Abstract
The EphA5 receptor is well established as an axon guidance molecule during neural system development and plays an important role in dendritic spine formation and synaptogenesis. Our previous study has showed that EphA5 is decreased in the developing brain of congenital hypothyroidism (CH) and the EphA5 promoter methylation modification participates in its decrease. c-Fos, a well-kown transcription factor, has been considered in association with brain development. Bioinformatics analysis showed that the EphA5 promoter region contained five putative c-fos binding sites. The chromatin immunoprecipitation (ChIP) assays were used to assess the direct binding of c-fos to the EphA5 promoter. Furthermore, dual-luciferase assays showed that these three c-fos protein binding sites were positive regulatory elements for EphA5 expression in PC12 cells. Moreover, We verified c-fos positively regulation for EphA5 expression in CH model. Q-PCR and Western blot showed that c-fos overexpression could upregulate EphA5 expression in hippocampal neurons of rats with CH. Our results suggest that c-fos positively regulates EphA5 expression in CH rat model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Congenital hypothyroidism (CH) is the most frequent congenital endocrine disease with a high morbidity of 1:3000 to 1:4000 newborns (Kang et al. 2017; Schoenmakers et al. 2015). It is usually caused by tyrosine hydroxylase (TH) deficiency during brain development and is often accompanied by neurodevelopmental disabilities (Ahmed 2015). To date, clinical intervention with early levothyroxine replacement therapy can improve neurological and health outcomes to some extent. However, a portion of CH children still have a poor prognosis with cognitive or motor sequelae (Lavado-Autric et al. 2003; Rovet 2002). Therefore, exploring its pathogenesis may contribute to identifying additional therapeutic strategies for CH.
The EphA5 receptor plays an important role in the nervous system development, especially in the initiation of the early phase of synaptogenesis (Akaneya et al. 2010; Cooper et al. 2009; Martínez et al. 2005, St John et al. 2000). Abnormal EphA5 expression is responsible for neurodevelopmental abnormalities in CH, especially synaptogenesis disorders (Wu et al. 2013, 2015). We also found that hypermethylation of the EphA5 promoter causes EphA5 downregulation, and demethylation with 5-aza-2′-deoxycytidine (5-aza-dC) treatment can upregulate EphA5 expression and ameliorate neuronal damage caused by hypothyroidism (Wu et al. 2015). In this study, we explored the regulatory mechanism of transcription factors for EphA5 downregulation in CH.
In our preliminary study, we analyzed the EphA5 promoter region with AliBaba2.1 software to identify new transcription factors that might regulate EphA5 expression. Five putative c-fos binding sites in the EphA5 promoter region with well-recognized DNA motifs were predicted, two of which were located in the methylated promoter region.
c-Fos, a well-known immediate early response proto-oncogene (Curran and Morgan 1995), has two main functions: a lipid synthesis activator (Velazquez et al. 2015a, b; Caputto et al. 2014) and a classical transcription factor that contributes to the formation of the transcription factor activator protein 1 (AP-1) (Velazquez et al. 2015a, b). c-Fos can heterodimerize with members of the Jun family (c-Jun, JunB, or JunD) to form the transcription factor AP-1, which mediates diverse cellular functions, ranging from proliferation, differentiation, growth and apoptosis to malignant cell transformation (Hess et al. 2004; Healy et al. 2013; Wang et al. 2016; Ye et al. 2014). In the central nervous system, c-fos is involved in spatial learning and memory abilities (Gandolfi et al. 2017; Fleischmann et al. 2003; Liu et al. 2013; Opazo et al. 2008; Vanelzakker et al. 2011). c-Fos is also a good biological marker for detecting the pathogenesis of central nervous system disorders (Ahn et al. 2015), and aberrant c-fos expression is highly associated with anxiety, depression and fear (Kung et al. 2010; Meloni et al. 2016; Nestler 2015). In recent years, Liu et al. has reported that iodine deficiency or hypothyroidism during the critical period of brain development can downregulate c-fos and c-jun expression in the rat hippocampus (Liu et al. 2013). However, whether c-fos participates in EphA5 decrease in CH model has not been reported. In this study, We used in vivo and in vitro approaches to demonstrate the role of c-fos in regulating EphA5 expression. The hippocampus and hippocampal neurons were chosen because of its importance for learning and memory processes (Kesner et al. 2000) and its susceptibility to perinatal TH deficiency (Wu et al. 2013, 2015). We chose P7 as the time point because the EphA5 expression decreases most prominently during rat brain development in CH (Wu et al. 2013), and the P7 rat brain is considered developmentally equivalent to the human brain at birth (Anderson et al. 2003). We hope that our research can provide effective theoretical support for the development of drugs to restore neurocognitive impairment in children with CH.
Materials and methods
Induction of the CH model
The CH model was invoked according to Wu et al. (2013). Briefly, pregnant Sprague-Dawley (SD) rats were fed 0.02% 2-mercapto-1-methylimidazole (MMI) (Sigma-Aldrich, #M8506) in drinking water beginning at gestational day 9 (G9) until the pups were sacrificed on postnatal day 7 (P7). The hippocampus of P7 pups were dissected out for quantitative polymerase chain reaction (qPCR), Western blot and immunofluorescence assays. Hippocampal tissues from untreated, normal control pups were used in parallel experiments.
Quantitative real-time PCR (qPCR)
Total RNA of the hippocampus or hippocampal neurons was isolated by using Trizol reagent (Life Technologies Ltd., Paisley, UK). cDNA was synthesized using PrimeScript RT reagent Kit (Takara) and qPCR was performed on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, USA) by using SYBR Green Real-Time PCR Master Mix (Toyobo, Japan). The primers used in this study were follows for c-fos: forward primer 5′-CAAACCGACCTACTGTCCC-3′, reverse primer 5′-ACCAACAACCTTGTCGTCATAT-3′; for EphA5: forward primer 5′-AAGCCAGATTCCCATCATTG-3′, reverse primer 5′-ATCCTGCTTTGCTTT GCTGT-3′; for GAPDH: forward primer 5′-GGCATCCTGGGCTACACT-3′, reverse primer 5′-CCACCACCCTGTTGCTGT-3′. GAPDH was used for normalization. All quantitative PCR reactions were performed in triplicate.
Western blot
Protein extracts were prepared from the hippocampus or hippocampal neurons using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, China). Equal amounts of protein were subjected to SDS-PAGE and electrotransferred to PVDF membranes (Bio-Rad, USA). The membranes were blocked with 5% non-fat milk in TBS for 2 h then incubated with a rabbit anti-EphA5 polyclonal antibody (Santa Cruz, USA; 1:100), a mouse anti-c-fos polyclonal antibody (Santa Cruz, USA; 1:100), or a mouse anti-β-actin monoclonal antibody (CST, USA; 1:2,000) at 4 °C overnight. The bound antibodies were then detected by HRP-labeled goat anti-rabbit immunoglobulin G (IgG) (Beyotime, China, 1:1000) or HRP-labeled goat anti-mouse IgG (Beyotime, China, 1:1000) followed by enhanced chemiluminescence (Pierce Chemical Company, USA).
Immunofluorescence staining
The P7 brains were harvested, post-fixed, and sectioned. Sections were incubated with the mouse anti-c-fos polyclonal antibody (Santa Cruz, USA; 1:50) at 4 °C for 24 h. The sections were further reacted with a FITC-labeled secondary antibody goat anti-mouse IgG (Gibco, 1:400) at 4 °C overnight followed by observation under a fluorescence microscope (Leica, Germany).
Primary culture of hippocampal neurons
The meninges of hypothyroid embryonic day (E) 17 Sprague-Dawley (SD) rat brains were stripped off and the hippocampus was dissected under an anatomical microscope. The hippocampus was cut into small pieces and digested with 0.125% trypsin for 15 min at 37 °C. Then, the digestion was neutralized with DMEM (HyClone, USA) containing 10% FBS (Gibco, USA), 1% GlutaMAX-I and 1% penicillin–streptomycin solution. After centrifugation at 1000 rpm for 5 min, the cells were resuspended and seeded onto pre-coated plastic plates at a density of 1–2 × 105 cells/ml. After culture for 4 h, cells were further subjected to B27 serum-free medium. About half of the medium was changed once every 3 days. On the 7th day of the culture, the hypothyroidism hippocampal neurons were divided into two groups: (1) Hypo-group, the hypothyroidism hippocampal neurons were infected with control lentivirus. (2) c-fos group, the hypothyroidism hippocampal neurons were infected with c-fos over-expressing lentivirus. After treatment for 72 h, hippocampal neurons were harvested for qPCR, Western blot. Normal hippocampal neurons infected with control lentivirus were regarded Control group.
Chromatin immunoprecipitation (CHIP) assays
PC12 cells were purchased from the Chinese Academy of Sciences at Shanghai Institutes for Biological Sciences Cell Resource Center and cultured in DMEM containing 10% FBS, 1% GlutaMAX-I and 1% penicillin–streptomycin solution. ChIP assays were performed using a SimpleChIP® Enzymatic ChIP kit (CST, USA) according to the manufacturer’s instruction. PC12 cells infected with the c-fos over-expressing lentivirus for 72 h (Lv-c-fos group) and PC12 cells infected with a control lentivirus for 72 h (Lv-Control group) were used in the ChIP assays with an anti-c-fos polyclonal antibody (Santa Cruz, USA; 1:50) or an anti-mouse IgG antibody (negative control). The chromatin fraction isolated by the ChIP kit was analyzed by qPCR with specific oligonucleotide primers for the promoter region of EphA5. The primers used were listed in Table 1.
Bioinformatics analysis
The promoter region spanning from − 2000 to + 1000 bp upstream the transcriptional start site of the rat EphA5 gene was analyzed by UCSC Genome Browser (http://genome.ucsc.edu/) and the putative binding sites for transcription factors were predicted by AliBaba2.1 software (http://gene-regulation.com/pub/programs/alibaba2). Meanwhile, the EphA5 promoter methylation was analyzed by MethPrimer Tools and Databases (http://www.urogene.org/methprimer/). The criteria of methylation prediction: island size > 100, GC% > 50.0, Obs/Exp > 0.6.
Plasmid constrction, transfection, and dual-luciferase assay
The full-length promoter region of EphA5 spanning from − 2000 to + 1000 bp upstream the transcriptional start site was cloned into a basic pGL3 luciferase reporter vector (pGL3-EphA5-WT). Meanwhile, The nucleotides corresponding to site2, site3 and site4 (listed in Table 1) in pGL3-EphA5-WT were deleted singly to generate three mutated promoter constructs. The mutated promoter constructs were termed pGL3-EphA5--828, pGL3-EphA5--100, and pGL3-EphA5-483 respectively. The recombinant plasmid and the c-fos over-expressing lentivirus were co-transfected into PC12 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. The cell lysates were analyzed for luciferase activity using a dual luciferase assay kit. The relative luciferase activity was evaluated by the ratio of firefly luciferase activity to renilla luciferase activity and luciferase vector pGL3-basic values were used as negative control.
Statistical analysis
SPSS 16.0 statistical software was used to analyze the experimental data, and the results were presented as the mean ± SD. An unpaired Student’s t test was used for statistical comparisons between two groups. Multiple groups (≥ 3) were calculated and analyzed by one-way analysis of variance (ANOVA). A p value less than 0.05 was considered statistically significant (*p < 0.05).
Results
Analysis of potential c-fos protein binding sites within the basal EphA5 promoter
Both EphA5 mRNA and protein expression were decreased in the developing brains of CH rats, and the greatest decrease occurred at P7 in the hippocampus (Wu et al. 2013). Methylation of the EphA5 promoter was responsible for its downregulation (Wu et al. 2015). To further understand the regulation of transcription factors for EphA5 downregulation, we analyzed the EphA5 promoter region (− 2000 to + 1000 bp upstream the transcriptional start site) by using AliBaba2.1 software to predict putative transcription factors. The result showed that 706 segments in this promoter sequence were identified as putative binding sites with Min mat. Conservation > 70%, among which five were the putative binding sites for c-fos protein (Fig. 1). These putative c-fos protein binding sites were located at − 958 to − 947 bp (site1), − 828 to − 817 bp (site2), − 100 to − 91 bp (site3), + 483 to + 493 bp (site4) and + 582 to + 591 bp (site5) upstream the transcriptional start site. The corresponding sequences were agaaagagtc (site1), ccagactcag (site2), tttaagatga (site3), ctggacgtgc(site4), tggatgggtg (site5), respectively. c-Fos, a well-known transcription factor, plays an important role during the nervous system development and takes part in regulating neuronal differentiation, survival and neurogenesis (Velazquez et al. 2015a, b; Zhang 2002). c-Fos was also reported to be aberrant by iodine deficiency or hypothyroidism (Liu et al. 2013; Dong et al. 2005). Take all these factors, we chosed c-fos as a transcription factor to study the transcriptional regulation of EphA5.
A diagram of the prediction of c-fos protein binding to the rat EphA5 promoter. Blue-shaded regions represent the EphA5 promoter hypermethylation region (CpG islands). The red lines represents the input sequence (− 2000 to + 1000 bp upstream the transcriptional start), and the blue lines represent c-fos binding sites on the EphA5 gene. (Color figure online)
c-fos expression was downregulated in the hippocampus of P7 CH rats
In this part, we explored two questions: whether c-fos mediates EphA5 dysregulation in this process and how c-fos regulates EphA5 expression. First, we detected c-fos expression at both the mRNA and protein levels. The results showed decreased mRNA and protein expression in the hippocampus of P7 CH rats compared to that in the normal controls (Fig. 2a–c). Immunofluorescence staining for c-fos protein showed decreased levels of c-fos in the hippocampus of P7 CH rats (Fig. 2d). These results suggest that c-fos is downregulated in the hippocampus of P7 CH rats.
c-fos expression is downregulated in the hippocampus of the P7 CH rats. a qPCR detection of c-fos mRNA expression in different groups. *p < 0.05 versus control group at the same time point. b Western blot bands of c-fos protein expression in different groups. c Grayscale analysis of Western blot bands by image J. *p < 0.05 versus control group at the same time point. d Immunofluorescence staining of c-fos expression in different groups. (a) c-fos expression in hippocampus of the P7 hypothyroid rats. (b) Hoechst 33,258 staining of nucleuses in hippocampus of the P7 hypothyroid rats. (c) the mergence of a and b. (d) Magnification of local region in c. (e) c-fos expression in hippocampus of the P7 normal rats. (f) Hoechst 33,258 staining of nucleuses in hippocampus of the P7 normal rats. (g) the mergence of e and f. (h) Magnification of local region in g. Original magnification for a, b, c, e, f and g, ×50; d and h, ×200
C-fos positively regulated EphA5 expression in PC12 cells
To test the notion that c-fos was a transcription factor for regulating EphA5 expression in CH, we over-expressed c-fos protein in PC12 cells with c-fos over-expressing lentivirus and detected EphA5 expression at both the mRNA and protein levels by qPCR and Western blot, respectively. PC12 cells showed endogenous c-fos expression, and the ectopically expressed c-fos increased EphA5 expression at both the mRNA and protein levels (Fig. 3). To further identify the cis-element that could recruit transcription factors, we performed ChIP assays using a c-fos antibody with nuclear fractions from c-fos over-expressing PC12 cells. Then qPCR was used to analyze the chromatin fraction isolated by immunoprecipitate, showing significant enrichments at site2, site3 and site4 in c-fos-immunoprecipitate compared with IgG control (Fig. 4a).
a Localisation of c-fos in transfected PC12 cells using fluorescence microscopy (×100). Lentivirus vectors encoding eGFP-alone (a) and c-fos-eGFP (b) were transfected into PC12 cells grown on 24 well plates. Left: Fluorescence images; Right: bright field images. b qPCR detection of c-fos mRNA expression in different groups. *p < 0.05 versus Lv-control group at the same time point. c qPCR detection of EphA5 mRNA expression in different groups. *p < 0.05 versus Lv-control group at the same time point. d Western blot detection of c-fos and EphA5 protein expression in different groups. Left: representative Western blot bands for c-fos and EphA5; Middle: Grayscale analysis of bands for c-fos by image J; Right: Grayscale analysis of bands for EphA5 by image J. *p < 0.05 versus Lv-control group at the same time point
The determination of c-fos binding sites for the EphA5 promter in PC12 cells. a ChIP assays with anti-c-fos antibody or control mouse IgG indicated that site2, site3 and site4 of the EphA5 promter were c-fos binding sites in PC12 cells, but not site1 or site5. The products of qPCR after ChIP assays were verificated by agarose gel electrophoresis. b the EphA5 promter (EphA5-WT) and deleted mutation of target sites (EphA5--828, EphA5--100, EphA5-483) were cloned into the pGL3 luciferase reporter vectors, respectively. c Relative luciferase activity in PC12 cells co-transfected with pGL3-EphA5-WT/--828/--100/-483 plasmids and c-fos over-expressing lentivirus. Luciferase expression levels were normalized to the luciferase activity of internal Renilla control. One-way analysis of variance (ANOVA) was used for statistical analysis. *p < 0.05 versus other group at the same time point
To test whether site2, site3 and site4 of the EphA5 promter are functional targets for c-fos protein, we constructed four recombinant plasmids (pGL3-EphA5-WT, pGL3-EphA5--828, pGL3-EphA5--100, and pGL3-EphA5-483) to perform luciferase assays in PC12 cells (Fig. 4b). The results showed that the overexpression of pGL3-EphA5-WT plasmid had a high transcriptional activity, while other three mutant constructs decreased the transcriptional activity to varying degrees (Fig. 4c). Therefore, the binding sequences of site2, site3 and site4 are positive regulatory elements which can bind with c-fos protein to upregulate EphA5 gene expression.
Overexpression of c-fos upregulated EphA5 expression in hippocampal neurons of rats with hypothyroidism
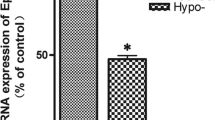
Given the c-fos positve regulation for EphA5 expression in PC12 cells, we speculated about a same effect in hippocampal neurons of rats with hypothyroidism. Subsequently, we performed an in vivo experiment to verify the role of c-fos for EphA5 expression. Hypothyroidism hippocampal neurons infected with the c-fos over-expressing lentivirus for 72 h (c-fos group), hypothyroidism hippocampal neurons infected with a control lentivirus for 72 h (Hypo group), normal hippocampal neurons infected with a control lentivirus (Control group) were used to detecte EphA5 expression at both the mRNA and protein levels by qPCR and Western blot, respectively. EphA5 expression in Hypo group decreased sharply at both the mRNA and protein levels comparing with Control group (Fig. 5). After overexpression of endogenous c-fos in c-fos group for 72 h, the EphA5 expression in c-fos group increased significantly at both the mRNA and protein levels comparing with Hypo group, but still lower than Control group (Fig. 3). These results indicate that c-fos is a positve regulator for EphA5 expression in hippocampal neurons of rats with hypothyroidism.
Overexpression of c-fos for 72 h upregulated EphA5 expression in hippocampal neurons of rats with CH. a Localisation of c-fos in transfected hippocampal neurons using fluorescence microscopy (×100). (a) lentivirus vectors encoding eGFP-alone were transfected into normal control hippocampal neurons (Control group). (b) lentivirus vectors encoding eGFP-alone were transfected into hippocampal neurons of rats with hypothyroidism (Hypo- group). (c) lentivirus vectors encoding c-fos-eGFP were transfected into hippocampal neurons of rats with hypothyroidism (c-fos group). b qPCR detection of EphA5 mRNA expression in different groups. *p < 0.05 versus other group at the same time point. c Western blot bands of EphA5 protein expression in different groups. d Grayscale analysis of Western blot bands by image J. One-way analysis of variance (ANOVA) was used for statistical analysis. *p < 0.05 versus other groups at the same time point
Discussion
We have previously shown that EphA5 is decreased in the developing rat brain of CH, and the largest decline occurs in the P7 hippocampus. However, the regulation of transcription factors for EphA5 downregulation is poorly understood.
In our prelimary study, bioinformatics analysis identified multiple candidate transcription factors that may regulate EphA5 expression. The most striking of these candidates was c-fos. c-Fos is an important transcription factor for regulating target gene expression by binding to its promoter or enhancer (Hess et al. 2004) and participates in diverse physiological and pathological processes. Some previous studies pointed out that c-fos expression in the rat hippocampus or CA1 hippocampal area is significantly decreased by iodine deficiency or hypothyroidism (Liu et al. 2013; Dong et al. 2005). In this study, we reconfirmed a significant decrease of c-fos expression in the P7 hypothyroidism hippocampus. c-Fos plays an important regulatory role in normal neuronal differentiation, neuronal excitability, survival and neurogenesis (Velazquez et al. 2015a, b; Zhang et al. 2002), and its dysregulation has been associated with abnormal development. For example, the brain size of c-fos−/− mice is significantly smaller compared to c-fos+/+ mice, and the c-fos−/− brain has decreased neurogenesis during embryonic development (Velazquez et al. 2015a, b). Numerous studies also had shown that the depletion of c-fos can cause important deficits in memory formation and the underlying neural plasticity in the hippocampus (Countryman et al. 2005; Niessen et al. 2013; Yasoshima et al. 2006). Therefore, c-fos decrease may be related to abnormal brain development in CH. Considering the importance of c-fos as a transcription factor during neurodevelopment (Velazquez et al. 2015a, b; Caubet 1989), we boldly speculates that c-fos is an effective transcription factor for EphA5 expression in CH.
To verify our speculation, we firstly investigated whether the change in c-fos expression was associated with EphA5 expression in PC12 cells. PC12 cell is a widely used neuronal cell line and has a endogenous c-fos expression. c-Fos, known as a neuronal marker (Makino et al. 2017), is required for normal cell turnover and its knockout caused a reduction in both cell proliferation and an increase in apoptosis (Shandilya et al. 2016). In this study, we observed a significantly increased EphA5 expression in PC12 cells after c-fos overexpression, indicating that c-fos may be a regulator for EphA5 expression. Subsequently, ChIP assays confirmed the interaction between c-fos and EphA5 (called c-fos/EphA5 signal axis) and specific binding sites (site2, site3 and site4) on the EphA5 promoter. Luciferase assays showed a positive regulation of c-fos for EphA5 expression in PC12 cells. Then, the c-fos/EphA5 signal axis was further detected in the hypothyroidism cell model. Expectedly, treatment of hypothyroidism hippocampal neurons with c-fos over-expressing lentivirus induced a significant increase in EphA5 exprssion, indicating the c-fos positive regulation for EphA5 expression in CH model. Hence, it is possible that c-fos actives EphA5 expression and its downregulation is related to the synaptogenesis disorder induced by EphA5 decrease during the hippocampus development in CH. But the EphA5 expression could not return to normal level after c-fos overexpression, suggesting that there are some other regulatory mechanisms for EphA5 decrease. For example, DNA methylation and histone deacetylation are two most common epigenetic alterations for EphA5 downrelation. The EphA5 promoter was barely methylated in hBMSCs while histone deacetylation could partially suppress EphA5 expression in early-passage cultures (Yamada et al. 2016). Wu et al. also suggested a DNA methylation modification in the regulation of EphA5 expression in CH. DNA methylation affects the interaction between the promoter region and transcription factors owing to the changes of chromatin structure or DNA conformation. In our prediction, we found that site2 and site3 are located near the CpG Island 1 region while site4 is located in the region of CpG island 2. Whether there is some relationship between CpG islands hypermethylation and c-fos binding capacity for regulating EphA5 expression in CH still needs to be explored.
In conclusion, we have firstly shown that c-fos binds to the EphA5 promoter and positvely regulates EphA5 expression. c-Fos can be considered a regulator of synaptogenesis through regulating EphA5 expression and its downregulation is responsible for EphA5 decrease in CH. Additional in vivo experimental studies with c-fos overexpression are necessary to further reveal the c-fos positive regulation for EphA5 expression during nervous system development in CH.
References
Ahmed RG (2015) Hypothyroidism and brain developmental players. Thyroid Res 8:2
Ahn JY, Tae HJ, Cho JH, Kim IH, Ahn JH, Park JH, Kim DW, Cho JH, Won MH, Hong S, Lee JC, Seo JY (2015) Activation of immediate-early response gene c-fos protein in the rat paralimbic cortices aftermyocardial infarction. Neural Regen Res 10(8):1251–1257
Akaneya Y, Sohya K, Kitamura A, Kimura F, Washburn C, Zhou R, Ninan I, Tsumoto T, Ziff EB (2010) Ephrin-A5 and EphA5 interaction induces synaptogenesis during early hippocampal development. PLoS ONE 5(8):e12486
Anderson GW, Schoonover CM, Jones SA (2003) Control of thyroid hormone action in the developing rat brain. Thyroid 13(11):1039–1056
Caputto BL, Cardozo Gizzi AM, Gil GA (2014) c-fos: an AP-1 transcription factor with an additional cytoplasmic, non-genomic lipid synthesis activation capacity. Biochim Biophys Acta 1841:1241–1246
Caubet JF (1989) c-fos proto-oncogene expression in the nervous system during mouse development. Mol Cell Biol 9:2269–2272
Cooper MA, Crockett DP, Nowakowski RS, Gale NW, Zhou R (2009) Distribution of EphA5 receptor protein in the developing and adult mouse nervous system. J Comp Neurol 514(4):310–328
Countryman RA, Kaban NL, Colombo PJ (2005) Hippocampal c-fos is necessary for long-term memory of a socially transmitted food preference. Neurobiol Learn Mem 84:175–183
Curran T, Morgan JI (1995) Fos: an immediate-early transcription factor in neurons. J Neurobiol 26:403–412
Dong J, Yin H, Liu W, Wang P, Jiang Y, Chen J (2005) Congenital iodine deficiency and hypothyroidism impair LTP and decrease c-fos and c-jun expression in rat hippocampus. Neurotoxicology 26(3):417–426
Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, Wagner EF, Gass P (2003) Impaired long-termmemory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-fos in the CNS. J Neurosci 23(27):9116–9122
Gandolfi D, Cerri S, Mapelli J, Polimeni M, Tritto S, Fuzzati-Armentero MT, Bigiani A, Blandini F, Mapelli L, D’Angelo E (2017) Activation of the CREB/c-fos pathway during long-term synaptic plasticity in the cerebellum granular layer. Front Cell Neurosci 11:184
Healy S, Khan P, Davie JR (2013) Immediate early response genes and cell transformation. Pharmacol Ther 137:64–77
Hess J, Angel P, Schorpp-Kistner M (2004) AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117(25):5965–5973
Kang HS, Kumar D, Liao G, Lichti-Kaiser K, Gerrish K, Liao XH, Refetoff S, Jothi R, Jetten AM (2017) GLIS3 is indispensable for TSH/TSHR-dependent thyroid hormone biosynthesis and follicularcell proliferation. J Clin Investig pii:94417. https://doi.org/10.1172/JCI94417
Kesner RP, Gilbert PE, Wallenstein GV (2000) Testing neural network models of memory with behavioral experiments. Curr Opin Neurobiol 10(2):260–265
Kung JC, Chen TC, Shyu BC, Hsiao S, Huang AC (2010) Anxiety- and depressive-like responses and c-fos activity in preproenkephalin knockout mice: oversensitivity hypothesis of enkephalin deficit-induced posttraumatic stress disorder. J Biomed Sci 17(1):29
Lavado-Autric R, Auso E, García-Velasco JV, Arufe Mdel C, Escobar del Rey F, Berbel P, Morreale de Escobar G (2003) Early maternal hypothyroxinemia alters histogenesis and cerebral cortex cytoarchitecture of the progeny. J Clin Investig 111(7):1073–1082
Liu Y, Zhang L, Li J, Shan Z, Teng W (2013) Maternal marginal iodine deficiency affects the expression of relative proteins during brain development in rat offspring. J Endocrinol 217:21–29
Makino H, Seki S, Yahara Y, Shiozawa S, Aikawa Y, Motomura H, Nogami M, Watanabe K, Sainoh T, Ito H, Tsumaki N, Kawaguchi Y, Yamazaki M, Kimura T (2017) A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci Rep 7(1):16983
Martínez A, Otal R, Sieber BA, Ibáñez C, Soriano E (2005) Disruption of ephrin-A/EphA binding alters synaptogenesis and neural connectivity in the hippocampus. Neuroscience 135(2):451–461
Meloni EG, Venkataraman A, Donahue RJ, Carlezon WA Jr (2016) Bi-directional effects of pituitary adenylate cyclase-activating polypeptide (PACAP) on fear-related behavior and c-Fos expression after fear conditioning in rats. Psychoneuroendocrinology 2(64):12–21
Nestler EJ (2015) ∆FosB: a transcriptional regulator of stress and antidepressant responses. Eur J Pharmacol 753:66–72
Niessen NA, Balthazart J, Ball GF, Charlier TD (2013) c-fos down-regulation inhibits testosterone-dependent malesexual behavior and the associated learning. Eur J Neurosci 38(9):3325–3337
Opazo MC, Gianini A, Pancetti F, Azkcona G, Alarcón L, Lizana R, Noches V, Gonzalez PA, Marassi MP, Mora S, Rosenthal D, Eugenin E, Naranjo D, Bueno SM, Kalergis AM, Riedel CA (2008) Maternal hypothyroxinemia impairs spatial learning and synaptic nature and function in the offspring. Endocrinology 149(10):5097–5106
Rovet JF (2002) Congenital hypothyroidism: an analysis of persisting deficits and associated factors. Child Neuropsychol 8(3):150–162
Schoenmakers N, Alatzoglou KS, Chatterjee VK, Dattani MT (2015) Recent advances in central congenital hypothyroidism. J Endocrinol 227(3):R51–R71
Shandilya J, Gao Y, Nayak TK, Roberts SG, Medler KF (2016) AP1 transcription factors are required to maintain the peripheral taste system. Cell Death Dis 7(10):e2433
St John JA, Tisay KT, Caras IW, Key B (2000) Expression of EphA5 during development of the olfactory nerve pathway in rat. J Comp Neurol 416(4):540–550
Vanelzakker MB, Zoladz PR, Thompson VM, Park CR, Halonen JD, Spencer RL, Diamond DM (2011) Influence of pre-training predator stress on the expression of c-fos mRNA in the hippocampus, amygdala, and striatum following long-term spatial memory retrieval. Front Behav Neurosci 5:30
Velazquez FN, Caputto BL, Boussin FD (2015a) c-Fos importance for brain development. Aging (Albany NY) 7(12):1028–1029
Velazquez FN, Prucca CG, Etienne O, D’Astolfo DS, Silvestre DC, Boussin FD, Caputto BL (2015b) Brain development is impaired in c-fos −/− mice. Oncotarget 6(19):16883–16901
Wang S, Lu J, You Q, Huang H, Chen Y, Liu K (2016) The mTOR/AP-1/VEGF signaling pathway regulates vascular endothelia cell growth. Oncotarget 7(33):53269–53276
Wu YJ, Xu MY, Wang L, Sun BL, Gu GX (2013) Analysis of EphA5 receptor in the developing rat brain: an in vivo study in congenital hypothyroidism model. Eur J Pediatr 172(8):1077–1083
Wu Y, Song H, Sun B, Xu M, Shi J (2015) DNA methylation of the EphA5 promoter is associated with rat congenital hypothyroidism. J Mol Neurosci 57(2):203–210
Yamada T, Yoshii T, Yasuda H, Okawa A, Sotome S. Dexamethasone regulates EphA5, a potential inhibitory factor with osteogenic capability of human bone marrow stromal cells. Stem Cells. 2016:1301608. https://doi.org/10.1155/2016/1301608
Yasoshima Y, Sako N, Senba E, Yamamoto T (2006) Acute suppression, but not chronic genetic deficiency, of c-fos gene expression impairs long-term memory in aversive taste learning. Proc Natl Acad Sci USA 103(18):7106–7111
Ye N, Ding Y, Wild C, Shen Q, Zhou J (2014) Small molecule inhibitors targeting activator protein 1 (AP 1). J Med Chem 57:6930–6948
Zhang J, Zhang D, McQuade JS, Behbehani M, Tsien JZ, Xu M (2002) c-fos regulates neuronal excitability and survival. Nat Genet 30:416–420
Acknowledgements
This study was supported by National Natural Science Youth Fund (81700696), China Postdoctoral Science Foundation (2016M591899), Postdoctoral Science Foundation of Jiangsu Province (1601094C), Nature Science Foundation of Nantong (MS12015002), Youth Key Talent of Medical Foundation of Jiangsu Province (QNRC2016684), 226 high-level talents training project of Nantong (2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All the authors report no disclosures relevant to the manuscript. Animal experiments were performed in strict accordance with the Institutional Animal Care guidelines.
Rights and permissions
About this article
Cite this article
Song, H., Zheng, Y., Cai, F. et al. c-Fos downregulation positively regulates EphA5 expression in a congenital hypothyroidism rat model. J Mol Hist 49, 147–155 (2018). https://doi.org/10.1007/s10735-018-9754-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-018-9754-7