Abstract
The aim of the investigation reported here was to assess the role of gibberellin in cotton fiber development. The results of experiments in which the gibberellin (GA) biosynthesis inhibitor paclobutrazol (PAC) was tested on in vitro cultured cotton ovules revealed that GA is critical in promoting cotton fiber development. Plant responses to GA are mediated by DELLA proteins. A cotton nucleotide with high sequence homology to Arabidopsis thaliana GAI (AtGAI) was identified from the GenBank database and analyzed with the BLAST program. The full-length cDNA was cloned from upland cotton (Gossypium hirsutum, Gh) and sequenced. A comparison of the putative protein sequence of this cDNA with all Arabidopsis DELLA proteins indicated that GhRGL is a putative ortholog of AtRGL. Over-expression of this cDNA in Arabidopsis plants resulted in the dwarfed phenotype, and the degrees of dwarfism were related to the expression levels of GhRGL. The deletion of 17 amino acids, including the DELLA domain, resulted in the dominant dwarf phenotype, demonstrating that GhRGL is a functional protein that affects plant growth. Real-time quantitative PCR results showed that GhRGL mRNA is highly expressed in the cotton ovule at the elongation stage, suggesting that GhRGL may play a regulatory role in cotton fiber elongation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phytohormone gibberellin (GA) is an essential plant hormone involved in regulating many aspects of plant growth and development (Harberd et al. 1998; Richards et al. 2001). Studies on plants lacking enzymes in the GA biosynthetic pathway have shown that GA is required during many different stages of the Arabidopsis thaliana life cycle, such as seed germination, hypocotyl growth, internode elongation, and the development of leaves, stamens, and petals (Koornneef and van der Veen 1980; Zeevaart and Talon 1992; King et al. 2001). The GA-deficient mutant ga1-3 is severely dwarfed, shows abnormal floral development, and requires exogenous GA for germination because the mutation blocks an early step in the GA biosynthetic pathway (Koornneef and van der Veen 1980; King et al. 2001).
The GA signaling pathway is repressed by the DELLA proteins. All of the DELLA proteins are characterized by their N-terminal conserved DELLA domain and VHYNP domain (Pysh et al. 1999; Willige et al. 2007). In the A. thaliana genome, there are five highly homologous DELLA protein repressors, including GA INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGL1 (RGA-like 1), RGL2, and RGL3 (Peng et al. 1997; Silverstone et al. 1998; Richards et al. 2001; Willige et al. 2007). GAI and RGA have been proven to have overlapping functions as repressors of plant elongation growth (Willige et al. 2007). RGL1 and RGL2 play a predominant role in controlling germination and floral development, respectively (Dill and Sun 2001; King et al. 2001; Lee et al. 2002; Cheng et al. 2004; Tyler et al. 2004). All of the DELLA repressors are inactivated in response to GA by ubiquitin proteasome-dependent protein degradation (Silverstone et al. 2001; Fu et al. 2002; Thomas and Sun 2004; McGinnis et al. 2003; Sasaki et al. 2003; Willige et al. 2007). A 17-amino acid deletion, including the conserved DELLA domain, which is the mutation present in the dominant Arabidopsis gai-1 mutant, renders mutant gai and rga proteins insensitive to GA-induced proteolysis, and plants expressing these mutant DELLA repressors are GA-insensitive, dark-green, late-flowering dwarfs (Peng and Harberd 1997; Dill and Sun 2001; Silverstone et al. 2001; Fleck and Harberd, 2002; Itoh et al. 2002; Dill et al. 2004; Willige et al. 2007). In DELLA proteins, in addition to the conserved DELLA domain and VHYNP domain, there is also a divergent Poly S/T region at the N-terminus. At the C-terminus, DELLA proteins also share multiple conserved sequence motifs, which include a putative nuclear localization signal (NLS), two LXXLL motifs, a conserved GRAS domain, and an src homology 2 (SH2)-like domain, similar to transcription factor, STATs (Peng et al. 1997, 1999; Richards et al. 2000).
Many DELLA proteins have also been discovered in other plants, such as height1 (Rht1) from wheat (Triticum aestivum), dwarf8 (d8) from maize (Zea mays), Slender1 (Sln1) from barley (Hordeum vulgare) (Gubler et al. 2002), Slr1 from rice (Oryza sativa), and Vvgai1 from grape (Vitis vinifera) (Peng et al. 1999; Ikeda et al. 2001; Boss and Thomas, 2002; Chandler et al. 2002; Muangprom and Osborn 2004). All of these DELLA proteins have the same function in repressing GA responses. The identification of GAI orthologues having the same function of repressing GA signaling in plants other than Arabidopsis has shown that components of the GA signal transduction pathway are conserved (King et al. 2001, Willige et al. 2007). In cotton, seven DELLA-like genes have been identified from an expressed sequence tag (EST) collection from immature cotton ovules (Yang et al. 2006; Wilkins and Arpat 2005). At least one of these, a GAI-like transcript, was present at higher levels in the immature ovules than in the other tissue samples.
Cotton fibers, which are highly elongated epidermal cells that grow from the seed integument, undergo a developmental program that includes cell fate determination, initiation, elongation, specialization and, finally, programmed cell death (Sun et al. 2004; Shi et al. 2006). The different stages of cotton fiber development are in part controlled by phytohormones (Beasley and Ting, 1974; Shi et al. 2006; Sun et al. 2004; Yang et al. 2006). Analysis of fiber development on cotton ovules grown in vitro in liquid media indicated that exogenous auxin (IAA) and GA are required for maximal fiber elongation (Beasley and Ting 1974; Shi et al. 2006; Sun et al. 2004; Yang et al. 2006). Our experiments using the GA biosynthesis inhibitor paclobutrazol (PAC) in the system of cotton ovule culture in vitro indicated that Gibberellic acid (GA3) plays a critical role in promoting cotton fiber development.
Materials and methods
Plant materials
Cotton plants (Gossypium hirsutum cv. Xuzhou 142) were grown in a soil mixture in a fully automated greenhouse. Arabidopsis seeds (A. thaliana ecotype Columbia) were obtained from the Nottingham Arabidopsis Stock Center, and plants were grown in potting soil at 22°C under a 16/8-h (light/dark) photoperiod with light supplied by fluorescent lamps. Cotton ovule culture was carried out essentially as described by Beasley and Ting (1974). For RNA extraction, ovules at −3 to 0 days after anthesis (DPA), 3 DPA, 5 DPA, and 10 DPA were excised from bolls on the cotton plant. Fibers at 15 DPA and 25 DPA were also collected. The dissected ovules and fibers were frozen and stored in liquid nitrogen immediately after harvest (Sun et al. 2004, 2005). For in vitro ovule cultures, fresh ovules (1 DPA) picked from the cotton plants were used. Harvested cotton ovaries were soaked in 70% ethanol for 1 min, rinsed in distilled and deionized water, and soaked again in 0.1% HgCl2 solution containing 0.05% Tween-80 for 20 min (sterilization). The ovules were immediately floated on liquid media supplemented with1.0 mg/l GA3 and 1.0 mg/l PAC in 50-ml flasks. The ovules were then incubated at 34°C in the dark. The GA3 (99.9%) was purchased from Qianxi Chemicals; GA biosynthesis inhibitor PAC was purchased from Jiafeng (China).
Cotton and Arabidopsis genomic DNA and RNA isolation
Cotton and Arabidopsis genomic DNA samples were isolated from leaf tissues using the DNeasy Plant Mini kit (Qiagen, Valencia, CA). For cotton total RNA isolation, samples were ground to powder in liquid nitrogen and stored at −80°C. Total cotton RNA was extracted using the method of Wan and Wilkins (1994). Arabidopsis RNA was isolated from entire plant tissues with the RNA Plant Extraction kit (TIANGEN, China).
Cloning DELLA Ortholog from cotton
A cotton nucleotide (GenBank accession no. AY208992) with high sequence homology to A. thaliana GAI (AtGAI) was identified in the NCBI GenBank database with the NCBI BLAST program. Gene-specific primers (forward: 5′-ATGAAGAGAGATCATCAAGA-3′, reverse: 5′-TCATTCACTCGTACATTCT G-3′) were synthesized according to the nucleotide sequence. A nucleotide was amplified from a cotton boll cDNA library by PCR. The amplified fragments were sequenced and compared to AtGAI. Fragments with high sequence similarity to AtGAI were selected for further primer design and PCR amplification to isolate a full-length GhRGL cDNA from the library. The putative GhRGL coding sequence was also amplified and cloned from cotton genomic DNA and completely sequenced.
Overexpression of GhRGL and Ghrgl in Arabidopsis
The Ghrgl mutant allele from G. hirsutum cv Xuzhou 142 differs from the GhRGL wild-type allele in that it contains a 51-bp deletion of coding sequence of DELLA domain. Both GhRGL DNA and Ghrgl DNA were respectively inserted into pBI121 in place of the GusA gene to create an expression cassette. First, pGEM-T containing GhRGL or Ghrgl was digested with SpeI and then with SacI. The resulting fragments were ligated, respectively, into pBI121 DNA, which was then digested with XbaI and SacI. Each of the ligated plasmids was transformed into Escherichia coli. Both the pBI121-GhRGL and pBI121-Ghrgl plasmids were identified by restriction analysis and then transformed into Agrobacterium tumefaciens GV3101 (C58). Agrobacterium tumefaciens containing the pBI121-GhRGL and pBI121-Ghrgl constructs were selected on LB plates containing kanamycin (50 mg/l), gentamycin (25 mg/l), and rifampicin (100 mg/l). The plasmids were isolated, and the insert was sequenced to confirm the accuracy of the GhRGL and Ghrgl transgene. The GhRGL and Ghrgl constructs were introduced into Arabidopsis plants (A. thaliana ecotype Columbia) separately by the Agrobacterium-mediated flower infiltration transformation method (Clough and Bent 1998). The plants in pots were placed in trays and covered with plastic wrap for 16–24 h to maintain conditions of high humidity. Plants were watered from the bottom of the tray, and watering was stopped as the seeds matured. Seeds were harvested from individual plants.
Arabidopsis seeds were sterilized with 75% ethanol for 1 min and 50% chlorine bleach solution for 10 min. The seeds were then rinsed with sterile water three to five times and sown on MS solid medium (Murashige and Skoog 1962) containing antibiotics (250 mg/l of carbenicillin and cefotaxime, respectively) and 50 mg/l kanamycin. The PCR analysis with primers specific for the CaMV 35S promoter and the transgene-encoding sequence was performed to confirm the insertion of the transgene in the plants with the following: 5′-ATTCCATTGCCCAGCT-3′ (forward) and 5′-CACTACCTGACTCAGTACCA-3′ (reverse).
The expression levels of GhRGL transgenic plants detected by semi-quantitative reverse transcription-PCR
For the semi-quantitative RT-PCR analysis, total RNA was extracted from 30-day-old transgenic Arabidopsis plants with the RNA Plant Extraction kit (TIANGEN, China). Total RNAs were treated with RNase-free DNase I (TaKaRa Biotechnology, Dalian, China) to remove residual DNA. The first strand cDNAs for each sample were generated using the cDNA Synthesis kit (TaKaRa Biotechnology). The reverse transcription (RT) product was used in semi-quantitative PCR reactions with primers for the GhRGL gene: 5′-GAGAAAGTTATGGGTACTGC-3′ (forward) and 5′-GA CAAGTTTCATAGAAGTGC-3′ (reverse). The semi-quantitative PCR cycling conditions consisted of a 10-min pre-denaturation at 94°C, followed by 24 cycles of 94°C (10 s), 52°C (10 s), and 72°C (30 s). An actin gene ACT2 was used as a control in the semi-quantitative PCR with specific primers (5′-ACGTGACCTTACTGATTACC-3′ and 5′-ACAATGTTACCGTACAGATC-3′).
GhRGL expression pattern analysis in cotton and the expression levels of GhRGL in transgenic Arabidopsis plants detected by real-time quantitative PCR
For detecting the expression levels of GhRGL in transgenic Arabidopsis plants, we selected an actin gene (Atact2) as an endogenous control; for the analysis of the GhRGL expression pattern in cotton, we selected a Histone 3 gene of G. hirsutum (Ghhis3) as an endogenous control. The primers and probes of Atact, Ghhis3, and GhRGL gene were designed by the on-line ProbeFinder ver. 2.40 program (https://www.roche-applied-science.com/sis/rtpcr/upl/acenter.jsp?id = 030000), Probe no. 31, 61, and 25 from the Universal ProbeLibrary of Roche (Indianapolis, IN) were selected for Atact2, Ghhis3, and GhRGL, respectively. Both forward and reverse primers for Atact2 (5′-GGAGAAGCTCTCCTTTGTTGC-3′ and 5′-GCTGGTCTTTGAGGTTTCCA-3′) were also from the Universal ProbeLibrary of Roche. Primers for Ghhis3 were 5′-TTCCAGAGGCTTGTTCGTG-3′ and 5′-TTCCAGAGGCTTG TTCGTG-3′, and GhRGL 5′-CAGCGGCGATGGGTATAG-3′ and 5′-CTTGTATGCCACCCAAGC AT-3′. The first strand cDNAs for each sample were generated using the PrimeScript RT reagent kit (TaKaRa Biotechnology). cDNAs from three biological samples were used for analysis, and all reactions were run in triplicate using a Stratagene Mx3000P QPCR System (Stratagene, La Jolla, CA). Reactions with no template and with only the RNA before RT were run as controls. Real-time RT-PCR was performed in a 25-μl reaction mixture containing 12.5 μl FastStart Universal Probe Master (ROX) (Roche), 0.25 μl hydrolysis probe (25 μM), 0.9 μl forward primer (50 μM), 0.9 μl reverse primer (50 μM), 9.45 μl ddH2O, and 2 μl of template cDNA (5 ng/μl). The PCR conditions were 10-min pre-denaturation at 94°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. The MxPro QPCR software ver. 3.00 (Stratagene) was used for data collection and analysis. Quantification results were expressed in terms of the cycle threshold (CT) value determined according to the manually adjusted baseline. Relative gene expressions in samples were determined using a method previously described (Zhang et al. 2007). Briefly, differences between the CT values of the target genes and control genes were calculated as ΔCT = CT target – CTcontrol, and the expression levels of target genes relative to control genes were determined as 2 -ΔCT. The PCR was repeated three times, and the average values of 2 -ΔCT were used to determine the difference in expression between samples and control (Zhang et al. 2007).
Results
GA plays key roles in cotton fiber elongation
Auxin and GA have long been known to play important roles in plant cell expansion or elongation (Phinney 1984; Evans 1985; Crozier et al. 2000; Shi et al. 2006), with Beasley and Ting (1974) being the first to report that the addition of IAA and GA3 to cotton ovule culture in vitro promoted the elongation of fibers on the ovule wall. We found that the presence of 1.0 mg/l GA3 alone in the ovule culture medium caused a significant increase in fiber length (Fig. 1). In contrast, the addition of 1.0 mg/l of a GA biosynthesis inhibitor, PAC, eliminated fibers from most parts of the ovule. The addition of both GA3 and PAC to the cultured ovules partially restored fiber development (Fig. 1), indicating that the inhibition of fibers by PAC is at least partially due to its inhibition of GA biosynthesis.
GhRGL protein has conserved motifs of DELLA proteins
In Arabidopsis, there are five DELLA proteins that are negative regulators of GA signaling, including GAI, RGA, GL1, GL2, and GL3 (Peng et al. 1997; Silverstone et al. 1998; Richards et al. 2001; Willige et al. 2007). The sequences of all of these genes are similar. According to BLAST search results, DELLA proteins from all plant species share a high degree of homology. We used the known DELLA proteins as queries to BLAST search GenBank and found a single cotton nucleotide (AY208992) with a strong sequence similarity to AtGAI. This 2041-bp nucleotide, which shares 59.5% identity to the overlapping region of AtGAI, is from a G. hirsutum L. cDNA library.
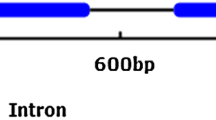
Based on the sequence of the nucleotide, we designed specific primers to amplify corresponding cDNAs from a cDNA library derived from cotton (G. hirsutum L.) boll mRNA. The resulting cDNA has a 1724-bp open reading frame (ORF) that encodes a derived polypeptide of 548 amino acids, as determined by Vector NTI software analysis. The coding sequence was also amplified and cloned from cotton genomic DNA. Our sequencing analysis results showed that the coding gene has no introns. The BLAST search revealed that the fragments have a high sequence similarity to AtGAI. We conclude that the cDNA is a putative ortholog of AtGAI.
The complete ORF of GhRGL has 63.5% nucleotide sequence identity to AtGAI. The amino acid sequence of GhRGL also has a high identity to all DELLA proteins in Arabidopsis, including GAI, RGA, RGL1, RGL2, and RGL3, showing identities of 60.5, 59.8, 63.1, 61.8, and 58.1%, respectively (Figs. 2, 3). The genes coding for DELLA proteins belong to a larger group of genes known as the GRAS family (Pysh et al. 1999). The DELLA proteins share a high homology with other GRAS family proteins at the C-terminus but not at the N-terminus. The GhRGL cloned in our study has a highly conserved DELLA domain, a VHYNP domain, and a divergent Poly S/T region that are characteristic of DELLA protein. This protein also shares multiple conserved sequence motifs which are inside the GRAS domain, including a putative nuclear localization signal (NLS) and two Leu zipper motifs.
Real-time PCR results suggest important roles for GhRGL in cotton fiber elongation
During cotton fiber development, protodermal cells of ovules undergo fiber initiation (−3 to 0 DPA), elongation (5–15 DPA), secondary cell-wall biosynthesis (15–25 DPA), and maturation (25–45 DPA), leading to mature fibers (Basra and Malik 1984; Tiwari and Wilkins 1995; Wilkins and Jernstedt 1999; Kim and Triplett 2001; Yang et al. 2006). In order to understand the expression pattern of the GhRGL gene during all stages of cotton fiber development, we employed the sensitive real-time PCR technique using gene-specific primers to quantify the transcript levels of GhRGL during the different stages of cotton fiber development. Total RNA samples isolated from different stages of cotton ovules and fibers were used for analysis, and six samples were selected for analyzing GhRGL gene expression pattern in cotton fiber development, including ovules at −3 to 0, 3, 5, 10, and 15 DPA, respectively, and fibers at 25 DPA. The GhRGL gene was expressed in all of the selected cotton samples (Fig. 4). Higher levels of GhRGL mRNA were detected in 5- and 10-DPA ovules, and the highest levels of GhRGL mRNA were detected in 10-DPA ovules. These results demonstrate that the GhRGL gene was expressed at the highest levels during the fiber elongation stages, suggesting that GhRGL has important roles in cotton fiber elongation.
GhRGL expression levels in cotton plants. GhRGL mRNA levels were assayed in total RNA samples using the TaqMan quantitative real-time PCR technique with GhHIS3 as a reference. The sample with the lowest ratio of GhRGL mRNA to GhHIS3 signal (−3 to 0 DPA ovule) was set at 1, and the values for the other samples are presented as a fold increase relative to that value. Values are means ± standard deviation of three independent assays
Overexpression of GhRGL and Ghrgl in Arabidopsis
In order to determine whether GhRGL encodes a functional DELLA protein, we separately introduced two recombinant genes, GhRGL and Ghrgl (17 amino acid deletion from GhRGL), placed under the control of a CaMV 35S promoter into Arabidopsis plants. Transgenic T1 seedlings were selected on kanamycin-containing media, and the phenotypes of the two transgenic Arabidopsis lines were examined. In transgenic lines expressing the 35S:GhRGL transgene, approximately 37% exhibited a dwarf phenotype (Fig. 5). The degree of dwarfism was different among different transgenic lines and ranged from severely dwarfed to mildly dwarfed and wild-type phenotypes (Fig. 5). We also examined the relationship between transgene expression and severity of the dwarf phenotype in these lines. Based on the results of the semi-quantitative RT-PCR, the transgene transcript was relatively abundant in lines that exhibited a severely dwarfed phenotype (e.g., RGL19) and less abundant in lines with taller stature (e.g., GAI11). Thus, there seemed to be a correlation between the severity of the dwarfing phenotype and the level of transgene expression. To confirm the results of semi-quantitative RT-PCR, we employed the sensitive real-time PCR technique using gene-specific primers to quantify transcript levels of GhRGL in different transgenic lines in the T3 generation. The height of six plants from six independent transgenic lines (T3-RGL1 to T3-RGL6) and of wild-type plants was measured after 30 days of growth (Fig. 6c). TaqMan assays were used to quantify the expression of GhRGL mRNA in these lines. The expression level of GhRGL mRNA was found to be negatively correlated with the height of the transgenic plants (Fig. 6a, b). The expression level of the GhRGL transgene in the shortest transgenic plant line (T3-RGL1) was nearly fivefold that in T3-RGL6 plants that showed an almost wild-type phenotype. Plant lines with intermediate heights showed intermediate levels of GhRGL expression.
Overexpression of GhRGL gene in Arabidopsis led to the dwarf phenotype. a Plants from three independent transgenic lines homozygous for a 35S:GhRGL transgene showing different degrees of dwarfism compared with a wild-type (WT) plant on the right. Transgenic lines (from left to right) are GhRGL19, GhRGL11 and GhRGL7. b Overexpression of GhRGL transgenic plants from three independent transgenic lines shown in (a) detected by semi-quantitative RT-PCR
GhRGL expression levels detected by quantitative (Q)PCR were negatively correlated with the height of GhRGL transgenic Arabidopsis plants. a Images of wild-type and third generation plants from six independent transgenic lines (T3-RGL1 to T3-RGL6). b GhRGL mRNA levels of the transgenic plants in a detected by QPCR. c The height of six independent transgenic Arabidopsis lines (T3-RGL1 to T3-RGL6) and of a wild-type plant
All transgenic lines expressing 35S:Ghrgl showed dwarf phenotypes (Fig. 7). These results indicate that Ghrgl can have a dominant effect over the endogenous DELLA protein coding gene in causing dwarfism.
Discussion
Bioactive GAs are important phytohormone that affect many aspects of plant development (King et al. 2001; Richards et al. 2001). The effect of GA on cotton fiber development has been documented in several studies (Beasley and Ting 1974; Sun et al. 2004; Yang et al. 2006; Shi et al. 2006). Davidonis (1990) reported that GA caused cell elongation in vitro in cotton ovule culture using both auxin-dependent and auxin-independent lines, although the elongation was more pronounced in the former. Exogenous application of IAA and GA to flower buds in planta and unfertilized ovules in vitro was found to produced an increased number of fibers (Gialvalis and Seagull 2001; Yang et al. 2006). Gokani and Thaker (2002) analyzed the effect of fiber length and dry weight in relation to endogenous GA levels and found that GA was one of the more important factors that determine fiber length. In our study, the addition of GA alone (without other plant hormones) to the culture medium promoted fiber elongation on fertilized ovules cultured in vitro, confirming earlier results. To further examine the GA dependence of cotton fiber development, we used a GA inhibitor, PAC, to block GA synthesis in vivo. The presence of PAC almost completely abolished fiber development, indicating that GA is required in fiber development. To the best of our knowledge, this is the first report of the effect of PAC on cotton fiber development. To test the possibility that this was through some pathways not mediated by GA, we added GA to the PAC treatment (combination treatment); our results showed that GA overcame the effect of PAC. Taking all these results together, it is clear that GA both promotes and is required for cotton fiber development.
Given that cotton fiber development is GA dependent and that DELLA proteins are known to be involved in GA signaling, we then looked at whether the cotton DELLA protein is involved in fiber development. We cloned a DELLA-like gene GhRGL from cotton. Bioinformatics analysis of the predicted polypeptide, GhRGL, revealed its high homology to DELLA proteins in Arabidopsis. This sequence homology and the presence of all of the important DELLA and GRAS function domains in GhRGL made it highly likely that GhRGL is a DELLA-type GA signal regulator in cotton.
To further demonstrate the role of GhRGL as a functional GAI-type DELLA gene, wild-type and mutant versions of GhRGL were overexpressed in Arabidopsis. Previous studies have shown that overexpression of wild-type AtGAI in Populus (Busov et al. 2006) produced wild-type plants, while dwarf transgenic plants were observed in rice (Fu et al. 2001) and tobacco (Hynes et al. 2003). Overexpression of GhRGL in Arabidopsis in our study produced plants with various degrees of dwarfism, which is in accordance with the results of AtGAI overexpression in rice and tobacco and suggests that GhRGL is functionally equivalent to AtGAI. Among the Arabidopsis transgenic lines overexpressing GhRGL, the degree of elongation was negatively correlated to the levels of GhRGL expression (Fig. 5). The level of GhRGL transcripts in the dwarf lines may have surpassed the capacity of endogenous GA to release the suppressing effect of GhRGL on plant height growth. In general, the higher the GhRGL transcript level, the more growth inhibition by GhRGL.
Mutations in the critical DELLA domain of AtGAI can render the resultant gai protein GA-insensitive, thus creating severe dwarf phenotypes in the plants that overexpress the mutant gai transcripts (Fu et al. 2001; Hynes et al. 2003; Busov et al. 2006). Ectopic expression of Ghrgl with a deletion in the DELLA domain of GhRGL in Arabidopsis resulted in severe dwarfs in most of the transgenic lines, further demonstrating that GhRGL is a functional equivalent of AtGAI.
To study the GhRGL gene expression pattern in cotton, we employed the sensitive real-time PCR technique using gene-specific primers to quantify transcript levels of GhRGL during different stages of cotton fiber development. The results showed that GhRGL had a higher level of expression in ovules at the fiber elongation stage, i.e, at 5 and 10 DPA, with the highest levels at 10 DPA. This result is in agreement with transcript profiles in immature cotton ovules obtained by Yang et al. (2006) using another DELLA protein-encoding gene in cotton. We also used HPLC methods to analyze the GA concentration in different tissues in cotton, and the results indicted a peak GA concentration in ovules at 5 and 10 DPA (data did not show). GhRGL represents a cotton DELLA protein that represses specific growth and development activities, and GA is thought to deactivate GhRGL-type DELLA proteins so that the repressing effect of GhRGL can be released. It is therefore puzzling that both the levels of GhRGL and GA increased in the ovules at the early to middle stages of cotton fiber development. One suggestion, based on results in Arabidopsis, is that certain levels of GAI are not subjected to GA regulation and therefore continue to suppress some of the GA promoted growth even in the presence of GA (Fleck and Harberd 2002). However, our data suggests that GhRGL is subjected to GA regulation because the mutant version of GhRGL with deletions in the GA-interacting DELLA domain, Ghrgl, was much more effective in producing dwarf phenotypes. Why the levels of a supposedly effector, GhRGL, and that of its supposedly repressor, GA, increase at the same development stages and in the same tissue remains to be determined.
References
Alamgir H, Peng J (2003) DELLA proteins and GA signalling in Arabidopsis. J Plant Growth Regul 22:134–140. doi:10.1007/s00344-003-0028-5
Basra A, Malik CP (1984) Development of the cotton fiber. Int Rev Cytol 89:65–113. doi:10.1016/S0074-7696(08)61300-5
Beasley C, Ting I-P (1974) The effect of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot 61:188–194. doi:10.2307/2441189
Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416:847–850. doi:10.1038/416847a
Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH (2006) Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins root growth and metabolite profiles in Populus. Planta 224:288–299. doi:10.1007/s00425-005-0213-9
Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiol 129:181–190. doi:10.1104/pp.010917
Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131:1055–1064. doi:10.1242/dev.00992
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi:10.1046/j.1365-313x.1998.00343.x
Crozier A, Kamiya Y, Bishop G, Yokota T (2000) Biosynthesis of hormones and elicitor molecules. In: Buchanan BB, Gruissem W, Jones RD (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 850–929
Davidonis GH (1990) Gibberellic acid-induced cell elongation in cotton suspension cultures. J Plant Growth Regul 9: 243–246
Dill A, Sun T-P (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159:777–785
Dill A, Thomas SG, Hu J, Steber CM, Sun T-P (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16:1392–1405. doi:10.1105/tpc.020958
Evans ML (1985) The action of auxin on plant cell elongation. Crit Rev Plant Sci 2:317–365
Fleck B, Harberd NP (2002) Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J 32:935–947. doi:10.1046/j.1365-313X.2002.01478.x
Fu X, Sudhakar D, Peng J, Richards DE, Christou P, Harberd NP (2001) Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13:1791–1802
Fu X, Richards DE, Ait-Ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14:3191–3200. doi:10.1105/tpc.006197
Gialvalis S and Seagull RW (2001) Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. J Cotton Sci 5:252–258
Gokani SJ and Thaker VS (2002) Role of gibberellic acid in cotton fibre development. J Agric Sci 138:255–260
Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV (2002) Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol 129:191–200. doi:10.1104/pp.010918
Harberd NP, King KE, Carol P, Cowling RJ, Peng JR, Richards DE (1998) Gibberellin: inhibitor of an inhibitor of…? Bioessays 20:1001–1008. doi:10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O
Hynes LW, Peng J, Richards DE, Harberd NP (2003) Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Res 12:707–714
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice a constitutive gibberellin response mutant is caused by a null mutation of the SLR1 gene an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57–70. doi:10.1105/tpc.010319
Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127:1361–1366. doi:10.1104/pp.010724
King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159:767–776
Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58:257–263. doi:10.1007/BF00265176
Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng JR (2002) Gibberellin regulates Arabidopsis seed germination via RGL2 a GAI/RGA-like gene whose expression is up-regulated following inhibition. Genes Dev 16:646–658. doi:10.1101/gad.969002
McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-P, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15:1120–1130. doi:10.1105/tpc.010827
Muangprom A, Osborn TC (2004) Characterization of a dwarf gene in Brassica rapa including the identification of a candidate gene. Theor Appl Genet 108:1378–1384. doi:10.1007/s00122-003-1551-2
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497
Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome deficient mutants of Arabidopsis. Plant Physiol 113:1051–1058
Peng JR, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205. doi:10.1101/gad.11.23.3194
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F et al (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261. doi:10.1038/22307
Phinney BO (1984) Gibberellin A1 dwarfism and the control of shoot elongation in higher plants. In: Crozier T, Hillman JR (eds) The biosynthesis and metabolism of plant hormones. Society for Experimental Biology Seminar Series 23 A. Cambridge University Press, London, pp 17–41
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROWLIKE genes. Plant J 18:111–119. doi:10.1046/j.1365-313X.1999.00431.x
Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52:67–88. doi:10.1146/annurev.arplant.52.1.67
Richards DE, Peng JR, Harberd NP (2000) Plant GRAS and metazoan STATs: one family? Bioassays 22:573–577. doi:10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H
Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299:1896–1898.
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling molecular biological and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18:651–664. doi:10.1105/tpc.105.040303
Silverstone AL, Ciampaglio CN, Sun T-P (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155–169
Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun T-P (2001) Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13:1555–1566
Sun Y, Fokar M, Asami T, Yoshida S, Allen RD (2004) Characterization of the Brassinosteroid insensitive 1 genes of cotton. Plant Mol Biol 54:221–232. doi:10.1023/B:PLAN.0000028788.96381.47
Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD (2005) Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol 46:1384–1391. doi:10.1093/pcp/pci150
Thomas SG, Sun T-P (2004) Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol 135:668–676
Tiwari SC, Wilkins TA (1995) Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot 73:746–757
Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135:1008–1019. doi:10.1104/pp.104.039578
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12. doi:10.1006/abio.1994.1538
Wilkins TA, Arpat AB (2005) The cotton fiber transcriptome. Physiol Plant 124:295–300. doi:10.1111/j.1399-3054.2005.00514.x
Wilkins TA, Jernstedt JA (1999) Molecular genetics of developing cotton fibers. In Basra AM (ed) Cotton fibers. Hawthorne Press, New York, pp 231–267
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann ESM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19:2140–2155
Yang S-S, Cheung F, Lee JJ, Ha M, Wei NE, Sze S-H, Stelly DM, Thaxton P, Triplett B, Town CD, Chen ZJ (2006) Accumulation of genome-specific transcripts transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47:761–775. doi:10.1111/j.1365-313X.2006.02829.x
Zeevaart J, Talon M (1992) Gibberellin mutants in Arabidopsis thaliana. In: Karssen CM, van Loon LC, Vreugdenhil D (eds) Progress in plant growth regulation. Kluwer, Dordrecht, pp 34–42
Zhang Y, Ni Z-F, Yao YY, Nie XL, Sun QX (2007) Gibberellins and heterosis of plant height in wheat (Triticum aestivum L). BMC Genet 8:40–50. doi:10.1186/1471-2156-8-40
Acknowledgments
The authors thank the Chinese Ministry of Science and Technology and The National Basic Research Program (project 2004CB117302) for the financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liao, Wb., Ruan, Mb., Cui, Bm. et al. Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum . Plant Growth Regul 58, 35–45 (2009). https://doi.org/10.1007/s10725-008-9350-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-008-9350-z