Abstract
Agrobacterium rhizogenes-transformed hairy root somaclones (rhizoclones) and regenerated shoots of a rare medicinal herb, Hybanthus enneaspermus (L.) F. Muell. were investigated as a source of coumarin. Transformed nature of rhizoclones was verified by PCR amplification of rolA-B and mas2 genes. Clonal fidelity among rhizoclones was revealed by RAPD analysis exhibiting DNA monomorphism. Of a total of 104 rhizoclones of A4 origin and 76 rhizoclones of 8196 origin, a sample of 16 PCR tested clones was selected for shoot regeneration including eight clones pertaining to each strain type on the basis of sustained root growth index. Opine gene expression was demonstrated in rhizoclones and regenerated shoots. Inter-rhizoclonal variations with respect to biomass proliferation and coumarin content were evident. HPLC-tested coumarin accumulation was ca. threefold higher in the superior rhizoclone of A4 origin (A4-HRL-2B7) compared to that in natural roots. Transformed leafy shoots derived from A4-HRL-2B7 had the maximum coumarin content (3.25 mg g−1 d.wt. extract) which was significantly higher than that of in natura aerial part samples. Genetic stability of selected fast-proliferating rhizoclones and their respective in vitro shoot regenerants in terms of RT-PCR based expression of rolB and rolC genes and coumarin content was sustained through 16 successive multiplication cycles. This investigation ushers the possibility of harnessing the genetically stable biosynthetic potential of hairy root clones and transformed shoot regenerants of H. enneaspermus in vitro towards sustainable production of coumarin and other medicinally important phytochemicals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hybanthus enneaspermus (L.) F. Muell. (Family—Violaceae) is a small suffrutescent perennial herb native to a few warmer regions of India. The plant grows 15–30 cm in height with many diffuse or ascending branches and is pubescent in nature (Kirtikar and Basu 1991). In Ayurveda, the most ancient Indian Traditional System of Medicine, the plant is known as ‘Madanmastak’ whose roots are traditionally used as an aphrodisiac in cases of erectile dysfunction and sterility in men. It is also used as demulcent, tonic, diuretic and also in urinary infections, diarrhoea, dysuria, leucorrhoea and gonorrhoea (Wealth of India 1959). The anti-hyperglycemic, anti-oxidative and anti-fertility activity in whole plant extracts of H. enneaspermus have recently been evaluated (Thenmozhi and Premalashmi 2011; Patel et al. 2011; Nathiya and Senthamil Selvi 2013).
Hybanthus enneaspermus is reported to contain various phyto-constituents such as dipeptide alkaloids, aurantiamide acetate, isoarborinol, β-sitosterol, coumarins, essential oils, triterpenoids, flavonoids, tannins, saponins, etc. (Amuthapriya et al. 2011; Krishnamoorthy et al. 2014). Coumarins, also known as benzopyrones, represent a class of heterocyclic compounds which are currently of considerable interest to research groups worldwide because of the unique diversity in their structure and biological activities. These compounds are used as drugs for various ailments such as asthma, lymphodema, Alzheimer’s disease and cancer. Though coumarin, a medicinally important phenylpropanoid, has been isolated from several higher plants and a few fungi too, the quest continues for discovery of new biological species as alternative source of novel coumarin derivatives. Interestingly, many additional structures related to coumarin have been developed via synthetic chemistry. A state-of-the-art in research relating to occurrence, synthesis and medicinal chemistry of simple coumarins and analogues has been aptly reviewed (Borges et al. 2005). During the past few years, biotransformation of coumarin towards production of novel derivatives of biological significance has emerged as an important area of research (Eisenbrand et al. 2003; Vassallo et al. 2003). In this context, Agrobacterium rhizogenes-transformed hairy root cultures (HRCs) are, indeed, relevant. In addition to being envisioned to serve as a sustainable source of bioactive raw material for pharmaceutical industry HRCs can be viewed as an effective biotechnological tool for biotransformation through which new avenues are expected to open up towards production of altered compounds of higher pharmaceutical relevance. There has been an increasing interest in utilizing HRCs as biocatalysts to transform substrates into value-added products through reactions involving glucosylation, glycosylation, esterification, oxidation, reduction, hydroxylation and condensation (Banerjee et al. 2012). For example, glucosylation and other types of conversions of seven coumarin and flavone derivatives were carried out by HRCs of Pharbitis nil (syn. Ipomoea nil; Family: Convolvulaceae) and the biotransformed products were detected in the root mass (Kanho et al. 2004). HRCs of Polygonum multiflorum (Family: Polygonaceae) have been shown to possess the ability of causing glycosylation of hydroxycoumarins and other phenolic compounds (Yan et al. 2007; Yu et al. 2008). A more recent study by Zhou et al. (2012) demonstrated biosynthesis of nine corresponding glycosides including four new compounds in HRCs of P. multiflorum via glycosylation of nine hydroxycoumarins (including the natural esculetin) with different substituted groups employed as substrates. Such type of investigations stimulated us to explore the possibility of extending biotransformation in the transformed root cultures of H. enneaspermus towards production of novel coumarin-derived phytocompounds, hitherto unknown for natural parts of this plant species. Fundamental to achieving this objective is to induce and establish in vitro-optimized HRCs of H. enneaspermus through genetic transformation mediated by selected strains of A. rhizogenes. It was also important to ascertain the clonal fidelity among the rhizoclones and, more importantly, the genetic stability of rhizoclones and regenerated shoots through successive multiplication cycles with a view to sustainably harnessing the biosynthetic potential of HRCs and transformed plants in respect of coumarin production.
Materials and methods
In vitro shoot cultures
Stem nodal explants of H. enneaspermus obtained from the experimental garden of the Post-Graduate Department of Botany, Utkal University (Bhubaneswar, Odisha, India) were inoculated in Murashige and Skoog’s (1962) medium (MS) augmented with 0.25 mg l−1 6N-benzyladenine (BA) in combination with 0.1 mg l−1 indole-3-acetic acid (IAA) and gelled with 8 g l−1 agar–agar (Hi-Media, Mumbai, India). Axillary proliferated shoots were multiplied in vitro by repetitive nodal culture and maintained as renewable axenic shoot cultures (25 ± 1 °C, 16 h photoperiod at 50 μmol m−2 s−1 photon flux density (PFD).
Transformation, establishment of rhizoclones and plant regeneration
Different strains of A. rhizogenes namely A4, A4T, 8196 and LBA 9402 used in the present study and their culture maintenance was as described earlier (Swain et al. 2012a). Leaf or internode explants from the outdoor-grown donor plant after surface-disinfection, as well as those from in vitro-grown axenic shoot cultures (7–8 week-old), were used for transformation. Apical portions of the cut internodal surfaces and leaf midribs were treated with 10–30 μl of an overnight suspension of A. rhizogenes (pre-cultured with 100 µM acetosyringone (Sigma-Aldrich, USA). For causing infection, explants were pricked with a sterile hypodermic needle and bacterial suspension was placed in form of a droplet at the site of prick (5 μl droplet/prick). Explant were blotted dry and transferred to agar-gelled MS medium without plant growth regulator supplement (MS0) for a co-cultivation period of 3 days, after which they were transferred to MS0 containing 500 μg ml−1 cefotaxime. Each treatment comprised of ten replicate culture vessels (4–6 explants per culture vessel) and experiments were repeated thrice. All the flasks/jars containing infected explants were kept inside the culture room (25 ± 1 °C) under dark/diffused light with a low PFD (10 μmol m−2 s−1).
Roots (1.0–1.5 cm) that were developed along the inoculated surface were individually excised and each transferred to a 50 × 11 mm transparent plastic Petri dish (TPX; Tarson, Mumbai) containing 3.0–4.0 ml of agar-gelled MS0. Proliferated root clones (rhizoclones) were replicated in 250 ml conical flasks (Borosil, India) each containing 50 ml MS0 medium supplemented with 500 μg ml−1 cefotaxime. Roots were incubated at 25 ± 1 °C and 20 μmol m−2 s−1 PFD and subcultured every 4 weeks. Subsequently, root cultures were maintained in MS0 free of the antibiotic and root growth index (RGI) was determined (Swain et al. 2012a, b). Each rhizoclone was replicated in ten culture flasks and experiments were repeated thrice. PCR-tested rhizoclones of either origin (A4/8196) were selected for regeneration studies and they were transferred to a higher illumination (30–40 μmol m−2 s−1 PFD). The organogenic green parts of the transformed root clump bearing adventitious shoot buds were transferred to MS medium augmented with cytokinins (BA/Kinetin) at a range of concentrations (0.25–1.0 mg l−1) for stimulating organogenesis and shoot elongation. Elongated shoots were subjected to successive nodal multiplication at 4 week intervals using MS + 0.25 mg l−1 BA and maintained in vitro as renewable shoot cultures. Each treatment comprised ten replicates and repeated thrice.
Molecular analysis of rhizoclones and regenerated shoots
Prior to plant regeneration, transformed nature of randomly selected fast-growing hairy root lines (rhizoclones) were verified by PCR amplification of A. rhizogenes pRi T-DNA-specific rolA-B and mas2 ORFs in root genomic DNA according to the published protocol (Sahu et al. 2014). Assay for opines in extracts of transformed roots and leaves from regenerated plants of selected transformed rhizoclones of A4/8196 origin was carried out by paper electrophoresis following the published procedure (Pal et al. 2013). The test of clonal fidelity was carried out by RAPD analysis using random oligonucleotide primers (10-mer) from OPERON Series C (1–19) (Operon Tech, Alameda, USA) that were selected for RAPD analysis using a PCR thermocycler (GeneAmp PCR System 9700, Applied Biosystems, Singapore). Genetic stability of the superior rhizoclones of A4/8196-origin was verified through different multiplication cycles (up to 16th culture passages) by PCR amplification followed by electrophoresis. Transgene stability with respect to rolB and rolC gene expression was verified by reverse transcription-polymerase chain reaction (RT-PCR) analysis. Total RNA were isolated from root tissues of selected rhizoclones (A4/8196 origin) and leaf tissues of plants, regenerated from respective rhizoclones, through 4–16 multiplication cycles (4, 8, 12, 16 subculture passages) using RNA isolation kit (RNeasy Mini Kit, Cat. No. 74104, Qiagen, USA). RT-PCR was conducted using the Quantitect reverse transcription kit (Qiagen, USA, Cat. No. 205310) following the manufacturer’s instructions. The primer sequences for amplifying rolB and rolC cDNA were the same used by Sahu et al. (2013) and those for the actin gene (house keeping gene used as loading control) were according to Swain et al. (2012b). The RT-PCR products were electrophoresed on 1.5 % agarose gel and gel-documented.
HPLC quantification of coumarin
The selected rhizoclones derived from A4/8196 strains and their respective shoot regenerants were harvested after 4 weeks of in vitro culture. Besides, the aerial parts/roots of H. enneaspermus were collected from garden-grown/in vitro-grown non-transformed plants. Samples were extracted in methanol, concentrated to dryness and the concentrate dissolved in HPLC-grade acetonitrile (100 mg ml−1), filtered and injected (25 μl) to HPLC column. Coumarin standard (Ebiochem, Sanghai, China) was prepared in acetonitrile at the concentration of 1 μg μl−1. An Agilent Technology 1200 Series HPLC system (Agilent Technology, USA) coupled with diode array detector (DAD; DE64264226) and consisting of a quarternary pump (DE62976370), a vacuum degasser (JP94177339) and EZ Chrom Elite software was used for the study. Chromatographic separation was achieved on a Agilent Zorbax Eclipse XBD-C18 semi preparative analytical column (250 mm × 9.4 mm, 5 μm Purosphere; Agilent, USA). The following gradient was used for separation: 0 min, 20:80 (A:B); 20 min, 80:20 (A:B), with flow rate of 1 ml min−1 (A, acetonitrile; B, water containing 0.01 % orthophosphoric acid). The detection was performed at 220 nm. Validation of the analytical method for coumarin was examined for specificity, linearity, accuracy, and precision, limit of detection (LOD) and limit of quantitation (LOQ) following methods as described by Su et al. (2009).
Statistical analysis
All transformation experiments were set up in a completely randomized design (CRD). Each treatment for induction of hairy roots consisted of ten replicate culture vessels (250 ml conical flask/500 ml jar) each containing 4–6 explants (leaf/internode). Each hairy root clone (rhizoclone) was replicated in ten culture flasks and each experiment was repeated thrice for evaluating root biomass growth. For estimating coumarin content data were pooled from a total of 6 replicates for each tissue sample (root/aerial part). Data were processed using analysis of variance (ANOVA) for a completely randomized design. Duncan’s new multiple range test (DMRT) (Gomez and Gomez 1984) was used to separate the mean of significant effect.
Results
Relative transformation efficiency
Axillary shoots were developed from nodal segments (ca. 90 % proliferation frequency, 38 shoots/explants) and in vitro shoot cultures were established through successive nodal multiplication. Among the two types of explants used for transformation experiments i.e. leaf and internode, the former was more responsive regardless of A. rhizogenes strains employed or explant source (Fig. 1a, b). Explants from garden-grown plants were less susceptible to Agrobacterium than in vitro-grown cultures on the basis of rhizogenic response and %transformation. In MS basal medium devoid of PGRs (MS0), hairy roots emerged at the pricked midrib sites of leaf explants from in vivo/in vitro sources within 6–8 days/5–7 days respectively for A4-treated explants, whereas those treated with 8196 strain required 10–12 and 8–10 days respectively for the same (Fig. 2a, b). In vivo internodes took 12–14 days (A4-treated) or 14–16 days (8196-treated) to develop roots. A relatively short period was taken for root emergence from in vitro internode explants i.e. 10–12 or 12–14 days in response to treatment with A4 or 8196 strain respectively. Leaves from in vitro cultures gave the best response compared to in natura leaves or internodes regardless of source and A. rhizogenes strain type. The percentage rhizogenic response of A4-treated in vivo leaf, in vitro leaf, in vivo internode and in vitro internode explants were 93.3, 100, 43.3 and 66.6 respectively. In comparison, explants treated with 8196 had at a lower percentage response i.e. 73.3, 80, 26.6 and 43.3 for respective explant categories.
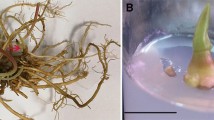
Plant regeneration from hairy root cultures of H. enneaspermus induced by A. rhizogenes. a Hairy root induction in a leaf midrib 10 days after infection with A4 strain. b Hairy root induction in a leaf midrib 14 days after infection with 8196 strain c growth of rhizoclone A4-HRL-2B7 on MS0 agar medium after 6 weeks of single root culture (cloning), d growth of rhizoclone 8196-HRL-3C6 on MS0 medium after 6 weeks of single root culture (cloning). e Proliferation of shoots from A4-HRL-2B7 rhizoclone after 6 weeks culture on MS + 0.2 mg l−1 BA. f Proliferation of shoots from 8196-HRL-3C6 rhizoclone after 6 weeks culture on MS + 0.2 mg l−1 BA
Establishment of transformed hairy root cultures as rhizoclones
Hairy roots developed in explant types (leaf/internode) from different sources (in vivo/in vitro) using A. rhizogenes strain types (A4/8196) were maintained separately as different somaclones (rhizoclones) in MS basal medium lacking plant growth regulators (MS0). The percentage survival was 100 % in all the transformed rhizoclones after subculture. A4-induced rhizoclones grew luxuriously with fast and extensive lateral branching within 5–6 weeks generating higher biomass compared to root clones incited by 8196 strain. The growth rate of A4 and 8196 induced hairy roots enhanced after subculture. A4 induced rhizoclones had the same growth rate during subsequent cycles of in vitro multiplication, each cycles being of 4 week period. However, for rhizoclones incited by 8196, growth efficiency declined after the 16th subculture. Of a total of 104 rhizoclones of A4 origin and 76 rhizoclone of 8196 origin, a sample of 16 clones were selected including eight clones pertaining to each strain type on the basis of sustained growth and biomass generation. Eight fast-proliferating rhizoclones from each strain i.e. A4 (A4-HRL-1A3, A4-HRL-2B7, A4-HRL-2B10, A4-HRL-4C7, A4-HRL-5E4, A4-HRL-6F3, A4-HRL-7G8, A4-HRL-3H2) and 8196 (8196-HRL-1A8, 8196-HRL-3C6, 8196-HRL-3C9, 8196-HRL-4D5, 8196-HRL-5E8, 8196-HRL-7F2, 8196-HRL-2G3, 8196-HRL-7H4) were further evaluated to identify the best rhizoclone of respective strain. Among these, A4-HRL-2B7 and 8196-HRL-3C6 exhibited the fastest growth and proliferation (Figs. 2c, d, 3). The hairy root somaclones derived from A4 grew luxuriously with fast and extensive lateral branching up to 5–6 weeks resulting in the highest biomass (rhizoclone A4-HRL-2B7, 15.5 g.f.wt.), whereas 8196 induced hairy root somaclones grew at a relatively slow rate (rhizoclone 8196-HRL-3C6, 9.5 g.f.wt.). During this period, the best two rhizoclones i.e. A4-HRL-2B7, 8196-HRL-3C6, attained maximum biomass displaying mean root growth index (RGI) values of 30 and 18 respectively. After 8 weeks of continuous growth (without subculture), hairy roots gradually changed their colour from white to brown and showed a decelerating growth rate associated with reduced branching ability irrespective of the strain type. Therefore, selected rhizoclones were subcultured at a regular interval of 4 weeks to ensure sustained growth.
Comparative growth and proliferation of selected rhizoclones incited by different A. rhizogenes strains (A4/8196) after 6 weeks of culture period (Rhizoclones represented by column bars, marked with a different alphabet are significantly different at p ≤ 0.05 as determined by Duncan’s new multiple range test: DMRT)
PCR detection of pRi T-DNA genes in rhizoclones
Genomic DNA isolated from several independent transformed hairy root clones (A4/8196), capable of showing auxin-independent growth, revealed the expected amplification product of 1540 bp specifying A4 rolA-B gene (Fig. 4a). This indicated the presence of the Agrobacterium ORFs (pRi T L -DNA) in the recipient plant genome via genetic transformation. Besides this, transformation was also ascertained by using the primer designed for mas2 gene which gave the predictable amplified products of 512 bp (Fig. 4b). Amplification product was not detected in DNA from untransformed plants when subjected to PCR amplification with the gene primers specific to the rolA-B or mas2 gene sequences.
PCR amplifications demonstrating T-DNA genes (rol/opine synthase genes) in selected rhizoclones resulted from transformation with A. rhizogenes A4/8196 strain. a PCR amplification of rolAB gene in A4-induced rhizoclones M: 1 kb ladder, PC: positive control (pRi A4), lane 1: Natural leaves from donor plant, lane 2–8: A4 rhizoclones (A4-HRL-1A3, A4-HRL-2B7, A4-HRL-2B10, A4-HRL-4C7, A4-HRL-5E4, A4-HRL-6F3, A4-HRL-2G3). b PCR amplification of mas2 gene; M: Medium range ruler, PC: Positive control (pRi 8196), lane 1: Natural leaves from donor plant, lane 2–8: 8196 rhizoclones (8196-HRL-1A8, 8196-HRL-3C6, 8196-HRL-3C9, 8196-HRL-4D5, 8196-HRL-5E8, 8196-HRL-7F2, 8196-HRL-2G3)
Plant regeneration from transformed rhizoclones
After 4–5 weeks of culture in MS0 the central portion of the transformed root clump of selected rhizoclones (A4-HRL-2B7 and 8196-HRL-3C6) turned green upon exposure to higher PFD (30–40 μmol m−2 s−1). 1 week later, shoot buds emerged but only a few of them developed into shoots (3–5 shoots per culture flask) at the end of the 8th week of culture. Following transfer of the organogenic greeny part of the rhizoclone with transformed shoot buds to MS medium augmented with 0.25 mg l−1 BA shoot proliferation was stimulated. About 30–40/10–15 transformed shoots were developed from each root clump of A4-HRL-2B7 or 8196-HRL-3C6 rhizoclones respectively within 6 weeks of transfer (Fig. 2e, f). Kinetin (0.5 mg l−1) augmented to MS medium also elicited shoots with fully-expanded leaves, though shoot number was less than that of BA (0.25 mg l−1). The primary transformed shoots were multiplied by in vitro culture of nodal explants via successive culture passages. The well-grown transformed shoots got rooted spontaneously without any supplement of auxins to the MS basal medium (MS0).
Opine gene expression in rhizoclones and transformed plants
Extracts of root tissues from the transformed rhizoclones and leaf tissues from regenerated plants subjected to paper electrophoresis developed silver nitrate positive blackish-brown spots along the electropherogram (Fig. 5). Root/leaf tissues of all examined rhizoclones contained a considerable amount of compounds which had the same mobility as authentic agropine and/or as additional compounds(s) with a lower mobility representing a combination of mannopine + mannopinic acid, the opine chain precursors of agropine. Agropine along with mannopine was evident in A4-induced rhizoclones and regenerated plants derived from them; whereas mannopine was the single predominating compound in all rhizoclones induced by 8196 strain or plants thereof. None of these compounds was evident in root extracts of a non-transformed in vitro regenerated plant.
Paper electropherogram showing opines in extracts of A4 and 8196 rhizoclones and in respective regenerated plants. S1, Agropine standard; NR, non-transformed roots (roots from in vitro non-transformed plant); lane 1–2: Root extracts of A4 rhizoclone (A4-HRL-2B7, A4-HRL-2B10); lane 3–4: Leaf extracts of plants regenerated from A4-HRL-2B7 and A4-HRL-2B10 rhizoclones respectively; lane 5: Root extract of 8196 rhizoclone (8196-HRL-3C6); lane 6: Leaf extract of a plant regenerated from 8196-HRL-3C6 rhizoclone, S2: Mannopine standard, A agropine, M mannopine, NS neutral sugar
Clonal fidelity of rhizoclones and shoot regenerants
To ascertain whether or not the rhizoclones induced by A4/8196 strains were genetically identical, random amplified polymorphic DNA (RAPD) marker analysis was used in this study. Of a total of 104 rhizoclones of A4 origin and 76 rhizoclones of 8196 origin, 40 randomly selected rhizoclones of either type were subjected to the test of clonal fidelity. Selected 19 different random decamer primers such as OPC01-OPC19 from OPC-series (Operon Tech, Alameda, USA) (Supplementary Table 1) were used on the basis of robustness of amplification, reproducibility and scorability of bands. A total of 165 scorable bands were generated by the length of amplification products ranging 150–2500 bp (Supplementary Table 1). The number of bands per primer varied from 5 to 17 with an average of 8.68 bands per primer. None of the primers was able to generate polymorphic bands in A4-induced rhizoclones. However, OPC-4 and OPC-19 produced DNA polymorphism in 2 different rhizoclones originated from 8196 source. The number of monomorphic bands was the highest i.e. 17 in case of primer OPC-16 (ranging from 150 to 1815 bp) and the lowest i.e. 5 in case of OPC-3 (ranging from 500 to 200 bp). Primer OPC-4 and OPC-19 gave rise to second highest of 12 monomorphic bands with fragment size ranging from 150 to 1815 and 400 to 2000 bp respectively whereas OPC-2 resulted 11 monomorphic bands ranging 150–1815 bp. The highest amplified band size of 2500 bp was recorded using OPC-12. The RAPD banding pattern showing absolute (100 %) DNA monomorphism (zero polymorphic DNA) was observed in all 16 fast-growing rhizoclones (8 each of A4/8196 origin) and plant respective plant regenerants indicating genetic integrity (Supplementary Table 2). The two deviant rhizoclones (8196-HRL-7H8 and 8196-HRL-5I4) showing polymorphic bands failed to grow further and eventually turned brown and necrotic (data not shown).
Coumarin content of rhizoclones and transformed shoots
Coumarin content of the transformed root samples and leafy shoots of regenerated plants of H. enneaspermus were determined by HPLC technique. These were compared with that of in vivo roots/aerial parts from naturally occurring plants and in vitro non-transformed roots/aerial parts (control) (Table 1). In vivo roots had the least coumarin content (0.282 mg g−1 d.wt. extract) among all other tissue samples analyzed. Aerial part samples of either origin (transformed/non-transformed plants) exhibited asuperiority over their respective root samples on the basis of significantly higher coumarin accumulation. In comparison with that of in vivo/in vitro non-transformed root samples coumarin content was 2.6- to 2.9-fold higher in A4-rhizoclone (A4-HRL-2B7), whereas the 8196-rhizoclone (8196-HRL-3C6) had only a marginally superior accumulation (Table 1). Transformed shoots of A4 origin exhibited the highest coumarin content (3.258 mg g−1 d.w. extract) which was significantly higher than that of aerial part samples of in vivo/in vitro origin.
Eight superior rhizoclones each of A4 and 8196 origin were selected on the basis of biomass production and their coumarin biosynthetic potential was compared. There was a distinct inter-rhizoclonal variation in coumarin content. It was observed that the fast-growing, late-maturing rhizoclones accumulated a higher coumarin content than the slow-growing and early maturing clones. Among the 16 selected rhizoclones, A4-HRL-2B7 with highest biomass growth had the maximum coumarin content (0.82 ± 0.022 mg g−1 d.wt. extract) among all root samples.
Genetic stability of rhizoclones and transformed plants
Genetic stability of the superior rhizoclones, one each of A4-origin (A4-HRL-2B7) and 8196-origin (8196-HRL-3C6), and their respective plant regenerants was indicated by RAPD amplification of DNA from root/leaf tissues obtained through 16 successive multiplication cycles displaying absolute homogeneity in the banding pattern. DNA monomorphism was revealed in rhizoclones and regenerated shoots through 4–16 cycles (4, 8, 12, 16 subculture passages) using the decamer primers OPC-4 (Fig. 6) and OPC-19 (Fig. 7) for A4 and 8196-derived plant materials (root/leaf tissues) respectively. Sustained RT-PCR amplification of rolB and rolC genes was demonstrated in the two selected rhizoclones and respective transformed plants through culture passages indicating transgene stability in terms of transcriptional activity of rol genes (Fig. 8 a, b).
Determination of genetic stability of A4-HRL-2B rhizoclone, and plants regenerated thereof, during successive multiplication cycles (4th–16th) using OPC-4 operon primer for RAPD analysis. M: low range DNA ruler Plus (GeNei-Merck, Mumbai, India), lane 0: DNA from primary transformed hairy root culture, lane 1–4: root DNA from respective subculture passages (4th, 8th, 12th, 16th); lane 5–8: leaf DNA of regenerated plants from respective subculture passages (4th, 8th, 12th, 16th)
Determination of genetic stability of 8196-HRL-3C6 rhizoclone, and plants regenerated thereof, during successive multiplication cycles (4th–16th) using OPC-19 operon primer for RAPD analysis. M: low range DNA ruler Plus (GeNei-Merck, Mumbai, India), lane 0: DNA from primary transformed hairy root culture, lane 1–4: root DNA from respective subculture passages (4th, 8th, 12th, 16th); lane 5–8: leaf DNA of regenerated plants from respective subculture passages (4th, 8th, 12th, 16th)
RT-PCR based rol gene expression in selected rhizoclones resulted from transformation with A. rhizogenes (A4/8196), and plants regenerated thereof, during successive multiplication cycles (4th–16th). a rolB gene (A4-HRL-2B rhizoclone) b rolC gene (8196-HRL-3C6 rhizoclone). Upper panel lanes 1–4: Root DNA from respective subculture passages (4th, 8th, 12th, 16th); Upper panel lanes 5–8: Leaf DNA of regenerated plants from respective subculture passages (4th, 8th, 12th, 16th). The lower panel lanes 1–8 shows the amplification of actin used as a loading control
The coumarin accumulation in A4 and 8196 induced rhizoclones (selected 8 each) and in vitro multiplied shoots in different culture passages (4th, 8th, 12th, 16th) covering a period of 4–16 months were analyzed. Both A4 and 8196 derived rhizoclones and shoots regenerated thereof showed a stable range of coumarin content through successive culture passages (Figs. 9, 10). Accumulation of coumarin did not vary significantly among the culture passages although A4-elicited rhizoclones maintained their superiority over those incited by 8196 in respect of coumarin content. There was no significant difference in respect of coumarin content in rhizoclone-derived in vitro shoots through corresponding multiplication cycles (4th–16th).
Genetic stability among selected rhizoclones incited by A. rhizogenes strains (A4/8196) in respect of coumarin production through successive culture passages (CP) each of 4 weeks duration. Data pooled from a total of six replicates for root tissue sample of each rhizoclone (column bars, each representing a culture passage viz. 4th/8th/12th/16th of each rhizoclone, marked with the same alphabet are not significantly different at p ≤ 0.05 as determined by Duncan’s new multiple range test: DMRT)
Genetic stability among shoot regenerants from selected rhizoclones incited by A. rhizogenes strains (A4/8196) in respect of coumarin production through successive culture passages (CP) each of 4 weeks duration. Data pooled from a total of six replicates for shoot tissue sample of each rhizoclone (column bars, each representing a culture passage viz. 4th/8th/12th/16th of each rhizoclone, marked with the same alphabet are not significantly different at p ≤ 0.05 as determined by Duncan’s new multiple range test: DMRT)
Discussion
Agrobacterium rhizogenes–mediated transformation efficiencies (based on days to root emergence, %rhizogenesis, roots per explant) of H. enneaspermus varied with respect to explant type and explant source. Leaf explants were found to be markedly more susceptible compared to stem internodal segments irrespective of explant source or Agrobacterium strains used. Although this corroborates with that reported for Gentiana macrophylla (Tiwari et al. 2007) and Picrorhiza kurroa (Verma et al. 2007), it disagrees with that for Clitoria ternatea (Swain et al. 2012a) and Amaranthus spinosus (Pal et al. 2013) in which internodal explants were more responsive. In our experiments with H. enneaspermus in vitro explants (leaf/internode) exhibited a superior response than those obtained from naturally grown plant sources. The varying susceptibility among plant species or between different explant types was ascribed to the auxin and sucrose content (Nilsson and Olsson 1997). The underlying mechanism of how the gene expression, age and endogenous hormonal balance of host tissue may affect the transformation frequencies is not well understood at present and warrants further investigation.
In H. enneaspermus, the agropine-type A4 strain induced a higher frequency of transformation compared to the manopine-type 8196 strain on the basis of earlier emergence and growth of hairy roots. The superior pathogenicity of A4 over other strains, observed in the present study, is in consonance with earlier reports on Pelargonium sp. (Saxena et al. 2007), P. kurroa (Verma et al. 2007) and Clitoria ternatea (Swain et al. 2012a); albeit contrary to that on Torenia fournieri (Tao and Li 2006) and Hyoscyamus sp. (Akramian et al. 2008). Differential expression of chromosomal virulence genes chvD-E of Agrobacterium and of the virulence regulon on the Ti/Ri plasmid are important internal factors influencing the infecting ability of A. rhizogenes (Tiwari et al. 2007). A constraint often met in Agrobacterium-mediated plant transformation perhaps owes to a difficult-to-predict pattern of T-DNA integration in the host genome and subsequent gene expression. The focal basis of inconsistency in T-DNA gene expression in plants could be attributed to the site of T-DNA integration into the plant genome (position effect) and/or copy number (Koprek et al. 2001; Kohli et al. 2003).
A. rhizogenes pRi T-DNA rolA, rolB and rolC oncogenes have been considered to be modulators of plant growth and cell differentiation. Though each of these genes, on its own, is capable to promote root formation yet they differ in their efficiency; for example, rolB is more efficient than rolA or rolC (Bulgakov 2008). In the present study, transformed nature of resulting rhizoclones of H. enneaspermus was ascertained by positive PCR amplification of rolA-B genes borne on pRi T-DNA segment. Positive PCR-based detection of T-DNA borne rol genes in hairy root genome have been reported in several plants of pharmaceutical or nutraceutical relevance (Sudha et al. 2013; Majumdar et al. 2012; Pal et al. 2013). Besides, an opine synthase gene, particularly mas2 coding for mannopine synthase has been detected in HRCs of H. enneaspermus as in a few others plant species including C. ternatea (Swain et al. 2012a, b) and Plumbago zeylanica (Nayak et al. 2015).
Production of transformed plants from HRCs has been reported in a few plant species, in which plant regeneration occurred either spontaneously or induced by PGRs. Similar to our observation in H. enneaspermus, spontaneous induction of shoot buds was also recorded in many plant species such as Taraxacum platycarpum (Lee et al. 2004), Centaurium erythraea (Subotic et al. 2003), Tylophora indica (Chaudhuri et al. 2006), Pelargonium graveolens cv. Hemanti (Saxena et al. 2007), Plumbago indica (Gangopadhyay et al. 2010), Bacopa monnieri (Majumdar et al. 2011) etc. In the present study, exogenous supplement of 0.25 mg l−1 BA not only enhanced shoot bud production frequency (100 %) but also, more importantly, stimulated shoot elongation. Shoot bud development from hairy root cultures (HRCs) was reported using MS with BA as the sole PGR supplement at varied concentrations viz. 1.0 mg l−1 (Taraxacum platycarpum, Lee et al. 2004), 2.0 mg l−1 (Plumbago rosea, Satheeshkumar et al. 2009), 10 mg l−1 (Ajuga reptans, Uozumi et al. 1996). A combination of 1.0 mg l−1 BA and 0.1 mg l−1 NAA (Rauwolfia serpentina, Mehrotra et al. 2013) or 2.0 mg l−1 zeatin (Amaranthus spinosus, Pal et al. 2013) was stimulatory for direct plant regeneration from hairy roots.
It was interesting to observe in the present study on H. enneaspermus that the coumarin content, as quantified by HPLC technique, was ca. threefold higher in one of the A4-derived superior rhizoclone compared to that in natural roots. Furthermore, the leafy shoots regenerated from the selected rhizoclone had a significantly higher coumarin accumulation than aerial parts of naturally occurring plants. Anticipated advances especially by designing and establishing efficient, cost-effective bioreactor configurations, should lead to commercialization via scaling-up of productivity of H. enneaspermus HRCs, thereby enabling them to offer their fullest potential as a sustainably exploitable source of natural coumarin group of compounds or their novel derivatives following biotransformation.
Verification of genetic integrity of Agrobacterium-induced rhizoclones is critically important to ensure sustained growth and production of root-specific bioactive compounds, thereby accounting for a consistent biotechnological application of transformed HRCs. Our results with H. enneaspermus demonstrated not only an absolute clonal fidelity among the rhizoclones but more importantly genetic stability of superior rhizoclones and their plant regenerants from respective cycles of cultures, using RAPD technique. In addition, transgene stability in respect of rolB and rolC gene expression was also sustained in the selected rhizoclones and respective transformed plants through 16 multiplication cycles. PCR amplification of rol genes has recently been used as an indicator of long-term expression stability in a hairy root clone of Rauvolfia serpentina (Pandey et al. 2014). Genetic stability of plants derived from encapsulated hairy roots of P. kurrooa has been demonstrated indicating a low frequency of DNA polymorphism i.e. 5.2 and 3.6 % using RAPD and ISSR primers respectively (Rawat et al. 2013). Perhaps, the apparent absence of genetic distortion through culture passages owes to rapid proliferation rate of HRCs and regenerated shoots in simple nutrient media without/with a low PGR supplement. The absence of significant variation in coumarin accumulation in rhizoclones through successive culture passages over a period of 16 months reflects long-term stability of rol gene expression.
Conclusion
In essence, the paper describes establishment of Agrobacterium rhizogenes-mediated transformed hairy root somaclones (rhizoclones) of a rare medicinal herb, Hybanthus enneaspermus, and regenerated shoots thereof, which were exploited as a renewable source of the health-proactive compound, coumarin. Further, we have demonstrated the genetic stability of rhizoclones associated with a sustained retention of their biosynthetic potential for coumarin production over a prolonged period of time through successive in vitro multiplication cycles (culture passages). Such an endeavour will help ensure long-term sustainable production of bioactive phytochemicals implicated in human healthcare.
References
Akramian M, Tabatabaei SMF, Mirmasoumi M (2008) Virulence of different strains of Agrobacterium rhizogenes on genetic transformation of four Hyoscyamus species. Am Euras J Agric Environ Sci 3:759–763
Amuthapriya D, Ranganayaki S, Suganya D (2011) Phytochemical screening and antioxidant potential of Hybanthus enneaspermus: a rare ethnobotanical herb. J Pharm Res 4:1497–1502
Banerjee S, Singh S, Rahman LU (2012) Biotransformation studies using hairy root cultures—a review. Biotechnol Adv 30:461–468
Borges F, Roleira F, Milhazes N, Santana Uriarte E (2005) Simple coumarins and analogues in medicinal chemistry: occurrence, synthesis and biological activity. Curr Med Chem 12:887–916
Bulgakov VP (2008) Functions of rol genes in plant secondary metabolism. Biotechnol Adv 26:318–324
Chaudhuri KN, Ghosh B, Tepfer D, Jha S (2006) Spontaneous plant regeneration in transformed roots and calli from Tylophora indica: changes in morphological phenotype and tylophorine accumulation associated with transformation by Agrobacterium rhizogenes. Plant Cell Rep 25:1059–1066
Eisenbrand G, Otteneder M, Tang W (2003) Synthesis of N-acetyl-S-(3-coumarinyl)-cysteine methyl ester and HPLC analysis of urinary coumarin metabolites. Toxicology 190(3):249–258
Gangopadhyay M, Chakraborty D, Bhattacharyya S, Bhattacharya S (2010) Regeneration of transformed plants from hairy roots of Plumbago indica. Plant Cell Tissue Organ Cult 102:109–114
Gomez KA, Gomez AA (1984) Statistical procedure for agricultural research. Wiley, New York, p 680
Kanho H, Yaoya S, Itani T, Nakane T, Kawahara N, Takase Y (2004) Glucosylation of phenolic compounds by Pharbatis nil hairy roots. Biosci Biotechnol Biochem 68:2032–2039
Kirtikar KR, Basu BD (1991) Indian Medicinal Plants. Vol 4, B/S Bishen Singh Mahendra Pal Singh, Dehradun
Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E, Christou P (2003) Transgene integration, organization and interaction in plants. Plant Mol Biol 52:247–258
Koprek T, Rangel S, McElroy D, Louwerse JD, Williams-Carrier RE, Lemaux PG (2001) Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol 125:1354–1362
Krishnamoorthy BS, Nattuthurai N, Logeshwari R, Dhaslima Nasreen H, Syedali Fathima I (2014) Phytochemical study of Hybanthus enneaspermus (Linn.) F. Muell. J Pharmacogn Phytochem 3(1):6–7
Lee MH, Yoon ES, Jeong JH, Choi YE (2004) Agrobacterium rhizogenes-mediated transformation of Taraxacum platycarpum and changes of morphological characters. Plant Cell Rep 22:822–827
Majumdar S, Garai S, Jha S (2011) Genetic transformation of Bacopa monnieri by wild type strains of Agrobacterium rhizogenes stimulates production of bacopa saponins in transformed calli and plants. Plant Cell Rep 30:941–954
Majumdar S, Garai S, Jha S (2012) Use of the cryptogein gene to stimulate the accumulation of bacopa saponins in transgenic Bacopa monnieri plants. Plant Cell Rep 31:1899–1909
Mehrotra S, Goel MK, Rahman LU, Kukreja AK (2013) Molecular and chemical characterization of plants regenerated from Ri-mediated hairy root culture of Rauwolfia serpentina. Plant Cell Tissue Organ Cult 114:31–38
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15:473–497
Nathiya S, Senthamil Selvi R (2013) Anti-fertility effect of Hybanthus enneaspermus on endosulfan induced toxicity in male rats. Int J Med Biosci 2(1):28–32
Nayak P, Sharma M, Behera SN, Thirunavoukkarasu M, Chand PK (2015) High-performance liquid chromatographic quantification of plumbagin from transformed rhizoclones of Plumbago zeylanica L.: inter-clonal variation in biomass growth and plumbagin production. Appl Biochem Biotechnol 175:1745–1770
Nilsson O, Olsson O (1997) Getting to the root: the role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol Plant 100:463–473
Pal A, Swain SS, Mukherjee AK, Chand PK (2013) Agrobacterium pRi TL-DNA rolB & TR-DNA opine genes transferred to the spiny amaranth (Amaranthus spinosus L.)—a nutraceutical crop. Food Technol Biotechnol 51:26–35
Pandey P, Kaur R, Singh S, Chattopadhyay SK, Srivastava SK, Banerjee S (2014) Long-term stability in biomass and production of terpene indole alkaloids by hairy root culture of Rauvolfia serpentina and cost approximation to endorse commercial realism. Biotechnol Lett 36:1523–1528
Patel DK, Kumar R, Prasad SK, Sairam K, Hemalatha S (2011) Antidiabetic and in vitro antioxidant potential of Hybanthus enneaspermus (Linn) F. Muell in streptozotocin-induced diabetic rats. Asian Paci J Trop Biomed 1:316–322
Rawat JM, Rawat B, Mehrotra S (2013) Plant regeneration, genetic fidelity, and active ingredient content of encapsulated hairy roots of Picrorhiza kurrooa Royle ex Benth. Biotechnol Lett 35:961–968
Sahu L, Jena S, Swain SS, Sahoo S, Chand PK (2013) Agrobacterium rhizogenes-mediated transformation of a multi-medicinal herb, Boerhaavia diffusa L.: optimization of the process and anti-microbial activity against bacterial pathogens causing urinary tract infections. Front Life Sci 7:197–209
Sahu L, Ray DK, Chand PK (2014) Proton induced X-ray emission (PIXE) technique for determining multi-element composition of transformed hairy root cultures of Boerhaavia diffusa L.: an important medicinal herb. J Radioanal Nucl Chem 300:345–354
Satheeshkumar K, Jose B, Soniya EV, Seeni S (2009) Isolation of morphovariants through plant regeneration in Agrobacterium rhizogenes induced hairy root cultures of Plumbago rosea L. Indian J Biotechnol 8:435–441
Saxena G, Banerjee S, Rahman L, Verma PC, Malavarapu GR, Kumar S (2007) Rose-scented geranium (Pelargonium sp.) generated by Agrobacterium rhizogenes mediated Ri-insertion for improved essential oil quality. Plant Cell Tissue Organ Cult 90:215–223
Su J, Zhang C, Zhang E, Shen Y-H, Li H-L, Liu R-H, Zhang X, Hu X-L, Zhang W-D (2009) Qualitative and quantitative determination of the major coumarins in Zushima by an high performance liquid chromatography with diode array detector and mass spectrometry. J Chromatogr A 1216:2111–2117
Subotic A, Budimir S, Grubišic D, Momcilovic I (2003) Direct regeneration of shoots from hairy root cultures of Centaurium erythraea inoculated with Agrobacterium rhizogenes. Biol Plant 47:617–619
Sudha CG, Sherina TV, Anand VPA, Reji JV, Padmesh P, Soniya EV (2013) Agrobacterium rhizogenes mediated transformation of the medicinal plant Decalepis arayalpathra and production of 2-hydroxy-4-methoxy benzaldehyde. Plant Cell Tissue Organ Cult 112:217–226
Swain SS, Sahu L, Pal A, Barik DP, Pradhan C, Chand PK (2012a) Hairy root cultures of butterfly pea (Clitoria ternatea L.): Agrobacterium × plant factors influencing transformation. World J Microbiol Biotechnol 28:729–739
Swain SS, Rout KK, Chand PK (2012b) Production of triterpenoid anti-cancer compound taraxerol in Agrobacterium-transformed root cultures of butterfly pea (Clitoria ternatea L.). Appl Biochem Biotechnol 168:487–503
Tao J, Li L (2006) Genetic transformation of Torenia fournieri L. mediated by Agrobacterium rhizogenes. South Afr J Bot 72:211–216
Thenmozhi DC, Premalashmi V (2011) Antioxidant effect of Hydroethanolic extract of Hybanthus on paracetamol induced oxidative stress in albino rats. Int J Pharm Biomed Sci 2(3):1285–1287
Tiwari RK, Trivedi M, Guang ZC, Guo GQ, Zheng GC (2007) Genetic transformation of Gentiana macrophylla with Agrobacterium rhizogenes: growth and production of secoiridoid glucoside gentiopicroside in transformed hairy root cultures. Plant Cell Rep 26:199–210
Uozumi N, Ohtake Y, Nakashimada Y, Morikawa Y, Tanaka N, Kobayashi T (1996) Efficient regeneration from gus-transformed Ajuga hairy root. J Ferment Bioeng 81:374–378
Vassallo JD, Morrall SW, Fliter KL, Curry SM, Daston GP, Lehman-Mckeeman LD (2003) Liquid chromatographic determination of the glutathione conjugate and ring-opened metabolites formed from coumarin epoxidation. J Chromatogr B 794:257–271
Verma PC, Rahman LU, Nagi AS, Jain DC, Khanuja SPS, Banerjee S (2007) Agrobacterium rhizogenes-mediated transformation of Picrorhiza kurroa Royle ex Benth.: establishment and selection of superior hairy root clone. Plant Biotechnol Rep 1:169–174
Wealth of India (1959) A dictionary of Indian raw materials and industrial products (vol 5). Publication and information Directorate, Council of Scientific and Industrial Research (CSIR), New Delhi
Yan CY, Yu RM, Zhang Z, Kong LY (2007) Biotransformation of 4-Hydroxybenzen derivatives by hairy root cultures of Polygonum multiflorum Thunb. J Integr Plant Biol 49(2):207–212
Yu RM, Zhou LB, Yan CY, Duan GY, Zhao Y (2008) Two new coumarin glucosides biosynthesized by transgenic hairy roots of Polygonum multiflorum. Chin Chem Lett 19(1):76–78
Zhou L, Tian T, Xue B, Song L, Liu L, Yu R (2012) Biosynthesis of coumarin glucosides by transgenic hairy roots of Polygonum multiflorum. Biosci Biotechnol Biochem 76(5):1008–1010
Acknowledgments
We thank Dr. David Tepfer of Laboratoire de Biologie de la Rhizosphere, Institut National de la Rechereche Agronomique, Versailles, Cedex, France) and Dr. M. R. Davey of School of Biological Sciences, University of Nottingham, Nottingham, UK for having kindly provided the Agrobacterium rhizogenes strains. Facility for HPLC analysis was kindly provided by Dr. S. Peruncheralathan, Reader-F and the Director, National Institute of Science Education and Research (Department of Atomic Energy, Government of India), Bhubaneswar-751 005, India. Funding support by the University Grants Commission (UGC), New Delhi, India through a UGC Research Award Project to PKC is gratefully acknowledged. P. R. B. acknowledges National Medicinal Plant Board, New Delhi, India for the receipt of a research fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Behera, P.R., Jena, R.C., Das, A. et al. Genetic stability and coumarin content of transformed rhizoclones and regenerated plants of a multi-medicinal herb, Hybanthus enneaspermus (L.) F. Muell.. Plant Growth Regul 80, 103–114 (2016). https://doi.org/10.1007/s10725-016-0145-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0145-3