Abstract
Mesenchymal stem cells (MSCs) have several features that make them an attractive option for potentiating cartilage repair. Synovium-derived (SMSCs) have been recently recognized as an excellent source. SRY-related HMG-box (Sox) family plays an important role in the proliferation and differentiation of SMSCs. However, the role of Sox4 in human SMSCs remains elusive. In the present study, we investigated the role of Sox4 in SMSCs through gain-of-function studies and found that Sox4 promoted cell proliferation and chondrogenesis. Furthermore, Sox4 could directly bind to the promoter of long noncoding RNA DANCR and increased its expression. Finally, knockdown of DANCR could reverse the stimulative effect of Sox4 on the proliferation and chondrogenesis of SMSCs. Taken together, our data highlights the pivotal role of Sox4 in the proliferation and differentiation of SMSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Articular cartilage absorbs an enormous mechanical load and allow almost frictionless motion between the articulating surfaces of diarthrodial joins (Marmotti et al. 2015; Scotece and Mobasheri 2015). However, cartilage tissue has very limited self-healing capacity for lack of vascular. Recently, various approaches applied in clinical practice, such as surgical interventions, have led to some positive results (Getgood et al. 2009; Hunziker 2002). But it is still unsatisfactory for its surgical injury to normal articular cartilage. Mesenchymal stem cells (MSCs), which are multipotent stromal cells that can differentiate into a variety of cell types, have been reported to be an attractive option for potentiating cartilage repair (Ham et al. 2015; Petrella et al. 2015; Trounson and McDonald 2015). MSCs can be isolated from various sources, such as umbilical cord, blood, bone marrow, adipose tissue, synovium, and other tissues (Friedenstein et al. 1968; Katz et al. 2005; Kern et al. 2006; Tepliashin et al. 2005; Wang et al. 2015). Recently, Synovium-derived (SMSCs) have been recognized as an excellent source, owing to their superior chondrogenic potential (Fan et al. 2009). SMSCs could be isolated from normal or pathological synovium and successfully proliferate (Trounson and McDonald 2015; Wu et al. 2013). In addition, synovial tissue is conveniently accessible for the acquisition of autologous SMSCs via undergoing an ordinary examination (De Bari et al. 2003).
SRY-related HMG-box (Sox) family plays an important role in proliferation and differentiation of MSCs (Liu and Lefebvre 2015; Sun et al. 2015). Sox4 is a member of the Sox family with a critical role in embryonic development and cell-fate determination during organogenesis and differentiation (Jafarnejad et al. 2013; Parvani and Schiemann 2013). Increased Sox4 activity contributes to cellular transformation. For example, Sox4 directly modulates key cellular regulators, including epidermal growth factor receptor (EGFR), heat shock protein 70 (Hsp70), frizzled homolog 5 (Fzd5), Delta-like 1 (Dll1), and Patched-1 (Ptch1) and the transcriptional regulators Foxa1 (Scharer et al. 2009). However, the role of Sox4 in human SMSCs remains elusive. Here, we demonstrated a pivotal role of Sox4 in chondrogenic differentiation and proliferation of human SMSCs by directly regulating the expression of long noncoding RNA DANCR (anti-differentiation noncoding RNA). DANCR was fist identified in hepatocellular carcinoma (HCC) (Yuan et al. 2015). DANCR is overexpressed in stem-like HCC cells and markedly increased stemness features to promote tumorigenesis. In addition, the function of DANCR relies largely on regulation of β-catenin transcription (Yuan et al. 2015). However, whether DANCR contribute to the differentiation and proliferation of human synovium-derived stem cells is still unknown.
Materials and methods
Isolation and culture of SMSCs
SMSCs were obtained from the synovium of osteoarthritis patients. SMSCs were isolated following a standardized procedure (De Bari et al. 2001). Briefly, Synovial tissue was digested in 0.2 % collagenase (Sigma) at 37 °C for 1 h. Single cell suspensions were centrifuged and resuspended in high glucose DMEM supplemented with 10 % fetal bovine serum (FBS) (Gibco), and then plated onto a culture dish.
Sox4 overexpression in human SMSCs
Full-length human Sox4 and DANCR cDNA was cloned from normal human tissue RNA pools by RT-PCR. Full-length Sox4 or DANCR cDNA was inserted into pcDNA3.1 mammalian expression vector. Stable cells were established by hygromycin selection.
RNAi-mediated knock down of DANCR
SiRNA sequences directed against DANCR were designed and synthesized by the Shanghai Genechem Company, China. The sequence was: CTACAGGCACAAGCCATTG. For transfection, siRNA or control in Lipofectamine™2000 (Invitrogen) was transfected into cells according to manufacturer’s instructions.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen) and reverse-transcribed using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara) according to the manufacturer’s instructions. cDNA was quantified by qRT-PCR using an ABI 7500 System (Applied Biosystems). PCR was performed using SYBR Green reagents (Takara) according to the manufacturer’s instructions. GAPDH was used as an internal control. Primers used for qRT-RCR are listed followed: Sox4: F: CCAAATCTTTTGGGGACTTTT, R: CTGGCCCCTCAACTCCTC; DANCR: F: GCGCCACTATGTAGCGGGTT, R: TCAATGGCTTGTGCCTGTAGTT; GAPDH F: GGTCTCCTCTGACTTCAACA, R: GTGAGGGTCTCTCTCTTCCT; Col2: F: CTGTGACTGTAGTTCCCGTTTAT, R: GAGGCTGTTAGGTGTAGGATTG; AGC1: F: GTTGCAGTGAGCCGAGATTA, R: CTCTGGGAGAGAATTGGACTTG; Sox9: GCAGCGAAATCAACGAGAAAC, TCCAAACAGGCAGAGAGATTTAG.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore), according to its manual. Briefly, crosslinked chromatin was sonicated into 200–1000 bp fragments. The chromatin was immunoprecipitated using anti-Sox4 (Abcam) antibody. Normal mouse immunoglobulin G (IgG) was used as a negative control. qRT-PCR was conducted using SYBR Green Mix (Takara). Primers are listed followed: F: TACTGCATGAGCCACACCTA, R: AGCCAATCCCGGGAAGAT.
In vitro differentiation assay
Empty-vector-transfected or Sox4-transfected SMSCs were cultured to obtain adequate cells for 25 days. After expansion, these cells were trypsinized and seeded at 5 × 106 cells/mL in 15-mL polypropylene tubes. The cells were centrifuged for 10 min at 2000 rpm for use in an aggregate culture system. The cell aggregates were prepared and cultured in chondrogenesis medium that consisted of insulin-transferrin-selenium premix (Gibco BRL), 50 mM ascorbate-2-phosphate (Sigma) and 100 nM dexamethasone (Sigma) at 37 °C. The aggregates formed into a free-floating mass within the first 24 h of culture. The medium was replaced every 3 days for 14 days. The volumes were calculated by the formula \( {\text{V}} = \raise.5ex\hbox{$\scriptstyle 1$}\kern-.1em/ \kern-.15em\lower.25ex\hbox{$\scriptstyle 2$} \, \left( {{\text{L }} \times {\text{W}}^{2} } \right) \), where L is the length (longest dimension) and W is the width (shortest dimension).
Histology
Aggregates were fixed in 10 % paraformaldehyde and embedded in paraffin and cut into 5-μm sections. Sections were stained with toluidine blue and Safranin-O stain according to routine protocols. Samples were examined via light microscopy (Leica).
Viability assay
The cells were seeded at a density of 3 × 103 cells per well in 96-well plates. Cell proliferation was determined by the Cell Counting Kit-8 (CCK-8) assay. Each well was incubated with WST-8 solution (Vazyme) for 3 h, and the absorbance of each well was measured at 450 nm using a spectrophotometer.
Measurement of glycosaminoglycans (GAG) content
The GAG content was measured as previously described (Kim et al. 2014).
Statistical analysis
All data are presented as the mean ± SD. Comparisons of two groups were performed by Student’s t test. Differences were considered statistically significant at p < 0.05. All data were analyzed using the SPSS software.
Results
Overexpression of Sox4 enhances cell proliferation
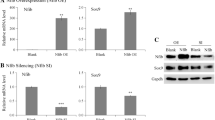
To investigate the immunophenotype change of Sox4-overexpressed SMSCs, we characterized MSC specific markers via qRT-PCR analysis. The results revealed that the surface marker genes expression (CD14−,CD19−, CD73+, CD90+, CD105+) of MSCs were not significantly different whether they were transfected with or without Sox4 (Fig. 1a). To characterize the influence of Sox4 overexpression on SMSCs viability, we measured cell viability using a CCK-8 assay. The CCK-8 assay revealed that cell numbers were not significantly different by day 3. However, cell viability of Sox4-overexpressed SMSCs gradually increased from day 4 to day 7 compared to control group (Fig. 1b). Therefore, Sox4 overexpression enhances SMSCs proliferation.
Overexpression of Sox4 enhances SMSCs proliferation. a For each group, the MSC-specific marker genes were determined by Real-time PCR. mRNA expression levels were normalized to GAPDH. b Cell viability was estimated at 1, 2, 3, 4, 5, 6 and 7 days after Sox4 transfection using the CCK-8 assay. Data are shown as mean ± SD; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test)
Sox4 promotes chondrogenic differentiation of SMSCs
To investigate the chondrogenic potential of Sox4-transfected SMSCs, we used the aggregate culture system. After Sox4 transfection for 14 days, aggregates of all groups were subjected to histological examination by toluidine blue (matrix proteoglycans) and Safranin-O staining. The aggregates formed by Sox4-overexpressed SMSCs had a greater amount of toluidine blue staining than control group (Fig. 2a). And, the size of aggregates from SOX4-transfected SMSCs was significantly increased compared with control group (Fig. 2b). Furthermore, we detected the expression of chondrogenic specific marker genes, such as Sox9, AGC1, and Col2. Consistent with histological results, these markers were significantly increased after transfection of Sox4 (Fig. 2c). Finally, we measured the accumulation of GAG in aggregates of all groups at day 14 after chondrogenic induction. GAG/DNA ratio of Sox4-overexpressed SMSCs were much higher than control group (Fig. 2d). These findings corresponded to the results of histological analysis and the expression of chondrogenic markers during chondrogenesis.
Chondrogenic differentiation of SMSCs in aggregate culture following Sox4 transfection. a Aggregates of all groups were stained with Safranin-O and toluidine blue for histological characterization. b Aggregate sizes for each group. c For each group, the chondrogenic-specific marker genes Aggrecan, Type II collagen and Sox9 were determined by Real-time PCR. mRNA expression levels were normalized to GAPDH. d Biochemical analysis for the assessment of chondrogenic differentiation index. After 14 days of chondrogenic induction, aggregates were digested in papain and sulfated glycosaminoglycans (S-GAG) and DNA content was measured. The GAG content was normalized to the total DNA content of each sample. Data are shown as mean ± SD; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test)
DANCR is a direct transcriptional target of Sox4
The important role of Sox4 in SMSCs motivated us to identify the target genes that were directly regulated by Sox4. It has been reported that lncRNA DANCR was closely associated with cell differentiation and cell transformation (Yuan et al. 2015). And we noticed that the DANCR promoter contained one consensus Sox4-binding motif (CAATGG). We hypothesized that DANCR was a direct target of Sox4. Interestingly, overexpression of Sox4 significantly increased DANCR level (Fig. 3a). ChIP assay also revealed the promoter of the DANCR gene to be directly bound by Sox4 in SMSCs (Fig. 3b). Finally, we cloned DANCR promoter (−500 to +200) and generated deletion mutant of promoter-luciferase construct based on the location of Sox4-binding motif (Fig. 3c). As determined by DANCR promoter luciferase reporter assay, DANCR promoter activity was increased upon Sox4 overexpression (Fig. 3d). When the region containing the Sox4-binding motif was deleted, we did not observe any changes in Sox4-mediated DANCR promoter-luciferase activation. These results indicate that lncRNA DANCR is a direct target of Sox4 in SMSCs.
DANCR is a direct transcriptional Target of Sox4. a DANCR expression was detected in SMSCs cells by Real-time PCR after transfection of Sox4 or the vector. b ChIP assay of Sox4 binding at the DANCR promoter. c DANCR promoter and deletion mutant of promoter luciferase construct. d Luciferase activity in SMSCs cotransfected with Sox4 and luciferase reporters containing DANCR promoter or its mutant. Data are shown as mean ± SD; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test)
Sox4 enhances chondrogenic differentiation and proliferation of SMSCs via upregulation of lncRNA DANCR
Next, we examined whether overexpressing DANCR could lead to the same phenotypes seen in SMSCs with Sox4 overexpression: chondrogenic differentiation and proliferation. Using CCK-8 assay, we found that overexpressing DANCR significantly promoted proliferation in SMSCs (Fig. 4a). In addition, we observed that DANCR significantly increased the size of aggregates formed by SMSCs compared with control group (Fig. 4b). This changes of phenotypes induced by DANCR was similar to which induced by Sox4.
Sox4 enhanced chondrogenic differentiation and proliferation of SMSCs via upregulation of lncRNA DANCR. a Cell viability was estimated at 1, 2, 3, 4, 5, 6 and 7 days after DANCR transfection using the CCK-8 assay. b Aggregate sizes for each group. c DANCR expression was detected in SMSCs cells by Real-time PCR after cotransfection of Sox4 and siRNA against DANCR. d Cell viability was estimated at 1, 2, 3, 4, 5, 6 and 7 days after DANCR siRNA and Sox4 transfection using the CCK-8 assay. e Aggregate sizes for each group. Data are shown as mean ± SD; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test)
To determine whether Sox4 functions upstream of DANCR in the regulation of chondrogenic differentiation and proliferation of SMSCs, we knocked down DANCR expression in Sox4-overexpressed SMSCs (Fig. 4c). We found that knocking down DANCR expression significantly decreased the proliferation and differentiation that were induced by the Sox4 overexpression (Fig. 4d, e). Together, these results demonstrated that Sox4 exerts its function at least in part via regulating DANCR expression.
Discussion
Transplantation of SMSCs possessing chondrogenic potential is now considered to be a new approach for repairing damaged articular cartilage. These SMSCs may be applied to clinical application due to its ability of differentiation into osteoblasts, chondrocytes, and adipocytes. It has been reported that the differentiation of SMSCs is closely associated with growth factors, such as TGF-β1. Previous studies have demonstrated that supplementation or overexpression of TGF-β1 promoted chondrogenic differentiation of SMSCs (Kim et al. 2014; Pei et al. 2008; Zhang et al. 2015). Sox4 is a known downstream of TGF-β1 pathway. In the present study, we demonstrated that overexpression of Sox4 is sufficient to promote proliferation and chondrogenic differentiation in the absent of TGF-β1 treatment. Although Sox family is well known to regulate various cellar processes, the function of Sox4 in chondrogenesis is largely unknown. Recent reports from other group suggested that Sox4 might play a role in chondrogenesis (Ezura et al. 2009). In this study, for the first time, we demonstrated that Sox4 overexpression enhancec chondrogenic differentiation and proliferation of SMSCs. However, the precise mechanisms by which Sox4 exerts its function in SMSCs remain unclear. One hypothesis is that Sox4 activates Wnt signaling pathway which is essential for SMSCs proliferation and differentiation through upregulation of β-catenin/TCF4 transcription (Naito et al. 2015; Saegusa et al. 2012). Another idea is that Sox4 increases the stability of β-catenin by directly binding to β-catenin and TCF family (Lee et al. 2011). A third possibility is that differentiation requires coordinated transcriptomic reprogramming via chromatin remodeling, and Sox4 increases and interacts with EZH2 which trimethylates histone 3 lysine 27 (H3K27me3) for gene repression (Koumangoye et al. 2015; Tiwari et al. 2013). The exact mechanisms of Sox4 on SMSCs proliferation and differentiation require further investigation.
lncRNAs have been established to participate in various biological processes that are crucial for development and differentiation. However, the roles of lncRNAs in the mechanisms of SMSCs differentiation are not completely understood. In the present study, ectopic expression of DANCR promoted chondrogenic differentiation and proliferation of SMSCs. DANCR also regulates Wnt pathway through interacting with CTNNB1 mRNA and stabilizing its stability (Yuan et al. 2015). We also found that DANCR is a target gene of Sox4. And Sox4 enhances chondrogenic differentiation and proliferation of SMSCs at least in part via upregulation of DANCR.
In conclusion, we demonstrated that Sox4 overexpression in SMSCs enhanced cell proliferation and chondrogenic potential via upregulation of lncRNA DANCR expression. The function of Sox4 on proliferation and chondrogenisis may through regulating Wnt pathway in diverse pattern. Additional studies are needed to illuminate the exact mechanisms, which will be helpful for the clinical application of SMSCs-based cartilage regeneration.
References
De Bari C, Dell’Accio F, Tylzanowski P, Luyten FP (2001) Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum 44:1928–1942. doi:10.1002/1529-0131(200108)44:8<1928:AID-ART331>3.0.CO;2-P
De Bari C, Dell’Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP (2003) Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol 160:909–918. doi:10.1083/jcb.200212064
Ezura Y, Sekiya I, Koga H, Muneta T, Noda M (2009) Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis Rheum 60:1416–1426. doi:10.1002/art.24472
Fan J, Varshney RR, Ren L, Cai D, Wang DA (2009) Synovium-derived mesenchymal stem cells: a new cell source for musculoskeletal regeneration tissue engineering part B. Reviews 15:75–86. doi:10.1089/ten.teb.2008.0586
Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP (1968) Heterotopic of bone marrow. Anal Precursor Cells Osteogenic Hematopoietic Tissues Transplant 6:230–247
Getgood A, Brooks R, Fortier L, Rushton N (2009) Articular cartilage tissue engineering: Today’s research, tomorrow’s practice? J Bone Jt Surg Br 91:565–576. doi:10.1302/0301-620X.91B5.21832
Ham O et al (2015) Therapeutic potential of differentiated mesenchymal stem cells for treatment of osteoarthritis. Int J Mol Sci 16:14961–14978. doi:10.3390/ijms160714961
Hunziker EB (2002) Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthr Cartil/OARS Osteoarthr Res Soc 10:432–463. doi:10.1053/joca.2002.0801
Jafarnejad SM, Ardekani GS, Ghaffari M, Li G (2013) Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci: CMLS 70:2677–2696. doi:10.1007/s00018-012-1187-y
Katz AJ, Tholpady A, Tholpady SS, Shang H, Ogle RC (2005) Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) cells. Stem Cells 23:412–423. doi:10.1634/stemcells.2004-0021
Kern S, Eichler H, Stoeve J, Kluter H, Bieback K (2006) Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 24:1294–1301. doi:10.1634/stemcells.2005-0342
Kim YI, Ryu JS, Yeo JE, Choi YJ, Kim YS, Ko K, Koh YG (2014) Overexpression of TGF-beta1 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cells. Biochem Biophys Res Commun 450:1593–1599. doi:10.1016/j.bbrc.2014.07.045
Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, Andl CD (2015) SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer 14:24. doi:10.1186/s12943-014-0284-y
Lee AK, Ahn SG, Yoon JH, Kim SA (2011) Sox4 stimulates ss-catenin activity through induction of CK2. Oncol Rep 25:559–565. doi:10.3892/or.2010.1091
Liu CF, Lefebvre V (2015) The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. doi:10.1093/nar/gkv688
Marmotti A, Rossi R, Castoldi F, Roveda E, Michielon G, Peretti GM (2015) PRP and articular cartilage: a clinical update. BioMed Res Int 2015:542502. doi:10.1155/2015/542502
Naito M, Ohashi A, Takahashi T (2015) Dexamethasone inhibits chondrocyte differentiation by suppression of Wnt/beta-catenin signaling in the chondrogenic cell line ATDC5. Histochem Cell Biol. doi:10.1007/s00418-015-1334-2
Parvani JG, Schiemann WP (2013) Sox4, EMT programs, and the metastatic progression of breast cancers: mastering the masters of EMT. Breast Cancer Res: BCR 15:R72. doi:10.1186/bcr3466
Pei M, He F, Vunjak-Novakovic G (2008) Synovium-derived stem cell-based chondrogenesis. Differ Res Biol Divers 76:1044–1056. doi:10.1111/j.1432-0436.2008.00299.x
Petrella F, Rizzo S, Borri A, Casiraghi M, Spaggiari L (2015) Current perspectives in mesenchymal stromal cell therapies for airway tissue defects. Stem Cells Int 2015:746392. doi:10.1155/2015/746392
Saegusa M, Hashimura M, Kuwata T (2012) Sox4 functions as a positive regulator of beta-catenin signaling through upregulation of TCF4 during morular differentiation of endometrial carcinomas. Lab Investig J Tech Methods Pathol 92:511–521. doi:10.1038/labinvest.2011.196
Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS (2009) Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res 69:709–717. doi:10.1158/0008-5472.CAN-08-3415
Scotece M, Mobasheri A (2015) Leptin in osteoarthritis: focus on articular cartilage and chondrocytes. Life Sci. doi:10.1016/j.lfs.2015.05.025
Sun D et al (2015) Constitutive L-Sox5 overexpression delays differentiation of ATDC5 cells into chondrocytes and correlates with reduced expression of differentiation markers. Mol Cell Biochem 401:21–26. doi:10.1007/s11010-014-2288-8
Tepliashin AS, Korzhikova SV, Sharifullina SZ, Chupikova NI, Rostovskaia MS, Savchenkova IP (2005) Characteristics of human mesenchymal stem cells isolated from bone marrow and adipose tissue. Tsitologiia 47:130–135
Tiwari N et al (2013) Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23:768–783. doi:10.1016/j.ccr.2013.04.020
Trounson A, McDonald C (2015) Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell 17:11–22. doi:10.1016/j.stem.2015.06.007
Wang Y, Yin Y, Jiang F, Chen N (2015) Human amnion mesenchymal stem cells promote proliferation and osteogenic differentiation in human bone marrow mesenchymal stem cells. J Mol Histol 46:13–20. doi:10.1007/s10735-014-9600-5
Wu B et al (2013) Lentiviral delivery of biglycan promotes proliferation and increases osteogenic potential of bone marrow-derived mesenchymal stem cells in vitro. J Mol Histol 44:423–431. doi:10.1007/s10735-013-9497-4
Yuan SX et al (2015) Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma via de-repression of CTNNB1. Hepatology. doi:10.1002/hep.27893
Zhang R et al (2015) BMP9-induced osteogenic differentiation is partially inhibited by miR-30a in the mesenchymal stem cell line C3H10T1/2. J Mol Histol 46:399–407. doi:10.1007/s10735-015-9628-1
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81301582), Postdoctoral Science Foundation of Jiangsu Province (Grant No. 1402003B) and China Postdoctoral Science Foundation funded project (Grant No. 2015M572812).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
None.
Additional information
Lei Zhang and Shuo Chen have been contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, L., Chen, S., Bao, N. et al. Sox4 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cell via activation of long noncoding RNA DANCR. J Mol Hist 46, 467–473 (2015). https://doi.org/10.1007/s10735-015-9638-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-015-9638-z