Abstract
In rice, an E-class gene, OsMADS1, acts to specify the identities of the lemma and palea. In this study, the OsMADS1 gene with a CaMV35S promoter was transformed into a japonica cultivar, Zhonghua 11. All transgenic plants successfully showed similar phenotypes, including dwarfism, distorted panicles, decreased numbers of branches and spikelets, and elongated sterile lemma. Histological analysis showed that the elongated sterile lemma developed with silicified epidermal and sclerenchymal cells, which were lacking in the wild-type sterile lemma, suggesting that the elongated sterile lemma had assumed the identity of the lemma or palea. Some marker genes were subjected to a detailed analysis of the distribution of their expression among the lemma, palea and sterile lemma. DROOPING LEAF (DL) and OsMADS6 genes were only expressed in the normal lemma or palea, respectively. In the elongated sterile lemma, a high level of DL gene expression was detected, while no expression of OsMADS6 was found, implying that the sterile lemma transformed into the lemma but not the palea. These results provide clues to elucidate the mechanism of evolution from lemma to sterile lemma in rice. qPCR analysis also suggested that the ectopic expression of OsMADS1 induced abnormal brassinosteroid and gibberellin acid activation, and then resulted in developmental defects in the stem and panicle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most flowers consist of four distinct organ types arranged in concentric whorls: sepals (whorl one), petals (whorl two), stamens (whorl three), and carpels and ovules (whorl four). The well-established ABCDE model, which is mainly based on genetic and molecular studies involving eudicots such as arabidopsis (Arabidopsis thaliana), snapdragons (Antirrhinum majus) and petunias (Petunia hybrida), explains how floral organ identity is coordinately defined by A-, B-, C-, D- and E-class MADS-box genes in eudicots (Coen and Meyerowitz 1991; Weigel and Meyerowitz 1994; Theissen and Saedler 2001; Ditta et al. 2004). Grass, one of the largest monocot families, includes many important crops such as barley (Hordeum vulgare), maize (Zea mays), rice (Oryza sativa) and wheat (Triticum aestivum). Grasses have floral architectures distinct from those of eudicots and even other monocots. Spikelets, the basal units of grass inflorescence, consist of glumes and between one and forty florets that contain lemma and palea (possibly homologous to sepals), lodicules (homologous to petals), stamens and pistils/ovules (Bommert et al. 2005; Itoh et al. 2005; Malcomber et al. 2006). Recently, several MADS-box genes specifying flower organ identity were characterised in rice, suggesting that the ABCDE model is also applicable to grass species, at least in part (Nagasawa et al. 2003; Yamaguchi et al. 2006; Dreni et al. 2007).
Among these genes, E-class genes show the greatest diversity in terms of their function and number. In Arabidopsis, the E-class genes SEPALLATA1/2/3/4 (SEP1/2/3/4) redundantly specify all floral organ identities and floral meristem determination (Pelaz et al. 2000; Ditta et al. 2004). In rice, five SEP-like genes are known (Malcomber and Kellogg 2004, 2005; Kater et al. 2006). OsMADS7 and OsMADS8 redundantly specify inner floral organ identities (Cui et al. 2010). Another SEP-like gene, OsMADS34/PANICLE PHYTOMER2 (PAP2), was found to control the development of branch, rudimentary glume, and sterile lemma (Gao et al. 2010; Kobayashi et al. 2010). OsMADS1/LEAFY HULL STERILE (LHS1) was the first SEP-like gene to be identified in rice. In several osmads1 mutants, the lemma and palea were transformed into leafy-like or glume-like organs, suggesting that OsMADS1 plays a key role in the specification of the identity of the lemma and palea (Jeon et al. 2000; Agrawal et al. 2005; Prasad et al. 2005; Chen et al. 2006).
The ectopic expression of genes can promote our understanding of gene function. In the wild type, OsMADS1’s expression was detected first in the floral meristem at an early stage, and then became predominantly localised in the palea and lemma (Prasad et al. 2001, 2005). In a previous study, the ectopic expression of OsMADS1 in rice under the control of the maize ubiquitin promoter caused a stunted panicle and the homeotic transformation of the sterile lemma into a lemma/palea-like organ (Prasad et al. 2001). However, the exact defects of vegetative organs in transgenic plants with the ectopic expression of OsMADS1 remain unclear. In addition, the details of the histology and molecular signatures of abnormal organs and the mechanisms behind the defects in transgenic plants are also unknown. In this study, ectopic expression of OsMADS1 under the control of the CMV 35S promoter was therefore performed in rice in order to investigate in detail its further functioning in the development of flowers and other organs. The transgenic plants displayed serious defects in the development of vegetative and reproductive organs, including dwarfism, distorted panicles, decreased numbers of branches and spikelets, and an elongated sterile lemma. qPCR analysis suggested that the ectopic expression of OsMADS1 induced abnormal brassinosteroid (BR) and gibberellin acid (GA) activation, and then resulted in developmental defects in the stem and panicle. Histological analysis suggested that the elongated sterile lemma had assumed the identity of the lemma or palea. Expression analysis of the marker genes DL and OsMADS6 then indicated that the sterile lemma had been transformed into the lemma but not the palea. These results suggest that the OsMADS1 gene plays an important role in the evolution from the lemma to the sterile lemma in rice.
Materials and methods
Plant materials
Transgenic plants that overexpressed the OsMADS1 gene were grown in experimental fields in Chongqing, China. Zhonghua 11 (ZH11) was used as the wild type.
Vector construction and transformation
RNA was isolated from the panicles of ZH11. The first-strand cDNA was synthesised from 2 µg of total RNA with oligo(dT)18 primers in a 25-µL reaction volume using the SuperScript III Reverse Transcriptase Kit (Invitrogen). To construct the OsMADS1-overexpression plasmids, OsMADS1 cDNA that contained the whole codon sequence and part of the untranslated region was amplified using the primers XbaI-M1F and BamHI-M1R (Supplemental Table 1). The PCR products were inserted into the expression cassette pCAMBIA1301-35S-NOS, which contains the beta-glucuronidase (GUS) reporter gene. The constructs were transferred into calli of japonica rice ZH11 by Agrobacterium-mediated T-DNA transformation, as described previously (Sang et al. 2012).
X-Gluc staining and Southern blot detection
To detect the expression of the reporter GUS, young leaf, stem, sheath and spikelet of transgenic plants were obtained and incubated for 0.5–3 h in X-Gluc staining dye at 37 °C. Then, these organs were observed using a Nikon E600 microscope and photographed.
For Southern blot detection, genomic DNA (15 µg) from the transgenic line and ZH11 was digested with HindIII. The OsMADS1 cDNA probe was amplified using the primers XbaI-M1F and BamHI-M1R and labelled using a DIG DNA Labelling Kit (Roche), in accordance with the manufacturer’s recommendations. Transmembrane were performed with the iBlot® Dry Blotting System. Pre-hybridisation, hybridisation, washing and immunological detection were performed in accordance with the manufacturer’s recommendations and as reported by Southern (1975).
Measurement of agronomic characteristics
At the maturation stage, plant height, internode length, panicle length, primary branch number, and secondary branch number were investigated in transgenic and wild-type plants.
Microscopy analysis
For paraffin sections, panicles were collected at the heading stage and fixed in FAA (50 % ethanol, 0.9 M glacial acetic acid and 3.7 % formaldehyde) overnight at 4 °C, dehydrated with a graded ethanol series, infiltrated with xylene and embedded in paraffin (Sigma). The 10-µm-thick sections were transferred onto poly-l-Lys-coated glass slides, deparaffinised in xylene, and dehydrated through an ethanol series. The sections were stained sequentially with 1 % safranin (Amresco) and 1 % Fast Green (Amresco), dehydrated through an ethanol series, infiltrated with xylene, and finally mounted beneath a coverslip. Light microscopy was performed using a Nikon E600 microscope.
For scanning electron microscopy (SEM), fresh samples were examined using a Hitachi SU3500 scanning electron microscope with a −40 °C cool stage and a low-vacuum environment. The stages of early spikelet development were the same as those defined previously (Ikeda et al. 2004).
qPCR analysis
RNA was isolated from leaf, stem, sheath and inflorescence in transgenic and wild-type plants using the RNAprep Pure Plant Kit (Tiangen) with DNase I treatment. The first-strand cDNA was synthesised from 2 µg of total RNA with oligo(dT)18 primers in a 25-µL reaction volume using the SuperScript III Reverse Transcriptase Kit (Invitrogen). Reverse-transcribed RNA (0.5 µL) was used as a PCR template with gene-specific primers (Supplemental Table 1). Actin was used as an endogenous control. The qPCR analysis was performed with an ABI Prism 7000 Sequence Detection System and the SYBR Supermix Kit (Bio-Rad). At least three replicates were performed and the mean values for the expression of each gene were obtained.
Results
Characterisation of transgenic plants
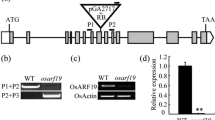
By Agrobacterium-mediated transformation, the recombinant plasmid 35S:OsMADS1 (Fig. 1a) was transformed into ZH11. Twelve T0 plants were obtained and five of these showed positivity for GUS staining (Fig. 1b). All of these five positive plants showed dwarfism and abnormal panicle development, while the negative plants showed no difference compared with the wild type. One of these lines was chosen for further analysis. Southern blot analysis showed that one copy of OsMADS1 had been inserted into this line (Fig. 1c). In the T1 population of this line, three types of plant were found: wild type, T0 type, and intermediate type, with a segregation ratio of 1:2:1. The latter group showed phenotypes between those of the other two, in terms of plant height and panicle development. In T2, the T0 type remained separate, while the intermediate type showed genetic stability. Thus, the T0 type consist of the heterozygote of ectopically expressed OsMADS1, while the intermediate type was the homozygote.
Characterisation of transgenic plants. a Ectopic expression vector of OsMADS1; b X-Gluc staining for GUS in the leaf, stem, sheath and spikelet of transgenic plants; c Southern blot detection of transgenic plants; d expression of OsMADS1 in the leaf, stem, sheath and spikelet of the homozygote, heterozygote and wild type. Error bars indicate SD. WT wild type, OE ectopic expression, hete heterozygote, homo homozygote
To examine the expression of OsMADS1 in the leaf, stem, sheath and spikelet of the homozygote, heterozygote and wild type, mRNA accumulation was measured using qPCR. OsMADS1 was expressed only in the spikelet of the wild type, but it was also expressed in the leaf, stem, and sheath of the transgenic plants. Surprisingly, the expression level of OsMADS1 in multiple tissues of the homozygote was significantly lower than that in the heterozygote (Fig. 1d). These results suggested that 35S:OsMADS1 inhibited the expression of the OsMADS1 gene in the homozygote.
Ectopic expression of OsMADS1 caused dwarfism and shorter panicle
The transgenic plants displayed clear dwarfism (Fig. 2a). At the heading stage, plant height differed significantly among the heterozygote, homozygote, and wild type. Average plant heights were 46.8, 73.9 and 114.8 cm, respectively (Fig. 2b). Further investigation revealed that the heterozygote had only three elongated internodes, the homozygote had four, and the wild type had five or six (Fig. 2c). Next, the lengths of elongated internodes and panicles were measured. There were significant differences in the lengths of panicles, first internodes, and second internodes among these three types of plant (Fig. 2d). These results indicated that dwarfism in these transgenic plants is caused by decreases in the panicle and internode lengths, as well as in the number of elongated internodes.
Agronomic traits of wild-type and transgenic plants. a Wild-type and transgenic plants at maturity; b height of wild-type and transgenic plants; c, d internode lengths of wild-type and transgenic plants. Error bars indicate SD. **Indicates a statistically significant difference (P < 0.01). WT wild type, OE ectopic expression, hete heterozygote, homo homozygote
In the transgenic plants, panicle development was severely disturbed. Compared with the wild type, the panicle was shorter and smaller in the transgenic plants (Fig. 3a, b). The numbers of primary and secondary branches differed significantly among the heterozygote, homozygote and wild type, in the following order: heterozygote < homozygote < wild type (Fig. 3c–e). These findings suggested that overexpression of OsMADS1 in the panicle inhibited branch differentiation and elongation.
Phenotypic analysis of inflorescences in wild-type and transgenic plants. a Spikelets at maturity; b panicles at maturity; c floral number; d primary branch number; e secondary branch number. Error bars indicate SD. **Indicates a statistically significant difference (P < 0.01). WT wild type, OE ectopic expression, hete heterozygote, homo homozygote
Abnormal expression of BR/GA-related genes in transgenic plants
To clarify the mechanism of dwarfism and shorter panicles in transgenic plants, the expression of some BR/GA related genes were investigated in stems and panicles. In stems, the mRNA expression levels of the positive regulators of the BR signalling pathway OsBRI1 and OsBZR1 were higher in transgenic plants (homozygote and heterozygote) than in the wild type; the expression of the BR biosynthetic gene OsGSR1 was higher in the heterozygote than in the wild type; and the expression of the negative regulatory factors in BR response OsGSK1, OsMADS22 and OsMADS55 was clearly down-regulated in the heterozygote (Fig. 4a). Additionally, the expression of OsGA2ox3, which encodes an inactivator of endogenous bioactive GA in vegetative tissues, was up-regulated in the stems of the heterozygote and homozygote (Fig. 4b). These results suggested that the ectopic expression of OsMADS1 induced an excess of active BR but inactivation of GA in the stem. In the panicle, most of the BR-related genes showed the opposite expression pattern compared with that in the stem. The expression level of the positive regulator OsBRI1 was down-regulated in the heterozygote, the negative regulators OsMADS22 and OsMADS55 were up-regulated in the heterozygote, and another negative regulator OsGSK1 was up-regulated in both the heterozygote and the homozygote (Fig. 4c). Additionally, the expression level of OsGA2ox1, another inactivator of endogenous bioactive GA in reproductive tissues, was up-regulated to nearly 73- and 253-fold in the transgenic panicles of the homozygote and heterozygote, respectively (Fig. 4d). These results suggest that the ectopic expression of OsMADS1 may induce BR deficiency and GA inactivation in the panicle.
Expression analysis of BR-related and GA-related genes between wild-type and transgenic plants. a The expression level of BR signalling pathway genes in stems. b The expression level of GA metabolic genes in stems. c The expression level of BR signalling pathway genes in panicles. d The expression level of GA metabolic genes in panicles. Error bars indicate SD
Ectopic expression of OsMADS1 led to lemma-like sterile lemma
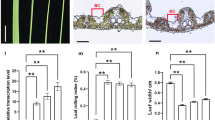
In the wild type, the spikelet contained a pair of rudimentary glumes, a pair of sterile lemmas, and a fertile floret. The floret had four whorls of floral organs, namely, one lemma and one palea joined together in whorl one, two lodicules on the lemma side in whorl two, and six stamens around a pistil with two stigmas in whorls three and four (Fig. 5a–e). The rudimentary glumes and the floret in the transgenic plants did not differ from those in the wild type. However, the sterile lemmas were clearly elongated (Fig. 5f–o). In the heterozygote, 76.1 % of spikelets had a pair of elongated sterile lemmas, 21.5 % of spikelets had an elongated inner sterile lemma and a normal outer sterile lemma, and only 2.4 % of spikelets developed a normal sterile lemma. In the homozygote, the proportions of these three spikelets were 61.6, 34.7 and 3.7 %, respectively (Fig. 5p).
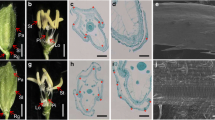
Phenotypes of spikelets in wild-type and transgenic plants. a, b Wild-type spikelet; c–e histological analysis of wild-type spikelet; f, g spikelet with only one sterile lemma elongated; h–j histological analysis of spikelet with only one sterile lemma elongated; k, l spikelet with both sterile lemmas elongated; m–o histological analysis of spikelet with both sterile lemmas elongated; p the types of spikelets in wild-type and transgenic plants; q–s expression analysis of OsMADS1, DL and OsMADS6 in the sterile lemma, lemma and palea of heterozygote, homozygote and wild type. Error bars indicate SD. le lemma, pa palea, osl outer sterile lemma, isl inner sterile lemma, losl elongated outer sterile lemma, lisl elongated inner sterile lemma, lsl0 normal sterile lemma, lsl1 elongated inner sterile lemma, lsl2 a pair of elongated sterile lemmas, WT wild type, OE OsMADS1 overexpression, hete heterozygote, homo homozygote. Bars 1000 μm (a, b, f, g, k, l), 500 μm (c, e, h, j, m, o) and 200 μm (a, b, f, g, k, l)
Further SEM and histological analysis revealed that the cells of the elongated sterile lemmas had identities matching those of the lemma. The wild-type lemma or palea consisted of four layers of cells: silicified epidermal cells, fibrous sclerenchymal cells, spongy parenchymal cells, and unsilicified epidermal cells. In the wild-type sterile lemmas, the epidermal cells were unsilicified and there was no sclerenchymal cell layer (Fig. 5d, e). In the transgenic plants, the elongated sterile lemmas had silicified epidermal cells, sclerenchymal, and parenchymal cells (Fig. 5i, j, n, o). These results suggested that the elongated sterile lemma had assumed the identity of the lemma or palea.
To clarify the identity of this elongated sterile lemma more precisely, the expression levels of OsMADS1, DL and OsMADS6 were determined in the sterile lemma, lemma and palea. Considering the OsMADS1 gene, in the wild type, its expression was only detected in the lemma and the palea. The expression of OsMADS1 in the lemma and palea of the heterozygote was more than five times higher than that in the wild type, while it was also clearly expressed in the sterile lemma. In the homozygote, the expression of OsMADS1 was detected in the sterile lemma, lemma and palea, but at a lower level than in the heterozygote (Fig. 5q). DL is a pistil identity gene in rice. It is strongly expressed in both pistil and lemma, but not in other floral organs, such as the palea and sterile lemma (Yamaguchi et al. 2004). In the present study, the expression of the DL gene was detected in the lemma but not in the palea or sterile lemma in the wild type. However, a strong signal of DL expression was found not only in the lemma but also in the sterile lemma in the transgenic plants (Fig. 5r). OsMADS6 plays a role in regulating the identities of the marginal region of the palea and the inner floral organs. It is expressed mainly in the palea and lodicule, but not in the lemma or the other floral organs (Ohmori et al. 2009; Li et al. 2010). In the present study, the signal of OsMADS6 expression was found in the palea of wild-type plants, but not in the lemma or sterile lemma. In the transgenic plants, the expression signal of OsMADS6 was not detected (Fig. 5s). These results indicate that the sterile lemma had transformed into the lemma, instead of the palea.
Scanning electron microscopy analysis of spikelet morphogenesis during early developmental stages [the spikelet 4 (Sp4)–Sp8 stage] was undertaken. In the wild type, the sterile lemma primordia were much larger than the newly formed lemma and palea primordia at the Sp4 stage. Subsequently, the rate of development of sterile lemma primordia was considerably slower than that of the lemma and palea primordia, which was followed by a period of rapid differentiation and proliferation. After the Sp7 stage, sterile lemma smaller than the lemma and palea were observed in the wild type (Fig. 6a–d). In the transgenic plants, no obvious difference from the wild type was observed, except abnormal development of the sterile lemma primordia. In some spikelets, the development of inner sterile lemma was abnormal, while no defect was found in the outer sterile lemma (Fig. 6e–h). In other spikelets, both inner and outer sterile lemma displayed developmental abnormality (Fig. 6i–l). Compared with the wild-type sterile lemma primordia, the abnormal sterile lemma primordia in transgenic plants always maintained rapid differentiation and proliferation during the Sp4–Sp8 stage (Fig. 6e–l). Then, lemma-like sterile lemma primordia were observed at the Sp8 stage in the transgenic plants (Fig. 6h, l). These results indicate that homeotic transformation from sterile lemma to lemma occurred at the primordial imitation and development stages in the transgenic plants.
Scanning electron micrographs of spikelet at the early flower development stage. a–d Wild-type spikelets; e–h spikelets with only one sterile lemma elongated in transgenic plants; i–h spikelets with both glumes elongated in transgenic plants. le lemma, pa palea, osl outer sterile lemma, isl inner sterile lemma, losl elongated outer sterile lemma, lisl elongated inner sterile lemma, fm floral meristem. Bars 100 μm
Discussion
The role of OsMADS1 in the evolution of sterile lemma
In rice, the spikelet contains only one top fertile floret and two pairs of lateral glume-like organs. The lower pair are organs that are homologous to glumes in other grasses, termed rudimentary glumes because of their extreme degeneration. The upper pair is termed the sterile lemma, with a size intermediate between those of lemma/palea and rudimentary glume. Recently, several genes that regulate the development of these organs have been identified. LONG STERILE LEMMA (G1)/elongated empty glume (ELE), encoding a DUF640 protein, was shown to control the development of sterile lemma. In the g1 mutant, the sterile lemma was transformed into a lemma-like organ (Yoshida et al. 2009; Hong et al. 2010). The loss of function of the OsMADS34 gene also led to elongation of the sterile lemma and its adoption of the appearance of a lemma or leaf-like organ (Gao et al. 2010; Kobayashi et al. 2010). In addition, O. grandiglumis, an Oryzeae species, contains a pair of long sterile lemmas that are the same as the lemma (Yoshida et al. 2009). These findings suggest that sterile lemma and lemma are homologous organs; in other words, that the sterile lemma simply originates from the rudimental lemma of a later floret.
In a previous study using stereomicroscopy and SEM, it was found that the sterile lemma was elongated and looked like a lemma/palea-like organ in transgenic plants that ectopically expressed the OsMADS1 gene (Prasad et al. 2005). In the present study, further histological analysis showed that elongated sterile lemma in OsMADS1-transgenic plants had the same cell structure as the lemma or palea in the wild type. In addition, qPCR analysis found that not only OsMADS1 but also the DL gene was ectopically expressed in the elongated sterile lemma. These results indicate that the ectopic expression of OsMADS1 led to a homeotic transformation from sterile lemma into lemma. Therefore, it can be speculated that when the two lateral florets degenerated into lemmas in ancestral rice, the pair of non-functional organs had negative effects on the fitness of the plant. Then, the expression of OsMADS1 was lost, resulting in the lemma further degenerating into a smaller and less obstructive organ, namely, the sterile lemma. This may have occurred in response to natural selection.
The sterile lemma appears as a lemma-like organ in the g1 mutant (Gao et al. 2010; Kobayashi et al. 2010). Thus it is probable that the ectopic expression of OsMADS1 in the sterile lemma of the g1 mutant led to its transformation into the lemma-like organ. However, OsMADS1 expression remains at a very low level in the g1 sterile lemma and shows no difference to that seen in the wild type (Supplemental Fig. 1). These results suggest that the sterile lemma may not have been transformed into a real lemma in the g1 mutant. Additionally, we also found that the expression of G1 was down-regulated in the sterile lemma of OsMADS1-transgenic plants (Supplemental Fig. 1). These results suggest that OsMADS1 specifying lemma identity required the expression of G1 to be restricted. In other words, the evolution of lemma to sterile lemma may have required both the absence of OsMADS1 expression and the acquisition of G1 function.
The function of OsMADS1 in differentiation of epidermal and sclerenchymal cells in hulls
In rice, the hull (lemma and palea) is considered to be the organ that is homologous to the sepal. In Arabidopsis, E-class genes SEP1/2/3/4 have a redundant function in the specification of the sepal (Ditta et al. 2004). However, in rice, the E-class genes appear to have undergone functional differentiation. OsMADS7/8 were regarded as having redundant function in the regulation of inner floral organ development. However, it is unclear whether they function in the specification of the hull (Cui et al. 2010). In an osmads34 single mutant, an elongated rudimentary glume was found, and no visible defects were observed in the hull. However, in an osmads34 osmads1 double mutant, the hull was longer and thinner than that in an osmads1 single mutant, also having a stoma structure, suggesting that OsMADS34 has a role in the specification of hull identity that is redundant with that of OsMADS1 (Lin et al. 2014). In previous studies, it was confirmed that OsMADS1 plays a key role in specification of the hull (Jeon et al. 2000; Agrawal et al. 2005; Prasad et al. 2005; Chen et al. 2006). In several osmads1 mutants, the hull was elongated and had the appearance of a leaf-like organ. Further histological analysis found that the numbers of both silicified epidermal and sclerenchymal cells were lower in the leafy hulls. These findings suggest that OsMADS1 functions in determining hull identity by regulating the differentiation of epidermal and sclerenchymal cells in hulls.
Our own results indicate that the ectopic expression of OsMADS1 led to a homeotic transformation from sterile lemma into lemma, as determined by stereomicroscopy, SEM, histology, and qPCR. In particular, histological analysis showed that the sterile lemma in OsMADS1-transgenic plants developed with silicified epidermal cells and sclerenchymal cells, which were lacking in the wild-type sterile lemma. It is well known that the main differences between sterile lemma and lemma are an unsilicified epidermis and the lack of a sclerenchymal cell layer in the sterile lemma. Therefore, the results suggest that the ectopic expression of OsMADS1 can lead to the differentiation of silicified epidermal and sclerenchymal cells on the basis of sterile lemma. Thus, it was further confirmed that OsMADS1 functions in the differentiation of epidermal and sclerenchymal cells in hulls.
Abnormal expression of BR/GA-related genes in transgenic plants
BR and GA are two positive hormones regulating plant cell elongation. In rice, mutations in the BR biosynthesis genes cause dwarfism (Tong and Chu 2012). However, an excess of active BR does not always induce stem elongation. For example, although OsMADS22 and OsMADS55 were shown to act as negative regulators of BR responses in stems, the reductions in stem number and length in the OsMADS55 RNAi plants and OsMADS55 + OsMADS22 double RNAi plants may have resulted from an excess of active BR. Therefore, the inhibition of stem growth could have been induced by either BR insensitivity or oversensitivity (Lee et al. 2008). It is clear that GA interacts with other phytohormones, including BR, to regulate plant growth and development (Weiss and Ori 2007). Under physiological conditions, BR promoted GA accumulation by regulating the expression of GA metabolic genes to stimulate cell elongation. When excessive active BR was applied, the hormone mostly induced GA inactivation through up-regulation of the GA inactivation gene GA2ox-3 and also suppressed BR biosynthesis, resulting in decreased hormone levels and growth inhibition (Tong et al. 2014).
In this study, the genes involved in the BR signalling pathway and GA metabolism showed abnormal expression levels in the transgenic plants compared with the wild type. In the stem, the expression level of BR/GA-related genes suggested that ectopic expression of OsMADS1 may have induced an excess of active BR, resulting in GA inactivation through up-regulation of the GA inactivation gene GA2ox-3. In the panicle of transgenic plants, the up-regulation of BR negative regulators suggest that BR deficiency had developed. Additionally, extreme up-regulation of the expression level of the GA inactivation gene GA2ox-1 in panicle may have resulted in decreased GA levels. Therefore, although an excess of active BR and a deficiency of BR may arise in the stem and panicle of transgenic plants, respectively, GA inactivation was induced in both the stem and the panicle. Although it is not clear how ectopic/over-expression of OsMADS1 induces inactive GA, an excess of active BR or a deficiency of BR, any one of these may result in growth inhibition in the stem or panicle of transgenic plants.
Co-suppression of OsMADS1 in homozygote plants
We initially found that the phenotype of the homozygote was less severely degraded than that of the heterozygote. We later found that the expression level of OsMADS1 in the homozygote was significantly lower than that in the heterozygote or even in the wild type. These results suggest that the expression levels of both endogenous and exogenous OsMADS1 were inhibited in homozygous transgenic plants. This phenomenon was initially observed in a study attempting to alter flower colours by overexpressing a gene encoding chalcone synthase in order to produce more flower pigmentation in petunias. However, white flowers with less pigmentation were found in some transgenic plants, and the endogenous gene and the transgene were down-regulated in the white flowers (Napoli et al. 1990). This phenomenon was termed the co-suppression of gene expression. There are two models to explain the molecular mechanism behind such co-suppression. The first proposes that the expression of highly similar genes is additive unless transgene transcript levels exceed a gene-specific threshold and consequently trigger post-transcriptional gene silencing (Schubert et al. 2004). The second suggests that improperly terminated, unpolyadenylated transcripts are highly susceptible to RDR6-mediated silencing (Luo and Chen 2007). In this study, the level of OsMADS1 transcript increased significantly but was not excessive in the heterozygous transgenic plants. However, the levels of the OsMADS1 transcript may have exceeded the gene-specific threshold in the homozygote and then triggered post-transcriptional gene silencing.
References
Agrawal KG, Abe K, Yamazaki M, Miyao A, Hirochika A (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of OsMADS1 gene. Plant Mol Biol 59:125–135
Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46:69–78
Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH (2006) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223:882–890. doi:10.1007/s00425-005-0141-8
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37. doi:10.1038/353031a0
Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61:767–781. doi:10.1038/353031a0
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14:1935–1940. doi:10.1016/j.cub.2004.10.028
Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk P, An G, Colombo L, Kater M (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52:690–699. doi:10.1111/j.1365-313X.2007.03272.x
Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153:728–740. doi:10.1104/pp.110.156711
Hong L, Qian Q, Zhu K, Tang D, Huang Z, Gao L, Li M, Gu M, Cheng Z (2010) ELE restrains empty glumes from developing into lemmas. J Genet Genom 37:101–115. doi:10.1016/S1673-8527(09)60029-1
Ikeda K, Sunohara H, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54:147–156. doi:10.1270/jsbbs.54.147
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46:23–47
Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G (2000) Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12:871–884
Kater M, Dreni L, Colombo L (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57:3433–3444. doi:10.1093/jxb/erl097
Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51:47–57. doi:10.1093/pcp/pcp166
Lee S, Choi SC, An G (2008) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54:93–105. doi:10.1111/j.1365-313X.2008.03406.x
Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20:299–313. doi:10.1038/cr.2009.143
Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theissen G, Meng Z (2014) The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol 202:689–702
Luo ZH, Chen ZX (2007) Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19:943–958. doi:10.1105/tpc.106.045724
Malcomber ST, Kellogg EA (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16:1692–1706. doi:10.1105/tpc.021576
Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10:427–435. doi:10.1016/j.tplants.2005.07.008
Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44:425–481
Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 130:705–718
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289. doi:10.2307/3869076
Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21:3008–3025. doi:10.1105/tpc.109.068742
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203. doi:10.1038/35012103
Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211:281–290. doi:10.1007/s004270100153
Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early acting regulator of inner floral organs. Plant J 43:915–928. doi:10.1111/j.1365-313X.2005.02504.x
Sang XC, Li YF, Luo ZK, Ren DY, Fang LK, Wang N, Zhao FM, Ling YH, Yang ZL, Liu YS, He GH (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol 160:788–807. doi:10.1104/pp.112.200980
Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16:2561–2572. doi:10.1105/tpc.104.024547
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Theissen G, Saedler H (2001) Plant biology: floral quartets. Nature 409:469–471
Tong H, Chu C (2012) Brassinosteroid signaling and application in rice. J Genet Genom 39:3–9. doi:10.1016/j.jgg.2011.12.001
Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26:4376–4393. doi:10.1105/tpc.114.132092
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209. doi:10.1016/0092-8674(94)90291-7
Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16:500–509. doi:10.1105/tpc.018044
Yamaguchi T, Lee DY, Miyao A, Hirochika H, An GH, Hirano HY (2006) Functional diversification of the two C-class MADS-box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18:15–28. doi:10.1105/tpc.105.037200
Yoshida A, Suzaki T, Tanaka W, Hirano HY (2009) The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA 106:20103–20108. doi:10.1073/pnas.0907896106
Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144:1240–1246
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31271304), the Natural Science Foundation Project of Chongqing (CSTC2012JJB80005) and by Fundamental Research Funds for the Central Universities (XDJK2012A001, XDJK2016A013).
Author information
Authors and Affiliations
Corresponding author
Additional information
Ling Wang and Xiao-Qin Zeng have contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Zeng, XQ., Zhuang, H. et al. Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice. Plant Growth Regul 81, 433–442 (2017). https://doi.org/10.1007/s10725-016-0220-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0220-9