Abstract
Allelopathy is the main cause of continuous cropping obstacles. Peanut wild species Arachis correntina (Burkart) Krapov. & W.C. Gregory is more resistant to continuous cropping obstacle than cultivated peanut, but its molecular mechanism in response to allelochemicals remains unknown. Benzoic acid (BA) and p-cumaric acid (PCA) are known allelochemical. To gain more insight into cellular response to BA and PCA, we applied high-throughput genetic sequencing to study the transcriptome changes of peanut cultivated variety Xiaobaisha and wild species A. correntina in the presence of 2 mM BA and PCA. The result showed that A. correntina resistance to BA and PCA stress was more weaker than Xiaobaisha. BA and PCA decreased significant shoot length and dry matter weight of A. correntina. Differentially expressed genes (DGEs) related to flavonoid biosynthesis, phenylpropanoid biosynthesis, plant–pathogen interaction and plant hormone signal transduction were transcribed less in A. correntina than Xiaobaisha under BA and PCA stress, including the less up-regulated genes involving in phenylpropane and flavonoid biosynthesis, detoxifying enzymes, Auxin responsive protein, WRKY family and Ef-hand, which might contribute to A. correntina’s weak resistance to BA and PCA stress. The results showed that the resistance to single allelopathic substance might not be the reason for A. correntina resistance to continuous cropping obstacle. In addition, DGEs of both A. correntina and Xiaobaisha were significantly enriched in the pathways associated with isoflavonoid biosynthesis and glutathione metabolism under BA and PCA stress. This was the first report on identification of DGEs under BA and PCA stress between peanut cultivated variety and wild species. The result would be great helpful to insight into the mechanisms of peanut response to allelochemicals stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peanut is one of the main oil crops in China. Continuous cropping obstacle positively affects the growth, yield and quality of peanut, which seriously restricts peanut production. The main reasons for peanut successive cropping obstacle include imbalance of soil nutrients, allelopathy, imbalance of microbial communities and increase of soil pathogenic microorganisms. Plant allelochemicals produced by the stem and leaf volatile, plant residue, leaf and root secretion release to the environment, which affect the surrounding plant growth and development (Rice 1984a). Allelochemicals in root secretion is the most important cause of continuous cropping obstacle (Aslam et al. 2017).
Peanut wild species have broader genetic diversity and many excellent traits, which cultivated peanut don’t have. Peanut wild species resistance to some diseases and insect pests, such as leaf spot disease, late blight and thrips, is significantly higher than that of cultivated peanut species (Krapovickas et al. 2007). Peanut wild relatives has a good application prospect in peanut genetic improvement (Leal-Bertioli et al. 2015; Ratnaparkhe et al. 2014; Bertioli et al. 2016). At present, there are few reports on the study of peanut resistance to continuous cropping obstacle. In the early stage, we carried out a study on screening and identification of wild germplasm resources, and preliminarily screened and identified a wild peanut species, Arachis correntina (Burkart) Krapov. & W.C. Gregory, which is more resistant to continuous cropping obstacle than cultivated peanut (Li et al. 2018). Arachis correntina also is one of the few wild peanut species that is compatible with cultivated peanut species. There are obvious differences in root exudates and allelopathic effects between A. correntina and peanut cultivars. A. correntina has less autotoxicitic compounds in root exudates than peanut cultivars (li et al. 2015). Rhizosphere substances of continuous cropping soil and hydroponic root exudates of peanut cultivar have significant inhibitory effects on plant height, root length, dry matter weight of peanut cultivar seedlings, while A. correntina show no significant inhibitory effects under the same treatment (li et al. 2015). We speculate that A. correntina resistance to continuous cropping obstacle was closely related to the specific genes of root response. Finding these genes with specific expression and understanding the role of these genes products in metabolism can reveal the molecular mechanism of peanut continuous cropping resistance. However, the molecular mechanism of A. correntina’s continuous cropping tolerance is still not completely clear, the number and species of genes isolated and identified are limited. The study on allelopathic effect of peanut wild species and cultivated variety provides the reference and theoretical basis to identify and evaluate their resistant abilities for continuous cropping obstacle.
Organic acids, phenols and terpenoids have been found to be the most common allelochemicals (Rice 1984b; Blum et al. 1999). Organic acids especially phenolic acids are typical kinds of allelochemicals such as salicylic acid, BA, ferulic acid and malic acid (Bezuidenhout et al. 2012; Chen et al. 2011; Uddin et al. 2012). BA has been isolated from cucumber root secretion, which is one of the main autotoxins in cucumber (Singh et al. 1999). BA also is isolated from ginseng rhizosphere soil, and significantly inhibit ginseng seed germination and growth (He et al. 2009; Li et al. 2011). In previous study, BA and PCA is isolated from peanut root secretions and its allelopathic inhibitory effects has been identified (Sène et al. 2000). Although BA and PCA has long been identified as allelochemicals, but molecular mechanism of plants response to BA and PCA is poorly understand. Transcription factors play an important role in the plants response to the abiotic stress (Thiruvengadam et al. 2016). With the development of tanscriptome technology, it is possible to study systematically plant adversity stress related to molecular mechanisms. The transcription factor of plant induced by allelochemicals stress can be united in wedlock with corresponding cis element, starting the corresponding gene transcription, regulation of a series of molecular response mechanism to reduce the adversity stress damage. Wild Peanut genome is published in 2016 (Bertioli et al. 2016), which allowes more precisely research related to peanut transcription factors induced by allelochemicals stress. Therefore this study intends to analyze peanut transcription factor expression changes under BA and PCA stress by using RNA-Seq methods, and preliminary clear the molecular mechanism of peanut response to BA and PCA.
Materials and methods
Plant materials and treatments
Peanut wild species A. correntina and cultivated variety Xiaobaisha were used for the experiments. Peanut wild species A. correntina was obtained from the national wild peanut germplasm nursery (Guangxi, China). Peanut cultivated variety Xiaobaisha was provided by Guangxi academy of agricultural sciences economic crops research institute (Guangxi, China). The experiment was carried out On March 18, 2018 in Guangxi crop genetic improvement and biotechnology lab, Nanning, Guangxi, China. Five peanut seeds were soak in a petri dish for 4 days (d) at 25 °C temperature in dark, treated with concentration of 2 mM BA and PCA, with sterile water as the control (cont4). Then five peanut seeds were sown in plastic pots (height 20 cm, width 13.3 cm) containing 450 mL MS liquid medium with plastic plates at 25 °C temperature light incubator cultured for 8d, which set up the same BA and PCA treatment, with no added BA and PCA as the control (cont8), six repeat each treatment. Then peanut seedling roots from BA and PCA stressed treatment and control were collected for determination of RNA extraction.
RNA extraction, library construction and RNA-sequencing
RNA extracted from the roots of per sample by total RNA Purification Kit (TIANDZ, Beijing, China) treated with DNase I (TaKaRaBiotechnology, Dalian, China). RNA concentration estimated was quantified by NanoDrop 2000 (Thermo). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 240 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Data analysis
Prior to DGEs analysis, raw reads were edited to filter out low-quality reads and containing adaptor sequence in order to obtain clean reads. These clean reads were then mapped to the reference genome sequence. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Hisat2 tools soft were used to map with reference genome. DEGs analysis of two samples was performed using the EBSeq R package (Leng et al. 2013). The resulting FDR (false discovery rate) were adjusted using the PPDE (posterior probability of being DE). The FDR < 0.05 & |log2(foldchange)| ≥ 1 was set as the threshold for significantly differential expression.
Gene functional annotation
Functional annotation of genes including DEGs were annotated against the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology).
qRT–PCR analyses
2 µg of total root RNA isolated as described above was reverse transcribed using M-MLV reverse transcriptase (Invitrogen, USA). Quantitative real-time PCR (qRT-PCR) were performed with three biological and technical replicates by using SYBR® Premix EX Taq™ (Takara, Tokyo, Japan). Primers were designed by used Premier3.0 primer design software online for qRT-PCR. The PCR amplification conditions were consist of one cycle of 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 20 s. The formula 2−ΔΔCT method was employed to calculat relative expression level of candidate genes (Livak and Schmittgen 2001).
Results
Effects of BA and PCA on peanut growth
Morphological and physiological characters related to plants growth and development had been widely used in the research on effects of allelopathic substances. The allelopathic inhibition of PCA and BA on Xiaobaisha was significantly lower than that of A. correntina. The result showned that both BA and PCA significantly inhibited shoot length and dry matter weight of A. correntina (p < 0.05), while there was no significant difference between treatments and control in Xiaobaisha (Table 1). BA and PCA could promote the root length of A. correntina, whereas slightly decreased that of Xiaobaisha.
Output statistics of sequencing data
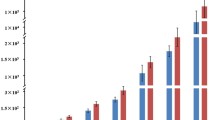
By sequencing, low-quality reads were removed and a total of 90.57 Gb clean data was obtained (Table 2). The Q30 bases percentage of each sample was not less than 90.83%. Total DGEs of two peanut varieties on the 8th day after BA and PCA stress was two times more than those on the 4th day (Fig. 1). Compared with the control, the number of DGEs in Xiaobaisha was the highest on the 8th day after BA treatment, with 6113 genes being differentially expressed, including 2752 genes being up-regulated and 3361 genes being down-regulated (Fig. 1). The number of DGEs in Xiaobaisha was relatively small on the 4th day after BA treatment, and there were 1897 DGEs, among which 1167 genes were up-regulated and 730 genes were down-regulated (Fig. 1).
DGEs analysis of peanut in response to BA
Gene ontology (GO) enrichment analysis of DEGs was implemented by the GO seq R packages based Wallenius non-central hyper-geometric distribution (Young et al. 2010), which could adjust for gene length bias in DEGs. Through the GO function annotation analysis, a total of 12,398 DGEs were found in A. correntina less than 56.5% that of Xiaobaisha (19,410). In molecular functions category, transcriptome analysis revealed more DGEs related to binding, catalytic activity and transporter activity in Xiaobaisha than A. correntina under BA stress (Supplemental Fig. S1; Supplemental Table S1). In the category of biological process, the transcription of DGEs associated with metabolic process, response to stimulus and single-organism process were higher in Xiaobaisha than in A. correntina (Supplemental Fig. S1; Supplemental Table S1). In the category of cell components, the number of DGEs associated with membrane, organelle, cell and cell part were significant less in A. correntina than in Xiaobaisha (Supplemental Fig. S1).
Different gene producted coordinate with each other to perform biological functions in plant. Pathway annotation analysis of DGEs was helpful to further understand the function of genes. KEGG (Kanehisa et al. 2008) was a database resource for understanding high-level functions and utilities of the biological system. We used KOBAS (Mao et al. 2005) software to test the statistical enrichment of DGEs in KEGG pathways. By KEGG class analysis, DGEs related to fatty acid degradation, flavonoid biosynthesis, phenylpropanoid biosynthesis, plant hormone signal transduction, plant–pathogen interaction and glycolysis/gluconeogenesis were more strongly transcribed in Xiaobaisha than in A. correntina (Supplemental Fig. S2). In contrast to control, there were significant differences for the DGEs of both Xiaobaisha and A. correntina in the pathways of flavonoid biosynthesis, isoflavonoid biosynthesis, phenylpropaboid biosynthesis, tryptophan metabolism, circadian rhythm-plant and photosynthesis-antenna proteins (Supplemental Fig. S2). 51 DGEs of both in Xiaobaisha and A. correntina were in response to BA stress at different treat times (Supplemental Fig. S3 c). Among them, 38 DGEs were annotated (Supplemental Table S2), involving in the pathway of carbohydrate transport and metabolism, signal transduction mechanisms, transcription, secondary metabolites biosynthesis, transport and catabolism.
DGEs analysis of peanut in response to PCA
By GO function annotation analysis, more DGEs responsed to biological process category, especially response to stimulus in Xiaobaisha than in A. correntina under PCA stress (Supplemental Table S3; Supplemental Fig. S4). In the category of molecular function, DGEs involved in associated with binding and catalytic activity were more strongly transcribed in Xiaobaisha than in A. correntina (Supplemental Table. S3). For the category of cell components, there were more DGEs related to membrane, organelle, organelle part and cell part in A. correntina compared with Xiaobaisha under PCA stress (Supplemental Table S3).
Base on KEGG class analysis, DGEs related to phenylpropanoid biosynthesis, plant hormone signal transduction, plant–pathogen interaction and diterpenoid biosynthesis were more strongly transcribed in Xiaobaisha than in A. correntina. Most strongly differentially transcribed DGEs of both Xiaobaisha and A. correntina were identified in the pathways of glutathione metabolism, phenylpropanoid biosynthesis, flavonoid biosynthesis, circadian rhythm-plant, linoleic acid metabolism, isoflavonoid biosynthesis, cysteine and methionine metabolism, stilbenoid, diarylheptanoid and gingerol biosynthesis, diterpenoid biosynthesis compared with the control (Supplemental Fig. S5). Xiaobaisha and A. correntina shared 28 DGEs response to PCA stress at different treat times (Supplemental Fig. S6; Supplemental Table S4), among which 26 DGEs were annotated (Supplemental Table S4), involving in phenylpropanoid biosynthesis, flavonoid biosynthesis, alpha-Linolenic acid metabolism, Isoflavonoid biosynthesis and glycerophospholipid metabolism.
DGEs analysis of peanut between BA and PCA treatment in response to BA and PCA
Under BA and PCA stress treatment, there were significant differences for the DGEs of both Xiaobaisha and A. correntina in the pathways of flavonoid biosynthesis, isoflavonoid biosynthesis, phenylpropaboid biosynthesis and circadian rhythm-plant (Supplemental Fig. S2 and Supplemental Fig. S4). DGEs related tophenylpropanoid biosynthesis (Supplemental Tables S5–S6), flavonoid biosynthesis (Supplemental Tables S7–S8), plant hormone signal transduction (Supplemental Tables S9–S10) and plant–pathogen interaction (Supplemental Tables S11–S12) were more strongly transcribed in A. correntina than in Xiaobaisha under BA and PCA stress. And this result was also shown in the heat map (Fig. 2), which showed changes in number of DGEs in A. correntina and Xiaobaisha under BA and PCA stress.
qRT-PCR validation of selected DGEs
To verify the accuracy of RNA-seq data, 8 DEGs were selected to design primers for qRT-PCR, including DGEs related to flavonoid biosynthesis, secondary metabolites biosynthesis (arahy.Tifrunner.gnm1.ann1.Z23BQJ), transport and catabolism (arahy.Tifrunner.gnm1.ann1.3G6AIR, arahy. Tifrunner.gnm1.ann1.ZRN1TD, arahy.Tifrunner.gnm1.ann1.LW6UI8), signal transduction mechanisms (arahy.Tifrunner.gnm1.ann1.3Z0Y0Z) and Transcription (arahy.Tifrunner.gnm1.ann1.4TST71) (Supplemental Table S2; Fig. 3a). Linear regression analysis showed that the RT-PCR data were significantly correlated with the RNA-seq data (R2 = 0.566, P < 0.001) (Fig. 3b), which indicated that the expression trend of DEGs between RNA-Seq and qRT-PCR was basically consistent. The results indicate that RNA-seq data in this study were reliable.
Discussion
Secondary metabolism provides a defense against abiotic stresses in plants. Secondary metabolism are the important sources of defensive substances including flavonoids, phenols, glucosinolates, terpenes and alkaloids, whose biosynthesis have the same core metabolic pathways such as phenylpropane metabolic pathways, tryptophan, tyrosine (Cramer et al. 2011; Dixon and Paiva 1995). Then, the specific metabolites are synthesized by using specific enzymes (Kliebenstein 2012). Due to its strong antioxidant effect, phenylpropane biosynthesis play an important role in plant response to oxidative stress caused by plant growth, disease resistance and abiotic stress (Yechun et al. 2011). Simple and complex phenolic compounds are thought to be antioxidants mainly derived from the phenylpropane pathway (Dixon and Paiva 1995). The accumulated material and reinforcement of cell wall organization can prevent exogenous substrates expansion and protect the integrity of the plant cells in infection site, these chemical components include alkaloids (Mao et al. 2011), lignin (József et al. 2007) and terpenoid substances (Łaźniewska et al. 2012). Phenylpropnaoid pathway is an important way of promoting the synthesis of plant secondary metabolites, and its mainly downstream branch pathway are biosynthesis of lignin, flavonoids and other benzene compounds, which are helpful to resist adversity stress for plants as phytoalexin, cell wall structure elements and signaling molecules (Dixon and Paiva 1995; Neale et al. 2000; Vogt 2010; Bonawitz et al. 2012). Flavonoid has a strong preventive effect on the oxidation of unsaturated fatty acids and can scavenge free radicals (Cao et al. 1997). In this study genes related to phenylpropane (Supplemental Table S5) and flavonoid pathway (Supplemental Table S7) were transcribed more stongly in Xiaobaisha than in A. correntina. The result implied mroe phenylpropane and flavonoid biosynthesis were available to Xiaobaisha, thereby could promote the accumulation of secondary metabolites such as terpenes, flavonoids and alkaloids, which were beneficial for plants to adapt to adversity stress (Ramakrishna and Ravishankar 2011). In addition enzyme genes related to secondary metabolites were more up-regulatein Xiaobaisha than in A. correntina under stress, including cytochrome p450, dioxygenase, oxidoreductase, dehydrogenase, oxygenas, oxidas and peroxidase (Supplemental Table S6 and Supplemental Table S8). These enzymes above had the functions of osmotic regulation and free radical scavenging (Stone and Walker 1995; Vranová et al. 2000; Tahtiharju and Palva 2001; Meskiere et al. 2003; Sheng 2003; Buchanan 2004), which were more helpful for Xiaobaisha roots against stress environment than A. correntina.
Signal transduction refers to a chemical or physical signals in the form of a series of molecular events through cell transfer, eventually leading to cell reaction process (Bradshaw and Dennis 2010). At the molecular level, these reactions include the change of gene transcription and translation, post-translational and conformational changes in proteins, as well as their location changes (Rodriguez et al. 1998; Lorenzo et al. 2002; Schweighofer and Meskiene 2004). These molecular events are the basic mechanisms controlling cell growth, proliferation, metabolism and many other processes (Sheen 1996; Krauss 2008). Plant hormone regulate plant response to stress (Bogatek and Gniazdowska 2007). The results of functional enrichment analysis showed that DEGs of two peanut varieties participated in the plant hormone signal transduction, including transcription, protein modification, hormonal regulation (Supplemental Table S9-S10). This suggested that the response of peanut root to BA and PCA stress involved in a very complex metabolic regulation network. Auxin plays its normal function of promoting the growth and development of plant roots. Aux/IAA family is a special kind of gene family that is induced by auxin (Worley et al. 2000), which has been found in pisumsativum, soybean and tobacco (Abel and Theologis 1995; Reed 2001). Studies have showed that the encoding product of Aux/IAA gene belongs to degradable nucleoprotein, because it plays the role of repressor protein in auxin signal transduction pathway and can negatively regulate the process of gene responsive auxin. The synthesis and degradation of these gens is the key step of auxin signal transduction process (Tiwari et al. 2001; Worley et al. 2000), thus caused the change of plant morphology and growth patterns (Liscm and Reed 2002). There were obivous different in plant hormone signal transduction between Xiaobaisha and A. correntina. The transcription of Aux/IAA and Auxin responsive protein were higher in Xiaobaisha than A. correntina, moreover most of Aux/IAA related genes were down-regulated and Auxin responsive protein were up-regulated in the former (Supplemental Tables S9–S10), which were bennifit for Xiaobaisha to the smooth transfer of auxin signal to the downstream, and could potentially reduce the harm of stress.
Genes expression of plant–pathogen interaction are induced in peanut under BA and PCA stress. With the recognition of pathogens and signal transduction, the downstream defense responses of plants are activated, including cell wall strengthening, accumulation of antimicrobial secondary metabolites and expression of pathogenesis-related genes (van Loon et al. 2006). There are two kinds of plant defense responses against pathogens, caused by plant pathogens in infection of host plants. PTI (PAMPs—triggered immunity) is the first defense reaction, which can make plants produce reactive oxygen species and the change in the amount of the expression of defense genes in plants (Sanabria et al. 2010; Dai et al. 2012; Jones and Dangl 2006). The second defense response is effector-triggered immunity (ETI), which is a kind of host immune response caused by the recognition of pathogen effectors (Jones and Dangl 2006). ETI can cause hypersensitive reactions near the infection site (Jones and Dangl 2006). Calcium (Ca2+) signaling is an important part of the PTI response, operating through Ca2+ sensing proteins (Luan et al. 2002; Gao et al. 2014; Chin et al. 2009; Saand et al. 2015). Structural the EF-hand Ca2+-binding proteins and calcium have been recognized as the key players in all aspects of cell function, starting with a cell’s birth during mitosis and ending with its apoptotic death (Jones and Dangl 2006). A malfunction in EF-hand proteins-signaling is considered the cause of many plant diseases. Functionally, EF-hand proteins can be divided into two general classes: the Ca2+ sensors and the Ca2+buffers (Kojetin et al. 2006; Brunet et al. 2005). The exceptional versatility of the EF-hand proteins is intimately associated with the diversity of the EF-hand motifs, such as discrepancy in conformations, domain organization, structural responses to calcium and so on. Here, the transcription of EF-hand was lower in A. correntina than in Xiaobaisha (Supplemental Table S11–S12), which indicated that Ef-hand genes were more activated and involved in the regulation of calcium ion signals in Xiaobaisha. With the recognition of pathogens and signal transduction, the downstream defense responses of plants are activated, including cell wall strengthening, accumulation of antimicrobial secondary metabolites and expression of pathogenesis-related genes (van Loon et al. 2006). It could potentially better protect the Xiaobaisha roots against abiotic stress. The multiple members of the WRKY family are involved in the response of plants to biological and abiotic stress. The promoter region contains binding sequences of multiple stress-related transcription factors (Ramamoorthy et al. 2008). There is a strong synergistic and regulatory relationship among WRKY family genes (Chujo et al. 2013; Qiu et al. 2007). In this study, WRKY transduction was more active in Xiaobaisha than in A. correntina (Supplemental Table 11), which implied that WRKY family might be closely related to Xiaobaisha resistant ability to stress.
Conclusions
The result suggested that Xiaobaisha had higher resistance to BA and PCA stress than A. correntina. Sequencing results showed that there were significantly differences in transcriptome level between A. correntina and Xiaobaisha at under BA and PCA stress. DGEs associated with fatty acid degradation, flavonoid biosynthesis, phenylpropanoid biosynthesis, plant hormone signal transduction, Plant–pathogen interaction and glycolysis/gluconeogenesis were more transcribed under BA stress in Xiaobaisha compared to A. correntina. Under PCA stress, DGEs related to phenylpropanoid biosynthesis, plant hormone signal transduction, plant–pathogen interaction and diterpenoid biosynthesis were more strongly transcribed in Xiaobaisha than in A. correntina. The transcription of DGEs associated with secondary metabolites, plant–pathogen interaction and plant hormone signal transduction were higher in Xiaobaisha than A. correntina uner BA and PCA stress. Moreover, the majority of these genes were closely related to flavonoid biosynthesis, phenylpropanoid biosynthesis, Aux/IAA and Auxin responsive protein, EF-hand and WRKY family.
References
Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin inducible proteins related to PSIAA4 from pea (Pisumsativum). Plant J 8:87–96
Aslam F, Khaliq A, Matloob A, Tanveer A, Hussain S, Zahir ZA (2017) Allelopathy in agro-ecosystems: a critical review of wheatalle- lopathy-concepts and implications. Chemoecology 27:1–24
Bertioli DJ, Cannon SB, Froenickel Froenicke L, Huang GD, Farmer AD, Cannon EKS, LiuX Gao DY, Clevenger J, Dash S, Ren LH, Moretzsohn MC, Shirasawa K, Huang W, Vidigal B, Abernathy B, Chu Y, Niederhuth CE, Umale P, Araújo ACG, Kozik A, Kim KD, Burow MD, Varshney RK, Wang XJ, Zhang XY, Barkley N, Guimarães PM, Isobe S, Guo BZ, Liao BS, Stalker HT, Schmitz RJ, Scheffler BE, Leal-Bertioli SCM, Xun X, Jackson SA, Michelmore R, Ozias-Akins P (2016) The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid an cestors of cultivated peanut. NatureGenet 48(4):438–446
Bezuidenhout SR, Reinhardt CF, Whitwell I (2012) Cover crops of oats, stooling rye and three annual ryegrass cultivars influence maize and Cyperus esculentus growth. Weed Res 52(2):153–160
Blum U, Shafer SR, Lehman ME (1999) Evidence for inhibitory allelopathic interactions involving phenolic acids in field soils: concepts vs an experimental model. Critical Rev Plant Sci 18(5):673–693
Bogatek R, Gniazdowska A (2007) ROS and phytohormones in plant–plant allelopathic interaction. Plant Signal Behav 2:317–318
Bonawitz ND, Soltau WL, Blatchley MR, Powers BL, Hurlock AK, Seals LA, Weng JK, Stout J, Chapple C (2012) REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J Biol Chem 287(8):5434–5445
Bradshaw RA, Dennis EA (2010) Handbook of cell signaling, 2nd edn. Academic Press, Amsterdam. ISBN 9780123741455
Brunet S, Scheuer T, Klevit R, Catterall WA (2005) Modulation of CaV1.2 channels by Mg2 + acting at an EF-hand motif in the COOH-terminal domain. J Gen Physiol 126(4):311–323
Buchanan BB (2004) Plant biochemistry and molecular biology. Science Press, Beijing, pp 563–569
Cao G, Sofic E, Prior RL (1997) Antioxidant and prooxidant behavior of flavonoids: structure - activity relationship. Free Radic Biol Med 22(5):749–760
Chen SL, Zhou BL, Lin SS, Li XY, Xue L (2011) Accumulation of cinnamic acid and vanillin in eggplant root exudates and the relationship with continuous cropping obstacle. Afr J Biotechnol 10(14):2659–2665
Chin K, Moeder W, Yoshioka K (2009) Biological roles of cyclic nucleotide-gated ion channels in plants: what we know and don’t know about this 20 member ion channel family. Botany 87:668–677
Chujo T, Miyamoto K, Shimogawa T, Shimizu T, Otake Y, Yokotani N, Nishizawa Shibuya N, Nojiri H, Yamane H, Minami E, Okada K (2013) OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol Biol 82(1/2):23–37
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163. https://doi.org/10.1186/1471-2229-11-163
Dai JC, Huang JG, Wang CL, Zhao KJ (2012) Pathogen conservation molecules and PAMP-triggered innate immunity in plants. Microbiol China 39(4):553–565 (in Chinese)
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Gao X, Cox KL, He P (2014) Functions of calcium-dependent protein kinases in plant innate immunity. Plants 3:160–176. https://doi.org/10.3390/plants3010160
He CN, Gao WW, Yang JX, Wu B, Zhang XS, Zhao YJ (2009) Identification of autotoxic compounds from fibrous roots of Panaxquinquefolium L. Plant Soil 318:63–72
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
József R, Ana MB, Dénes S (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144(4):1863–1877
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kliebenstein DJ (2012) Plant defense compounds: systems approaches to metabolic analysis. Annu Rev Phytopathol 50(1):155–173
Kojetin DJ, Venters RA, Kordys DR, Thompson RJ, Kumar R, Cavanagh J (2006) Structure, binding interface and hydrophobic transitions of Ca2 + -loaded calbindin-D(28K). Nat Struct Mol Biol 13(7):641–647
Krapovickas A, Gregory WC, Simpson WCE (2007) Supplemento || taxonomy of the genus Arachis (Leguminosae). Bonplandia 16:7–205
Krauss G (2008) Biochemistry of signal transduction and regulation. Wiley, New York, p 15. ISBN 978-3527313976
Łaźniewska J, Macioszek VK, Kononowicz AK (2012) Plant–fungus interface: the role of surface structures in plant resistance and susceptibility to pathogenic fungi. Physiol Mol Plant Pathol 78(78):24–30
Leal-Bertioli SCM, Santos SP, Dantas KM, Inglis PW, Nielen S, Araujo ACG, Silva JP, Cavalcante U, Guimarães PM, Brasileiro ACM, Carrasquilla-Garcia N, Penmetsa RV, Cook D, Moretzsohn MC, Bertioli DJ (2015) Arachis batizocoi:a study of its relationship to cultivated peanut (A. hypogaea) and its potential for introgression of wild genes into the peanut crop using induced allotetraploids. Ann Bot 115(2):237–249
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, Haag JD, Gould MN, Stewart RM, Kendziorski C (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29(8):1035–1043
Li Y, Huang XF, Ding WL (2011) Autotoxicity of Panax ginseng rhizosphere and non-rhizosphere soil extracts on early seedlings growth and identification of chemicals. Allelopathy J 28(2):145–154
Li Z, Jiang LG, Tang RH, Xiong FQ, Tang XM, Jiang J, He LQ, Zhong RC, Han ZQ (2015) Identification and allelopathy of chemical compositions of peanut root exudates. J South China Agric Univ 36(5):48–53 (in Chinese)
Li Z, Jiang LG, Tang RH, Guo WF (2018) Effects of long-term continuous peanut cropping on dry matter weight of different peanut varieties, soil nutrient contents and enzyme activities. Soils 50(03):491–497 (in Chinese)
Liscm E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49(3–4):387–400
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lorenzo O, Nicolas C, Nicolas G, Rodriguez D (2002) Molecular cloning of a functional protein phosphatase 2C (Fs PP2C) with unusual features and synergistically up-regulated by ABA and calcium in dormant seeds of Fagus sylvatica. Physiol Plant 114:482–490
Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14:S389–S400
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23(4):1639–1653
Meskiere I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rod-riguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitrogen-activated protein kinase. J Biol Chem 278:18945–18952
Neale AD, Biomstedt CK, Brorson P, Le TN, Guthridge K, Evans J, Gaff DF, Hamill JD (2000) The isolation of genes from the resurrection grass SporobolusStapfianus which are induced during severe drought stress. Plant Cell Environ 23:265–277
Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interactions 20(5):492–499
Ramakrishna A, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Signal Behav 6(11):1720
Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S (2008) A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol 49(6):865–879
Ratnaparkhe MB, Lee TH, Tan X, Wang X, Li J, Kim C, Rainville LK, Lemke C, Compton RO, Robertson J, Gallo M, Bertioli DJ, Paterson AH (2014) Comparative and evolutionary analysis of major peanut allergen gene families. Genome Biol Evol 6(9):2468–2488
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6:420–425
Rice EL (1984a) Allelopathy, 2nd edn. University of Oklahoma Press, Oklahoma, pp 1320–1344
Rice EL (1984b) Allelopathy, 2nd edn. University of Oklahoma Press, Oklahoma, pp 5–7
Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phos-phatase 2C involved in ABA signal transduction in Arabidopsis thaliana. FEBS Lett 421:185–190
Saand MA, Xu YP, Li W, Wang JP, Cai XZ (2015) Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front Plant Sci 6:303. https://doi.org/10.3389/fpls.2015.00303
Sanabria NM, Huang JC, Dubery IA (2010) Self/non-self perception in plants in innate immunity and defense. Self Nonself 1(1):40–54
SchweighoferA Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 19:236–243
Sène M, Doré T, Pellissier F (2000) Effect of phenolic acids in soil under and between rows of a prior sorghum (sorghum bicolor) crop on germination, emergence, and seedling growth of peanut (Arachis hypogea). J Chem Ecol 26(3):625–637
Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274:1089–1092
Sheng L (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54:63–92
Singh HP, Batish DR, Kohli RK (1999) Autotoxicity: concept, organisms, and ecological significance. Crit Rev Plant Sci 18:757–772
Stone JM, Walker JC (1995) Plant protein kinase families and signal transduction. Plant Physiol 108:451–457
Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 6:461–470
Thiruvengadam M, Baskar V, Kim SH, Chung IM (2016) Effects of abscisic acid, jasmonic acid and salicylic acid on the content of phytochemicals and their gene expression profiles and biological activity in turnip (Brassica rapa ssp. rapa). Plant Growth Regul 80:377–390
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell 1(3):2809–2822
Uddin MR, Li X, Won OJ, Park SU, Pyon JY (2012) Herbicidal activity of phenolic compounds from hairy root cultures of Fagopyrumtataricum. Weed Res 52(1):25–33
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20
Vranová E, Langebartels C, Van MM, Inzé D, van Camp W (2000) Oxidative stress, heat shock and drought differentially affect expres- sion of a tobacco protein phosphatase 2C. J Exp Bot 51:1763–1764
Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21:553–562
Yechun W, Steven C, Oliver Y (2011) Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol 91(4):949–956
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. https://doi.org/10.1186/gb-2010-11-2-r14
Acknowledgements
This study was funded by the Science Development Foundation of Guangxi Academy of Agricultural Sciences (Nos. 2018JZ35, 2017JZ15), and the National Natural Science Foundation of China (Nos. 31460115, 31660087, 31660428).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animals rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Xiong, F., Guo, W. et al. The root transcriptome analyses of peanut wild species Arachis correntina (Burkart) Krapov. & W.C. Gregory and cultivated variety Xiaobaisha in response to benzoic acid and p-cumaric acid stress. Genet Resour Crop Evol 67, 9–20 (2020). https://doi.org/10.1007/s10722-019-00859-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-019-00859-6