong HA, Manoharan R, Ha H (2011) B-MYB positively regulates serine-threonine kinase receptor-associated protein (STRAP) activity through direct interaction. J Biol Chem 286:7439–7456. doi:https://doi.org/10.1074/jbc.M110.184382
Subramanian A et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi:https://doi.org/10.1073/pnas.0506580102
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102. doi:https://doi.org/10.1093/nar/gkx247
Tao D, Pan Y, Lu H, Zheng S, Lin H, Fang H, Cao F (2014) B-myb is a gene implicated in cell cycle and proliferation of breast cancer. Int J Clin Exp Pathol 7:5819–5827
Tao D, Pan Y, Jiang G, Lu H, Zheng S, Lin H, Cao F (2015) B-Myb regulates snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cell. Med Oncol 32:412. doi:https://doi.org/10.1007/s12032-014-0412-y
Tarasov KV et al (2008a) B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One 3:e2478. doi:https://doi.org/10.1371/journal.pone.0002478
Tarasov KV et al (2008b) Linkage of pluripotent stem cell-associated transcripts to regulatory gene networks. Cells Tissues Organs 188:31–45. doi:https://doi.org/10.1159/000118787
Wang J, Li H, Liang Z (2019) circ-MYBL2 serves as a sponge for miR-361-3p promoting cervical cancer cells proliferation and invasion. Onco Targets Ther 12:9957–9964. https://doi.org/10.2147/OTT.S218976
Ye H, Li L, Guo H, Yin Y (2012) MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A 109:20142–20147. doi:https://doi.org/10.1073/pnas.1205232109
Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R (2015) MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals Autophagy 11:1081–1098 doi:https://doi.org/10.1080/15548627.2015.1040970
Zhan M et al (2012) The B-MYB transcriptional network guides cell cycle progression and fate decisions to sustain self-renewal and the identity of pluripotent stem cells. PLoS One 7:e42350. doi:https://doi.org/10.1371/journal.pone.0042350
Zhang X, Lv QL, Huang YT, Zhang LH, Zhou HH (2017) Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2 promotes progression of human glioma. J Exp Clin Cancer Res 36:105. doi:https://doi.org/10.1186/s13046-017-0573-6
Zhu W, Giangrande PH, Nevins JR (2004) E2Fs link the control of G1/S and G2/M transcription. EMBO J 23:4615–4626. doi:https://doi.org/10.1038/sj.emboj.7600459
Zhu Y et al (2018) Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat Commun 9:1595. doi:https://doi.org/10.1038/s41467-018-03951-0
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors have given consent for submission and publication of the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, X., Zhang, X., Wu, CC. et al. The role of MYB proto-oncogene like 2 in tamoxifen resistance in breast cancer. J Mol Histol 52, 21–30 (2021). https://doi.org/10.1007/s10735-020-09920-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-020-09920-6