Abstract
Low silicon rice gene 1 (Lsi1) belongs to a member of Nod26-like major intrinsic protein (NIP) subfamily and is thought to control silicon (Si) accumulation in rice. In order to further elucidate its regulatory mechanisms in the defense of rice plants to enhanced ultraviolet-B (UV-B) radiation stress, Lsi1 was subjected to suppressed and overexpressed treatments in a UV-B tolerance rice accession Lemont as well as to overexpressed treatment in a UV-B sensitive rice accession Dular. The results showed that transcript levels of Lsi1 increased in Lsi1-overexpressed transgenic lines of rice accessions Lemont and Dular, but down-regulated in Lsi1-RNAi transgenic line of Lemont in comparison with their wild types (WT). A similar tendency was found in the root Si uptake and the Si concentrations in leaves. Further, the different transgenic rice lines and their WT were exposed to enhance UV-B radiation, study by suppression subtractive hybridization (SSH) found that Lsi1 not only could regulate the expression of phenylalanine ammonia-lyase (PAL), 4-coumarate-CoA ligase (4CL) and photolyase (PL), but also could induce other signal transduction-, detoxification-, resistance-, and photosynthesis-related genes. The findings suggested that the regulation of silicon nutrient by enhancing/inhibiting expression of Lsi1 could effectively induce the transcription of the mRNAs relative to tolerance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Depletion of the stratospheric ozone (03) layer due to the emission of chlorofluorocarbons and other trace gases has resulted in increased solar ultraviolet-B (UV-B: 280–315 nm) radiation on the earth’s surface (Ganguly et al. 2009; Blumthaler and Ambach 1990), which would reduce aboveground biomass and plant height, and increased DNA damage, and the effects of UV-B radiation on plants showed that they responded similarly in Arctic, Antarctic and lower latitudes (Newsham and Robinson 2009). Thus, there is an urgent need to find out effective ways to alleviate such damages on plant growth and development
Silicon (Si) is the second most prevalent element in the soil. Although Si is not considered to be an essential element for higher plants, it is beneficial to plant growth and contributes to plant defense against many biotic and abiotic stresses, such as salt tolerance (Liang et al. 2006), drought tolerance (Gong et al. 2005) and tolerance to toxic metals in plant (Liang et al. 1994; Epstein 1999; Ma 2004; Ma and Yamaji 2006). Si is considered as a modulator in enhancing plant resistance. Improvement of Si nutrient could invoke plant produce phenolics and phytoalexins, which could increase plant’s capability to defend stressful factors (Fawe et al. 1998).The related mechanism was that Si might decrease the oxidative damage in plants subjected to environmental stresses (Saqib et al. 2008). The roles of Si in enhancing plant tolerance to biotic stresses (e.g. fungal diseases) are also attributed partly to Si-enhanced antioxidant defense capacity. It deposited beneath the cuticle to form a cuticle-Si double layer which acts as a physical barrier to avoid fungal penetration, and supplement with Si in a Magnaporthe grisea-infected susceptible rice resulted in accumulation of the momilactone phytoalexins (Rodrigues et al. 2003) and up-regulated glucanase, peroxidase and PR-1 transcripts (Rodrigues et al. 2005).
Si may also help alleviate physical stress caused by UV. Our previous study (Wu et al. 2007) has found that the amount of silica on the adaxial surface in UV-B tolerance rice Lemont was higher than that in the UV-B sensitive type Dular, and showed increasing tendency in the tolerance rice when exposed to UV-B radiation. The reverse was true in its counterpart Dular, suggesting that the characteristics of silicic accumulation might be one of the mechanisms for rice to defense enhanced UV-B radiation. Li et al. (2004) also found that rice plants grown in hydroponics with Si supplement could enhance UV-B resistance compared to that in Si-deficient condition, which was believed to be attributed to increased phenolic compounds in epidermis of rice plants in responding to silicon supplement. Si is taken up through plant roots in the form of silicic acid and transport to the shoot via the transpiration stream, and then polymerized and accumulated on the shoot tissues as silica (Takahashi and Hino 1978; Ma and Takahashi 2002). Ma et al. (2006, 2007) reported that the uptake of Si in rice is mediated by at least two transporters, low silicon rice 1 (Lsi1) and low silicon rice 2 (Lsi2). Lsi1 is an influx transporter, while Lsi2 is an efflux transporter of Si. Therefore, it suggests that Lsi1 functions in controlling Si accumulation in rice, meaning that inhibiting the gene expression would reduce the uptake of Si in rice, which in turn might result in decreased plant tolerance to many stresses. So, it is hypothesized that by regulating the transcript level of Lsi1 may increase plant tolerance to various stressful factors.
Based on the hypothesis, the present study was conducted to determine whether Lsi1 plays a role in regulating rice tolerance to UV-B stress by employing gene suppression and overexpression approaches. A comparison of molecular changes between transgenic lines including Lsi1 suppression and its overexpression lines, and their wide-types (WT) was conducted in the responses to enhanced UV-B radiation, and the molecular response network was further detected by using suppression subtractive hybridization (SSH) approach (Diatchenko et al. 1996).
Materials and methods
Plant materials
We used the rice accessions, Lemont (UV-B tolerant, introduced from USA) and Dular (UV-B sensitive, from India) in this study (Fang et al. 2009a). The grain of the rice accession, Lemont were surface-sterilized with NaClO for 30 min, placed in Petri dishes in a temperature-controlled growth chamber and then sown in separate seedling plates. At the 3-leaf stage, uniform seedlings from each plant were selected and transferred into a Styrofoam plate (with holes spaced at 5 × 6 cm2); the seedlings were affixed to the plate by inserting a cotton plug into each hole on the plate. The Styrofoam plate was allowed to float in a pot (45 × 35 × 15 cm3) filled with 10 l rice (Oryza sativa L.) culture solution (Si solution) and had the following composition: 482 mg (NH4)2SO4, 248 mg KH2PO4, 185 mg KNO3, 149 mg K2SO4, 864.3 mg Ca(NO3)2·4H2O, 1350.6 mg MgSO4·7H2O, 2,000 mg Na2SiO3·9H2O, 457 mg FeSO4·7H2O, 484.4 mg EDTA, 14.3 mg H3BO3, 0.4 mg CuSO4·5H2O, 1.1 mg ZnSO4·7H2O, 9.05 mg MnCl2·4H2O, 0.45 mg Na2MoO4·2H2O, which is minor optimization from Yoshida et al. (1976), and the solution was changed every week. The pot was continuously aerated and its outer side was painted black to prevent algal growth. The pH value of the solution was maintained at 5.5 throughout the experiment. After 7 days, the root tissue of Lemont was sampled and then immediately frozen in liquid nitrogen and stored at −80°C for the isolation of total RNA.
Total RNA from the roots of Lemont was isolated using the Trizol method (Invitrogen) according to the manufacturer’s instructions; the genomic DNA was digested using DNase I (Takara). Complementary DNA (cDNA) was synthesized using the ExScript RT reagent Kit (Takara). A 300 base pairs (bp) partial coding region from 900 bp down-stream of ATG of rice Lsi1 (Genbank accession No. AB222272) was cloned with a forward primer 5′-cgGGATCCtcgccgacttcttccctc-3′ and a reverse primer 5′-ggGGTACCatcgctccggtgaactgc-3′ containing BamH I and Kpn I sites (underlined), respectively. The same fragment was again amplified with the primers containing different recognition sites for Spe I and Sac I, respectively, in forward primer 5′-ggACTAGTatcgctccggtgaactgc-3′ and reverse primer 5′-cGAGCTCtcgccgactcttccctc-3′. The full-length coding region of rice Lsi1 was amplified with gene specific forward primer (5′-cgGGATCCatggccagcaacaactcg-3′) and reverse primer (5′-ggGGTACCtcacacttggatgttctccatc-3′) containing BamH I and Kpn I sites. Before construction of the stability vectors, all the gene fragments were subsequently cloned in pMD18-T vector (Takara) and transformed in Escherchia coli (strain DH5α) and sent to Shanghai Sangon Biological Engineering Technology and Service Co., Ltd., China for sequencing. The obtained DNA sequences were analyzed at NCBI (the National Center for Biotechnology Information, USA, www.ncbi.nlm.nih.gov) to further confirm the sequence similarity with Lsi1. Both fragments were inserted into pTCK303 to create a Lsi1-RNAi stability vector (pTCK303-Lsi1), and the ORF of rice Lsi1 was inserted into Ubiquitin-1301 to create a Lsi1-overexpression stability vector (Ubiquitin-1301-Lsi1).
Mobilization of constructs in Agrobacterium tumefaciens
The recombinant vectors pTCK303-Lsi1and Ubiquitin-1301-Lsi1 were, respectively, mobilized into A. tumefaciens disarmed strain EHA105 by freeze and thaw method (Höfgen and Willmitzer 1988). Transformants were selected on YEB-agar containing kanamycin (50 μg/ml) and rifampicin (20 μg/ml).
Transformation of rice plants
The protocols of A. tumefaciens-mediated transformation of rice plants were performed following the methods described by Supartana et al. (2005) with minor modification. The grain of UV-B tolerant rice, Lemont was sterilized with ethanol and sodium hypochlorite and soaked at 20°C for 36 h. Water was replaced once during soak. After 36 h of soak, the embryo region of the grain turned white. At this stage, neither shoots nor roots had appeared. To inoculate A. tumefaciens contained with the recombinant vector pTCK303-Lsi1 or Ubiquitin-1301-Lsi1 into the embryonic apical meristem in the soaked grain, respectively, a site of the husk where a shoot would later emerge was pierced to a depth of 1–1.5 mm with a needle (Φ0.70 mm; Kangyou Medical Instrument Co., Ltd. Jiangsu, China) that had been dipped in the A. tumefaciens inoculum. The inoculated grain were placed on filter papers on wet vermiculite in flasks covered with aluminum foil and incubated at 23°C in the dark for 5 days. Then, the germinated seedlings were transplanted in a Styrofoam plate (with holes spaced at 5 × 6 cm2) float in a pot (45 × 35 × 15 cm3) filled with 10 l culture solution which is described in section “Plant materials”. To further confirm that the overexpression of Lsi1 functions in enhancing rice UV-B tolerance, we inoculate A. tumefaciens contained with Ubiquitin-1301-Lsi1 onto the UV-B sensitive rice Dular following the same procedure as mentioned above. Simultaneously, either the strain EHA105 with a blank-pTCK303 vector or a blank-Ubiquitin1301 vector was transformed into the embryonic apical meristem in the soaked grain of Lemont, and only the strain EHA105 with a blank-Ubiquitin1301 vector into that of Dular as the controls of Lsi1 overexpressed transgenic lines.
Histochemical analysis of GUS gene expression in transgenic rice lines
GUS dyeing was carried out according to the method described by Jefferson (1989). Segments of leaves sampled from transgenic rice lines were incubated in a solution containing 100 mM Na3PO4(pH 7.0), 0.1%Triton X-100, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 1 mg·mL−1 5-bromo-4-chloro-3-indolyl-β-D-GlcUA(X-Gluc) at 37°C for 12 h. The stained tissues were then transformed into 95% ethanol for 24 h to remove chlorophylls. The leaves of the WT, e.g. the rice accessions, Lemont and Dular, were used for GUS dyeing as control. An 800-bp GUS gene contained in pTCK303 and Ubiquitin-1301 vectors was amplified from the positive transgenic rice, the sequence of GUS primers is 5′-gcatgttacgtcctgtagaaaccc-3′and 5′-caaagccagtaaagtagaacggt-3′. The stability of the transgenic plants was checked by germinating in a hygromycin B water solution.
Detection of Lsi1 gene expression, Si uptake and Si concentrations in transgenic rice lines
In this section, the identified transgenic rice lines and their WT were transplanted into the pots, each pot contained 0.5 l Si solution and one rice plant. After 7 days, total RNAs of transgenic rice lines and WT were isolated and cDNA was synthesized following the method as described in section “Plant materials”. Gene expression levels of Lsi1 were detected in both transgenic rice lines and WT, respectively, according to qRT-PCR methods. The forward primer 5′-tcgccgacttcttccctc-3′ and reverse primer 5′-atcgctccggtgaactgc-3′ were used. Further, the root uptake of Si and Si concentration in transgenic rice lines and WT leaves were determined by the colorimetric molybdenum blue method, following the protocols as describe by Ma et al. (2003).
UV-B treatment
The identified transgenic rice lines and their WT were then transplanted, respectively, into the new pots for recovery in the solution culture above. After 7 days, each transgenic rice line and WT were exposed to enhance UV-B radiation, the treatment was followed by the method as described by Fang et al. (2009a), the procedures were described as follows, fluorescent lamps (40 W, Beijing Electric Light Sources Research Institute, China) were used as the source of UV-B radiation, these lamps were suspended above the rice plants and UV-radiation of wavelength less than 280 nm (UV-C) was eliminated by wrapping the lamps in a 0.1-mm cellulose diacetate film (West Design Product Co., Ltd, United Kingdom).The distance between the rice canopy and the lamps was 30 cm and 5 lamps were used. The rice seedlings were irradiated with UV-B for a period of 1 week. The UV-B radiation was applied at the top of the leaf canopy for a period of 7 h per day (from 09:00 to 16:00 h) at an intensity of 18.6 kJ m−2 d−1(It equals to the enhance dose of UV-B radiation when 25% depletion of the stratospheric 03 layer occurred in the summer solstice in Fuzhou). Rice under solar radiation was taken as control. The second leaves from the apex (fully open leaf at that point of time) of the two treated rice accessions and their corresponsive controls were randomly sampled on days 7 after UV-B treatment. The obtained samples were immediately frozen in liquid nitrogen and stored at −80°C prior to SSH analyze. SSH was carried out using the PCR-Select cDNA Subtraction Kit (Clontech) according to the manufacturer’s introduction (Fang et al. 2009a, b; Song et al. 2008).
Statistical analysis
All data were subjected to analysis of variance (ANOVA) using SPSS. Means were tested by the least significant difference at P < 0.05 (LSD0.05).
Results
Lsi1 expression in transgenic rice lines
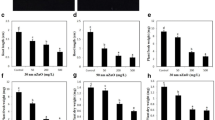
GUS dyeing and amplification of GUS gene were used to identify the transgenic rice (Fig. 1). To detect the expression level of Lsi1 in transgenic rice lines, qRT-PCR was performed on the roots of the transgenic rice together with WT. As expected, Lsi1 expression was inhibited in Lsi1-RNAi transgenic line of UV-B resistant rice Lemont, showing 2.10 times down-regulation in comparison with its WT; the reverse was true in the case of Lsi1-overexpressed transgenic line of Lemont, showing 2.25-folds up-regulation in Lsi1 transcript level. In terms of Lsi1-overexpressed transgenic line of UV-B sensitive rice Dular, it was up-regulated by 1.87 times in Lsi1 expression compared to its WT. However, there was no difference in Lsi1 expression between WT plants and blank vector control lines (Fig. 2). The sterilized T1–T3 grain were also confirmed germinating in a hygromycin B water solution (20 μg/ml), showing the stability of the transgenic plants (data not shown).The result confirmed that it was successful in Lsi1 suppression and its overexpression in rice with the aid of the molecular biological technique.
qRT-PCR analysis Lsi1 expression in transgenic rice plants and WT. Lemont, Dular: Rice cultivars; Lsi1-RNAi: Lsi1-RNAi transgenic rice line; Lsi1-overexpression: Lsi1-overexpression transgenic rice line; RNAi-Control: the control, RNAi vector transgenic rice line; Overexpression-Control: the control, overexpression vector transgenic rice line; The same below
Si uptake and Si concentrations in transgenic rice lines
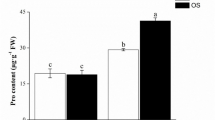
We also investigated the Si transport activity in transgenic rice lines and WT, the result showed that Si uptake was significantly reduced in Lsi1-RNAi transgenic line of Lemont compared with its WT. On 7 days, the accumulation of Si was 18.90 mg in 1 g dry root that sampled from Lsi1-RNAi transgenic line of Lemont in hydroponics with Si supplement, while 49.65 mg in that of WT. Si uptake capacity was significantly enhanced in Lsi1-overexpressed transgenic line of Lemont, the amount of Si uptake was 93.87 mg on 7 days. As far as the UV-B sensitive rice, Dular was concerned, the overexpression of Lsi1 also significantly increased Si transport capacity in the roots of the transgenic line of Dular, the result showed that the amount of Si uptake was 55.16 mg in the transgenic line on 7 days, showing much higher than that (28.38 mg) in WT (Fig. 3I). The same trend was found in Si concentrations in the leaves of rice lines, showing that 10.17 mg Si in 1 g leaf dry weight in Lsi1-RNAi transgenic line of Lemont, 21.08 mg g−1 in the WT and 38.32 mg g−1 in Lsi1-overexpressed transgenic line of Lemont; For Dular rice accession, Si concentration in Lsi1-overexpressed transgenic line was 22.34 mg g−1, which was significantly higher than that (12.15 mg g−1) in its WT (Fig. 3II). This result confirmed that Lsi1 functions indeed in controlling silicon accumulation in rice.
Root Si uptake and leaf Si concentration in transgenic rice and WT. Values are means ± SE based on three replicates. Different letters in each column indicate significant differences between means at P < 0.05 according to LSD. Small letters denote significant differences between Lsi1-overexpressed line of Dular and WT; Large letters designate the differences between Lsi1-RNAi, Lsi1-overexpressed lines of Lemont and WT (P < 0.05). Values with the same letter are not significantly different (P < 0.05). FW: Fresh weight
The result also found that the Lsi1-overexpressed transgenic lines of Lemont and Dular were not phenotypically different from WT, but the yellow specks on the leaves of Lsi1-RNAi transgenic lines of Lemont were detected as shown in Fig. 4, implying that Lsi1 might be involved in the defense to UV-B stress in some way.
Analysis of differential gene expression by using SSH
To unravel the regulatory network of Lsi1 in the defense response to UV-B irradiation, we isolated down-regulated genes in Lsi1-RNAi transgenic line of Lemont, as well as up-regulated genes in Lsi1-overexpressed transgenic lines of Lemont and Dular in the comparison with WT under enhanced UV-B radiation treatment using SSH. The positive gene clones were selected for reverse RNA blot after PCR (Fig. 5).
These clones were sequenced, and the cDNA sequences were searched against standard databases (www.ncbi.nlm.nih.gov). According to nucleic acid homologies and encoded protein sequences of the rice (O. sativa L.) database, the putative functions of the assigned expressed sequence tags (ESTs) are given in Table 1. The results indicated that several genes showed various expression trends in different transgenic rice lines. The identified genes were given as follow: A resistance-related non-coding RNA, which has been reported to be look like the inducible RNA in planthopper-infested O. sativa, was up-regulated in Lsi1-overexpressed transgenic lines of Lemont and Dular, but down-regulated in Lsi1-RNAi transgenic line of Lemont compared with WT when exposed to enhance UV-B radiation. The same was true in some genes, such as the gene encoding DNA repair associated enzyme putative CPD photolyase (PL, EC 4.1.99.3), the detoxification-related genes coding for metallothionein-like protein type 1 and glutathione S-transferase (GST, EC 2.5.1.18), and the genes for 4-coumarate-CoA ligase-like protein (4CL, EC 6.2.1.12) and phenylalanine ammonia-lyase (PAL, EC 4.3.1.5), which are associated with phenolics and flavonoids synthesis. However, some genes in rice plants in question showed different expression patterns in the responses to UV-B radiation, such as the genes encoding rice photosynthesis-related proteins, including photosystem I reaction centre subunit N, chlorophyll a-b binding protein 2, photosystem ii 10 kDa polypeptide, ribulose-bisphosphate carboxylase activase large isoform precursor protein and ribulose bisphosphate carboxylase/oxygenase activase appeared to decrease transcript levels in Lsi1-RNAi transgenic line of Lemont, reverse was true in Lsi1-overexpressed transgenic lines of Lemont and Dular. The genes coding for protein kinase domain containing protein, zinc-finger protein Lsd1, and dof zinc finger protein, which are all involved in signal transduction, were down-regulated in Lsi1-RNAi transgenic lines of Lemont, but up-regulated in Lsi1-overexpressed transgenic lines of Lemont and Dular. A down-regulated gene encoding constitutive photomorphogenic (COP8) homolog was detected only in Lsi1-RNAi transgenic line of Lemont, and the gene coding for NPH3 domain containing protein, which is a NPH1 photoreceptor-interacting protein, was up-regulated in Lsi1-overexpressd transgenic lines of Lemont and Dular. Some down-regulated genes which are related to cell physiology, such as cell elongation protein, peptide chain release factor 2 family protein and K+ potassium transporter family protein were detected only in Lsi1-RNAi transgenic line of Lemont, however, the genes encoding amino acid selective channel protein, 16 kDa membrane protein and membrane related protein-like were up-regulated in Lsi1-overexpressed transgenic line of Lemont and Dular. This result provided us the valuable information to outline the regulatory network how to increase the tolerant ability to defense UV-B radiation via the feedback regulation of Lsi1 to increase the activities of PAL and PL etc. in rice responding to UV-B radiation stress.
Discussion
The previous studies have shown that as a vital role involves in regulating plant growth and enhancing plant tolerance to abiotic and biotic stresses, accumulation of Si in rice plants is mediated by a member of Nod26-like major intrinsic protein (NIP) subfamily encoded by Lsi1, and the suppression of Lsi1 in rice would result in reduced Si accumulation (Ma et al. 2006). In this study, we suppressed Lsi1 transcript level in UV-B resistance rice Lemont and enhanced transcript level of Lsi1 in Lemont and UV-B-sensitive rice Dular using RNAi and overexpression technique, respectively. The results certified that the absorptive capability of Si was significantly increased in Lsi1-overexpressed transgenic lines of Lemont and Dular compared to WT. When the transgenic rice lines and WT were exposed to UV-B irradiation, we found that a resistance-related non-coding RNA (nc1) was up-regulated in Lsi1-overexpressed transgenic lines of Lemont and Dular, but down-regulated in Lsi1-RNAi transgenic line of Lemont compared to their corresponsive WT plants. It has been reported that resistance-related non-coding RNA (ncRNA1) was an inducible RNA detected in planthopper-infested O. sativa L. spp indica. The present result suggested that the regulation of rice UV-B tolerance by Lsi1 might be based on the differential expression ways of ncRNA, which acts as a regulator in rice to trigger off the activation of plant defense system in the tolerance response to UV-B radiation. The tentative cassette reaction might be to increase messenger RNA (mRNA) expression of the antioxidant enzymes glutathione S-transferase and metallothionein-like protein type 1, which was up-regulated in Lsi1-overexpressed transgenic lines of Lemont and Dular, in the comparison with WT, when they were exposed to UV-B radiation. It has been reported that the up-regulated expression of the genes coding for the two antioxidant enzymes may contribute to prevent from the oxidative damage caused by UV-B radiation etc. (Marrs 1996). The result was further confirmed in the expression pattern of the gene encoding glutathione S-transferase and metallothionein-like protein type 1 in Lsi1-RNAi transgenic rice, showing down-regulation in the mRNA expression level of the two enzymes in the Lsi1-suppressed transgenic line under UV-B radiation supplement.
The present study also showed that the overexpression of Lsi1 was able to significantly promote the up-regulation of PAL and PL in rice. As PAL is a key enzyme in plant phenylalanine metabolism pathway, it catalyzes the conversion of phenylalanine to cinnamate and initiates all secondary phenolic biosynthesis (Zon and Amrhein 1992), meanwhile, and also shows the activity to convert tyrosine into p-coumarate, following by 4CL converting p-coumarate into its coenzyme-A ester, which in turn triggers off the reaction with malonyl CoA in the flavonoid biosynthetic pathway starting with the condensation of one molecule of 4-coumaroyl-CoA and three ones of malonyl-CoA to produce naringenin chalcone (Bohm 1998; Harborne and Williams 2000), and the production of various secondary substances, which known as primarily UV-B absorbing compounds, participating in plant responses to enhanced UV-B radiation (Barabás et al. 1998). The analysis of the gene differential expression and its related metabolites showed that the up-regulated transcripts level of PAL and 4CL in Lsi1-overexpressed transgenic lines of Lemont and Dular resulted in significantly increased content of UV-B absorptive substances including flavonoids and total phenolics in the leaves of the two transgenic rice lines, and this in turn led to enhanced UV-B tolerance in rice (McClure 1975).
Our previous study has also reported that the activation of a metabolic response to promote the synthesis of UV-protective substances, flavonoids and phenolics, would in turn reduce the UV-induced damage or trigger the molecular processes involved in the repair of UV-B-damaged DNA, and the two steps involved in the process of the molecular regulation to up-regulate transcript level of PAL, 4CL and PL etc. as mentioned above, seems to be vital for the development of tolerance in the rice cultivar (Fang et al. 2009a). The present study certified that enhanced UV-B radiation also resulted in induced the up-regulation of the gene coding for PL, which functions in splitting the cyclobutane ring and repairs the damaged DNA caused by UV (Mees et al. 2004). It is therefore suggested that the activation of the mRNA expression level of PL would contribute to enhance DNA repairing capability, and consequently enhance rice resistance to UV-B radiation stress.
The present result suggested that Lsi1 affect rice photosynthesis and photomorphogenesis under UV-B radiation. One case in this study can be taken for the example. As we know, the ribulose bisphosphate carboxylase/oxygenase (Rubisco, EC 4.1.1.39), which involves the ATP-dependent carboxylation of the epsilon-amino group of lysine leading to the formation of a carbamate structure, is a key enzyme for photosynthesis in the leaves of rice. The detection indicated that it was significantly suppressed by elevated UV-B radiation at the transcription step during early leaf stages (Takeuchi et al. 2006); however, the present result showed that the mRNA expression of Rubisco in Lsi1-overexpressed lines of Lemont and Dular was higher than that of WT when exposed to UV-B radiation. The same tendency was found in the gene expression level of other photosynthesis-related proteins, such as photosystem I reaction centre subunit N, which functions in mediating the binding of the antenna complexes to the PSI reaction center and core antenna, and plays an important role in docking plastocyanin to the PSI complex (Haldrup et al. 1999), chlorophyll a-b binding protein 2 known as light-harvesting complex (LHC), which works as a light receptor, in capturing and delivering excitation energy to photosystems (Liu and Shen 2004), and photosystem II 10 kDa polypeptide, which is associated with the oxygen-evolving complex of photosystem II (Lautner et al. 1988). While in Lsi1-RNAi transgenic line of Lemont, the expression level of these genes was lower than that of WT when exposed to UV-B radiation. The results further supported the hypothesis that the higher Lsi1 expression level is beneficial for the enhancement of UV-B tolerance in rice.
With respect to photomorphogenic responses of different transgenic lines to the stressful condition, the different transgenic rice lines showed various sensitivities to the supplement of UV-B radiation. UV-B radiation induced photomorphogenic responses in Lsi1-overexpressed transgenic lines of Lemont and Dular by up-regulating the expression of nonphototropic hypocotyl 3 (NPH3) gene encoding a NPH1-interacting protein, which is a member of a large protein family, apparently specific to higher plants, and may function as an adapter or scaffold protein to bring together the enzymatic components of a NPH1-activated phosphorelay. In Arabidopsis thaliana seedlings, the phototropism is initiated by NPH1, a light-activated serine-threonine protein kinase (Motchoulski and Liscum 1999). However, in Lsi1-RNAi transgenic line of Lemont, the supplement of UV-B radiation inhibited the photomorphogenesis, which was attributed to down-regulation of constitutive photomorphogenic 8 (CSN complex subunit), a photo receptor in photomorphogenesis, which involves the expansion of leaves and accumulation of chlorophyll etc. (Kim et al. 1998).
It was also found that the overexpression of Lsi1 in rice could activate amino acid selective channel protein, membrane related protein, which contribute to strengthen the membrane structure in cell, protein kinase domain containing protein, and zinc-finger protein, which are associated with signal transduction in the pathway to increase UV-B tolerance in rice through the feedback regulation of Lsi1 function, but the reverse was true in Lsi1-RNAi transgenic line of Lemont when exposed to UV-B radiation. In addition, the inhibited expression of Lsi1 in rice also resulted in down-regulated mRNA transcripts level of cell elongation protein, peptide chain release factor 2 family proteins and K+ potassium transporter family protein, which are involved in cell physiology. These findings suggested that Lsi1 may play different roles in the feedback regulation through the positive or negative regulatory machinery of the gene.
In conclusion, rice UV-B tolerance could be regulated by Lsi1, and the overexpression of Lsi1 contributes to enhance rice UV-B tolerance and the resistance-related non-coding RNA acts as a regulator in the channel, the active effect involves enhancing gene expression level of PAL, 4CL etc. Increasing accumulation of Si in rice can also activate PL and the related antioxidant enzymes to increase DNA repair ability to reduce the damage caused by UV-B radiation. At the same time, the photosynthesis and detoxification-related pathways were also strengthened in the leaves of Lsi1-overexpressed lines of UV-B resistant rice Lemont and its counterpart Dular, and all of them contribute to enhanced rice defense to UV-B irradiation. Further investigation should be put on the changes in grain yield and nutrient content in the grain products of different transgenic rice accessions grown in the supplement of UV-B radiation to aim at further understanding the possibility to obtain rice accessions with the high yielding and strong resistance in plant breeding by using overexpression technique of functional genes.
Abbreviations
- 4CL:
-
4-coumarate-CoA ligase
- Lsi1:
-
Low silicon rice 1
- PL:
-
Photolyase
- PAL:
-
Phenylalanine ammonia-lyase
- Si:
-
Silicon
- SSH:
-
Suppression subtractive hybridization
- UV-B:
-
Ultraviolet-B
- WT:
-
Wild types
References
Barabás KN, Szegletes Z, Pestenácz A, Fülöp K, Erdei L (1998) Effects of excess of UV-B irradiation on the antioxidant defence mechanisms in wheat (Triticum aestivum L.)seedlings. J Plant Physiol 153:146–153
Blumthaler M, Ambach W (1990) Indication of increasing solar ultraviolet-B radiation flux in alpine regions. Science 248:206–208
Bohm BA (1998) Flavonoid functions in nature. In: Introduction to flavonoids. Chemistry and biochemistry of organic natural products, vol 2. Harwood Academic Publishers, Amsterdam, pp 339–364
Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Nat Acad Sci USA 93:6025–6030
Epstein E (1999) Silicon. Annu Rev Plant Physiol Plant Mol Biol 50:641–664
Fang CX, Wu XC, Zhang HL, Xiong J, Wu WX, Lin WX (2009a) UV-induced differential gene expression in rice cultivars analyzed by SSH. Plant Growth Regul 59:245–253
Fang CX, Xiong J, Qiu L, Wang HB, Song BQ, He HB, Lin RY, Lin WX (2009b) Analysis of gene expressions associated with increased allelopathy in rice (Oryza sativa L.) induced by exogenous salicylic acid. Plant Growth Regul 57:163–172
Fawe A, Abou Zaid M, Menzies JG, Belanger RR (1998) Silicon-mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology 88:396–401
Ganguly AR, Steinhaeuser K, Erickson DJ III, Branstetter M, Parish ES, Singh N, Drake JB, Buja L (2009) Higher trends but larger uncertainty and geographic variability in 21st century temperature and heat waves. Proc Nat Acad Sci USA 106:15555–15559
Gong HJ, Zhu XY, Chen KM, Wang SM, Zhang CL (2005) Silicon alleviates oxidative damage of wheat plants in pots under drought. Plant Sci 169:313–321
Haldrup A, Naver H, Scheller HV (1999) The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J 17:689–698
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Jefferson RA (1989) The GUS reporter system. Nature 342:837–838
Kim BC, Tennessen DJ, Last RL (1998) UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 15:667–674
Lautner A, Klein R, Ljungberg U, Reiläender H, Bartling D, Andersson B, Reinke H, Beyreuther K, Herrmann RG (1988) Nucleotide sequence of cDNA clones encoding the complete precursor for the ‘10-kDa’ polypeptide of photosystem II from spinach. J Biol Chem 263:10077–10081
Li WB, Shi XH, Wang H, Zhang FS (2004) Effects of silicon on rice leaves resistance to ultraviolet-B. Acta Bot Sin 46:691–697
Liang YC, Ma TS, Li FJ, Feng YJ (1994) Silicon availability and response of rice and wheat to silicon in calcareous soils. Commun Soil Sci Plant Anal 25:2285–2297
Liang YC, Zhang WH, Chen Q, Liu YL, Ding RX (2006) Effect of exogenous silicon (Si) on H+-ATPase activity, phospholipids and fluidity of plasma membrane in leaves of salt-stressed barley (Hordeum vulgare L.). Environ Exp Bot 57:212–219
Liu XD, Shen YG (2004) NaCl-induced phosphorylation of light harvesting chlorophyll a/b proteins in thylakoid membranes from the halotolerant green alga, Dunaliella salina. FEBS Lett 569:337–340
Ma JF (2004) Role of silicon in enhancing the resistance of plants to biotic and abiotic stresses. Soil Sci Plant Nutr 50:11–18
Ma JF, Takahashi E (2002) Soil, fertilizer, and plant silicon research in Japan. Elsevier Science, Amsterdam, pp 73–106
Ma JF, Yamaji N (2006) Silicon uptake and accumulation in higher plants. Trends Plant Sci 11:392–397
Ma JF, Higashitani A, Sato K, Takeda K (2003) Genotypic variation in silicon concentration of barley grain. Plant Soil 249:383–387
Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M (2006) A silicon transporter in rice. Nature 440:688–691
Ma JF, Yamaji N, Mitani N, Tamai K, Konishi S, Fujiwara T, Katsuhara M, Yano M (2007) An efflux transporter of silicon in rice. Nature 448:209–211
Marrs KA (1996) The functions and regulation of glutathione S transferases in plants. Annu Rev Plant Physiol Plant Mol Biol 47:127–158
McClure JW (1975) Physiology and function of flavonoids. In: Harborne JB, Mabry TJ, Mabry H (eds) The flavonoids, vol 2. Academic Press, New York, pp 970–1055
Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, Essen L (2004) Crystal structure of a photolyase bound to a CPD-Like DNA lesion after in situ repair. Science 306:1789–1793
Motchoulski A, Liscum E (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286:961–964
Newsham KK, Robinson SA (2009) Responses of plants in polar regions to UVB exposure: a meta-analysis. Glob Change Biol 15:2574–2589
Rodrigues FÁ, Benhamou N, Datnoff LE, Jones JB, Bélanger RR (2003) Ultrastructural and cytochemical aspects of silicon. Phytopathology 93:535–546
Rodrigues FÁ, Jurick WM II, Datnoff LE, Jones JB, Rollins JA (2005) Silicon influences cytological and molecular events in compatible and incompatible rice-Magnaporthe grisea interactions. Physiol Mol Plant Pathol 66:144–159
Saqib M, Zörb C, Schubert S (2008) Silicon-mediated improvement in the salt resistance of wheat (Triticum aestivum) results from increased sodium exclusion and resistance to oxidative stress. Funct Plant Biol 35:633–639
Song BQ, Xiong J, Fang CX, Qiu L, Lin RY, Liang YY, Lin WX (2008) Allelopathic enhancement and differential gene expression in rice under low nitrogen treatment. J Chem Ecol 34:688–695
Supartana P, Shimizu T, Shioiri H, Nogawa M, Nozue M, Kojima M (2005) Development of simple and efficient in planta transformation method for rice (Oryza sativa L.) using Agrobacterium tumefaciens. J Biosci Bioeng 4:391–397
Takahashi E, Hino K (1978) Silica uptake by plant with special reference to the forms of dissolved silica. J Sci Soil Manure 49:357–360
Takeuchi A, Yamaguchi T, Hidema J, Strid A, Kumagai T (2006) Changes in synthesis and degradation of Rubisco and LHCII with leaf age in rice (Oryza sativa L.) growing under supplementary UV-B radiation. Plant Cell Environ 25:695–706
Wu XC, Lin WX, Huang ZL (2007) Influence of enhanced ultraviolet-B radiation on photosynthetic physiologies and ultrastructure of leaves in two different resistivity rice cultivars. Acta Ecol Sinica 27:0554–0564 (in Chinese with English abstract)
Yoshida S, Forno DA, Cock JH (1976) Laboratory manual for physiological studies of rice, 3rd edn. The International Rice Research Institute, Manila, the Philippines, pp 1–83
Zon J, Amrhein N (1992) Inhibitors of phenylalanine ammonia lyase: 2-aminoindan-2-phosphonic acid and related compounds. Justus Liebigs Ann Chem 6:625–628
Acknowledgments
This work was supported by The National Natural Science Foundation of China (No. 30971737, 30571103), the Provincial Natural Science Foundation of Fujian, China (No. 2009J01055), National Research Foundation for the Doctoral Program of Higher Education of China (No. 20093515110009) and the National Innovation Experiment Program for College Students (No. 011469).
Author information
Authors and Affiliations
Corresponding author
Additional information
Chang-Xun Fang and Qing-Shui Wang contributes equally to this work.
Rights and permissions
About this article
Cite this article
Fang, CX., Wang, QS., Yu, Y. et al. Suppression and overexpression of Lsi1 induce differential gene expression in rice under ultraviolet radiation. Plant Growth Regul 65, 1–10 (2011). https://doi.org/10.1007/s10725-011-9569-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-011-9569-y