Abstract
Turnip (Brassica rapa L. em. Metzg. subsp. rapa), which is considered to be a primitive type of cultivated B. rapa, is cultivated worldwide as vegetable and fodder. To elucidate the phylogenetic relationships of Eurasian turnips, we examined their morphology and analyzed 6 cpSSR and 18 nuSSR loci in 87 accessions. Examination of seed coat mucilage and leaf hairs revealed existence of geographic distinctions. Two haplogroups were categorized among 12 haplotypes identified by the analysis of cpSSRs and two clusters were detected based on nuSSRs. These haplogroups and clusters were different between eastern and western Eurasia. Although morphological differences were detected between eastern and western Japan, no clear differences of haplotypes and clusters were found in Japanese turnips. Accessions from continental Asia showed various haplogroups and clusters and higher levels of genetic diversity than those from other regions. These results, in addition to previous studies suggest that central Asia is the sole geographic origin of turnips that Asian turnips did not originate as descendants of European turnips, and that almost all Japanese turnips were derived from central Asia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Turnip (Brassica rapa L. em. Metzg. subsp. rapa) is a diploid (2n = 20), and an annual or biennial plant cultivated worldwide as vegetable and fodder (Rakow 2004; Hammer et al. 2013). The wild form of B. rapa is distributed widely from Europe to central Asia (De-Candolle 1886; Sinskaia 1928; Mizushima and Tsunoda 1967; Prakash and Hinata 1980). De-Candolle (1886) proposed that turnips were cultivated in Europe around 2500–2000 BC and spread from there to Asia (Gomez-Campo and Prakash 1999). According to leaf traits and geographical distribution, Sinskaia (1928) classified turnips into seven geographic groups; (1) Teltow turnips, (2) West European turnips with dissected leaves, (3) Asia Minor and Palestine turnips, (4) Russian turnips of the Petrovsky type, (5) Asiatic Afghanistan turnips with glabrous leaves, (6) Japanese turnips with entire glabrous leaves, and (7) European entire-leaved turnips with pubescent leaves. She proposed that Teltow and Afghanistan turnips represent a primitive group of forms very near to the wild progenitors of turnips, but that the Japanese turnips are advanced forms. Based on these observations, two hypotheses were suggested for the origin of turnips; (1) they originated in Asia or (2) their origin is polyphyletic, with Asiatic and European turnips having developed independently in Asia and Europe.
In Japan, many landraces have been differentiated. Based on morphological traits such as seed coat mucilage and leaf hairs, the turnips of Japan are generally classified into three groups: (1) Japanese-type, which are characterized by seed coat mucilage and no leaf hairs; (2) European-type, characterized by non-seed coat mucilage and leaf hairs; and (3) intermediate-type, with segregation for seed coat mucilage and leaf hairs (Shibutani and Okamura 1954; Aoba 1958). Geographically, Aoba (1961, 1981a) reported that the European type is distributed in eastern Japan, the Japanese type in western Japan, and the intermediate type mainly in the Chubu district of central Japan. It has been suggested that Japanese-type turnips are closely related to turnips of Afghanistan, on the basis of their similar characters (Sinskaia 1928; Shibutani and Okamura 1954), whereas European-type turnips in eastern Japan, which have the same characters as European turnips, had immigrated by another route, such as northern China or Siberia, and that intermediate-type turnips in central Japan had originated from crosses between European- and Japanese-types (Aoba 1961, 1981a). Schebalina and Sazonova (1985), who classified turnips to three groups such as European, Iraqian and Asian, suggested that European group distribute widely in Eurasia from Europe to Japan.
Molecular markers are powerful tools for genetic analysis, plant systematics, and plant breeding. Among these markers, simple sequence repeat (SSR) markers have been developed for both chloroplast and nuclear genomes and have been used in various types of analysis in many crops (Kalia et al. 2011). Chloroplast SSRs (cpSSRs) show a high level of intraspecific variation and are suitable for evolutionary studies (Provan et al. 2001). Nuclear SSRs (nuSSRs) are also powerful tools for characterizing genetic differentiation in nuclear polymorphism (Wolfe and Liston 1998). Intra- and interspecific genetic relationships using SSRs have been reported in Brassica and related species (Flannery et al. 2006; Allender et al. 2007; Louarn et al. 2007; Yamane et al. 2009; Pino Del Carpio et al. 2011).
The purpose of the present study was to characterize the phylogenetic relationships among turnips in Eurasia based on cpSSRs and nuSSRs as well as morphology such as seed coat mucilage and leaf hairs, which have been used to classify turnips. Moreover, we attempted to test the hypothesis that the two groups of turnips in Japan (European-type in the east and Japanese-type in the west) are derived from different origins.
Materials and methods
Plant materials
Eighty-seven turnip cultivars and strains were used in this study, which were kindly provided from National Institute of Agrobiological Sciences (NIAS) in Japan, the Institute of Plant Genetics and Crop Plant Research (IPK) in Germany, the Dutch Crop Genetic Resources Center (CGN) in the Netherlands, and the Vavilov Research Institute of Plant Industry (VIR) in Russia (Table 1). These accessions were classified into six geographical groups (Europe, Russia, continental Asia, eastern Japan, central Japan, and western Japan) (Table 1).
Examinations of seed coat mucilage and leaf hairs
The presence or absence of seed coat mucilage was evaluated by easy distinction method (Yazawa et al. 1986). Twenty seeds per an accession were placed in 2 mL plastic tube containing 1 mL of distilled water for 2–3 h at room temperature, and then dipped into ethyl acetate for 30 s, air-dried for 5 min at room temperature, and dusted with 1 g cerite. A powder covering of cerite was considered as the “presence” and the absence of cerite was considered the “absence” of seed coat mucilage. Accessions with both seed types were also found. The leaf hairs of adult plants were scored at three levels: “absence,” “few,” and “many.”
Genotyping of SSR
Total DNA was extracted from the first true leaf of a single plant of each accession using a CTAB method (Rogers and Bendich 1988). Conserved 20 primer pairs that flank cpSSRs have been selected from several species in Brassicaceae (Chung and Staub 2003; Flannery et al. 2006; Allender et al. 2007). Of these 20 primer pairs, six showed polymorphisms in eight randomly selected turnip accessions of turnips and were used for subsequent analysis (Table 2). For nuSSRs analysis, 18 primer pairs were selected from a total of 30 pairs derived from the BRMS primer set (Suwabe et al. 2006), based on the linkage group, amplification strength, and polymorphism, and covered eight of the ten linkage groups of B. rapa (Table 2). All cpSSR and nuSSRs loci were amplified by PCR. Each reactions was performed in 10 μL of total volume containing 10 ng of template DNA, 10 mM Tris–HCl (pH 8.3), 20 mM KCl, 1.5 mM MgCl2, 100 mM dNTP, 0.05 mM forward primer with fluorescent label (HEX, FAM, VIC or TET), 0.05 mM reverse primer and 0.1 unit of Taq DNA polymerase. The thermal cycling conditions were 5 min at 94 °C, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min, and then 72 °C for 5 min. One microliter PCR product was diluted to 20 μL with Hi-Di Formamide and added 0.2 μL ROX 500 internal size standard (Applied Biosytstems) was added before loading on an ABI 310 DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions. Trace files from the sequencer were then scored using GeneMapper v3.5 software (Applied Biosystems).
Statistical analysis
Each accession was assigned a haplotype based on the combination of allelic information from six cpSSR loci. The relationships among haplotypes were analyzed with a median-joining network (Bandelt et al. 1999) using the NETWORK computer program (www.fluxus-engineering.com). Gene diversity H (Nei 1973) within geographic groups was estimated for both cpSSRs and nuSSRs using GenAlEx version 6 (Peakall and Smouse 2006). The software STRUCTURE 2.2 (Pritchard et al. 2000) was used to identify the population structure. The program was run using the admixture model with 200,000 replicates for burn-in and 200,000 replicates for analysis. The most likely number of K was estimated by calculating ΔK to identify the top level in the hierarchical structure (Evanno et al. 2005). Principal coordinate analysis of nuSSR data was performed with GenAlEx v. 6 program (Peakall and Smouse 2006). Neighbor-joining trees (Saitou and Nei 1987) based on Nei’s genetic distance (Nei et al. 1983) were constructed using Populations 1.2.30beta (Langella 2007).
Results
Seed coat mucilage and leaf hairs
All turnip accessions from Europe and Russia showed absence of seed coat mucilage and many leaf hairs (Table 1, Fig. 1A, B). In those from continental Asia, three types of seed mucilage (absence, presence, and segregation) were found, and three types of leaf hairs were also found, although most accessions were characterized by many leaf hairs. Especially, such phenomena were also found in accessions in central Asia. Japanese accessions classified to the three geographical regions showed distinct geographical distributions. In eastern Japan, accessions without seed coat mucilage and/or with leaf hairs were more frequent than other types, whereas in western Japan those with seed coat mucilage and/or without leaf hairs were the most frequent. The characteristics of accessions from central Japan were intermediate between those from western and eastern Japan (Fig. 1A, B).
In general, turnips lacking seed coat mucilage tended to possess leaf hairs and those with mucilage tended to lack leaf hairs. A Chi square test of independence using the 87 accessions showed that these two characters were not independent (χ2 = 32.226, p = 1.75 × 10−6).
Haplotypic polymorphism in cpSSR data
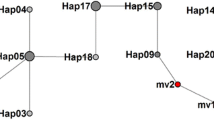
Analysis of six cpSSRs revealed a wide range of diversity among the turnips accessions. The numbers of alleles at cpSSR loci ranged from 2 to 4, and gene diversity H values ranged from 0.045 to 0.690 (Table 2). A total of 12 haplotypes were detected among the 87 accessions (Table 3). The median-joining network analysis showed that the haplotypes could be roughly categorized into two groups, haplogroup I (haplotypes A–F) and haplogroup II (haplotypes G–L) (Table 3, Fig. 2). All accessions from Europe and Russia had haplotypes belonging to haplogroup I, whereas almost all accessions from Japan had the haplotypes belonging to haplogroup II (Figs. 1C, 2, Table 1). The haplotypes of both haplogroups were detected in turnips of continental Asia, and also in those of central Asia. The gene diversity H of the geographical groups ranged from 0.073 to 0.363, with the highest value found in turnips from continental Asia and the lowest value in those from Europe (Table 4).
Median-joining network with branch lengths proportional to the number of mutational steps between 12 chloroplast haplotypes. The size of pie charts is proportional to the frequency of haplotype and the pie charts are separated by the colors, which mean the geographical subgroups. The mv1 and mv2 mean median vector
Phylogenetic analysis by nuSSR polymorphism
The number of alleles at 18 nuSSR loci in the 87 accessions varied from 3 to 17 per locus (Table 2). The gene diversity H ranged from 0.453 to 0.840 (Table 2). The genetic structure of the accessions based on STRUCTURE analysis is shown in Fig. 3. The model with K = 2 was selected as the most likely number of genetic clusters, because this model showed the highest ΔK [27.64 (K = 2), 0.84 (K = 3), and 0.12 (K = 4)]. All accessions from Europe and Russia were assigned to cluster 1, whereas all but two (Tomisato-kabu, Shogatsu-kabu) from Japan were assigned to cluster 2 (Figs. 1D, 3). Many accessions from continental Asia were assigned to cluster 1 and some to cluster 2. In a principal coordinate analysis, the first two coordinates explained 19.7 % (14.1 % by PC1 and 5.6 % by PC2) of the total variation (Fig. 4). All accessions from Europe and Russia were scattered in the second and third quadrants, whereas almost all Japanese accessions were scattered in the first and fourth quadrants. In the Japanese accessions no geographical differences (eastern vs. western) were detected. Accessions from continental Asia showed a wide distribution. The phylogenetic trees showed the similar results (Fig. 5). The gene diversity H of the geographical groups ranged from 0.562 to 0.691, higher than those for the cpSSR markers. Despite the small differences among geographic groups, the highest value was found for continental Asia (Table 4).
Discussion
Our morphological examination of 87 landraces of turnips showed geographical differences in variation of seed coat mucilage and leaf hairs. To our knowledge, this is the first report characterizing the seed coat mucilage of extensive accessions from Europe, Russia, and continental Asia. Our results reveal that European and Russian turnips had no genetic variations in comparison with other regions. Sinskaia (1928) indicated that turnips from Europe, Russia, and Asia Minor have pubescent leaves, whereas those from Afghanistan and Japan have glabrous leaves, with turnips from Afghanistan sometimes showing pubescent leaves. However, she investigated a limited number of Japanese turnips. Via examination of seed coat mucilage and leaf characters in extensive Japanese and some European accessions, Shibutani and Okamura (1954) and Aoba (1961, 1981a) reported that Japanese turnips were classified into three types, Japanese-type (with presence of seed mucilage and glabrous leaves, and distributed in western Japan), European-type (with absence of seed mucilage and pubescent leaves, and distributed in western Japan) and intermediate-type. Our results agree with these previous studies.
Considering that no accessions with seed coat mucilage were found except in western Japan, Aoba (1981b) speculated that the seed coat mucilage character of B. rapa occurred with the establishment of B. rapa L. em. Metzg. subsp. niposinica (Bailey) Hanelt in western Japan and was introduced to turnips (subsp. rapa). However, our results reveal that turnips with seed coat mucilage are present among accessions from central Asia, indicating that two types of seed coat mucilage were already present in turnips before their introduction to Japan.
As mentioned in the introduction, two hypotheses for the origin of turnips have been proposed: (1) the origin is monophyletic, with turnips having evolved either in Europe (De Candolle 1886; Prakash and Hinata, 1980) or in Asia (Sinskaia 1928) and (2) the origin is polyphyletic, with turnips having been cultivated in both Europe and Asia (Sinskaia 1928). Sinskaia (1928) reported that Teltow turnips (cultivated in Europe) and Afghan turnips had characters near those of the wild progenitor. Takuno et al. (2007) considered that turnips may be a primitive type of cultivated B. rapa that originated in central Asia or in Europe and spread to east Asia, Europe, and India. Our results of SSR analyses showed the presence of two distinct groups of haplotypes and population structures in Eurasian turnips, eastern and western, which are consistent with the results of a phylogenetic analysis of B. rapa using AFLP (Zhao et al. 2005; Takuno et al. 2007). Turnips in continental Asia included both types of the cpSSR haplogroup, and showed a higher level of genetic diversity than those in other regions. Wild form of B. rapa is distributed from Europe to central Asia (De-Candolle 1886; Mizushima and Tsunoda 1967; Gomez-Campo and Prakash 1999). These results suggest that central Asia is the sole geographical origin of turnips or one of its primary centers of origin. To correctly determine whether turnips are of monophyletic or polyphyletic origin, investigation of many accessions in central Asia and identification and comparison of the genes controlling swollen root formation in European and Asian turnips are needed.
Japan is considered as a center of turnip varietal development (Nishi 1980). Based on morphological examination, Japanese turnips are roughly divided into two types, Japanese- and European-type (Shibutani and Okamura 1954; Aoba 1961, 1981a; this study). Aoba (1961, 1981a) speculated that Japanese-type turnips distributed in western Japan migrated from Afghanistan to Japan via China or the Korean peninsula, whereas European-type turnips in eastern Japan came by way of Siberia or northern China (having originated in Europe) to eastern Japan. We expected that there are genetic differences in Japanese turnips between eastern and western Japan, and that eastern Japanese and European turnips have common genetic background. However, the analyses of cpSSR and nuSSR revealed no genetic divergence were found between eastern and western Japan, indicating that turnips in eastern Japan are different from European ones. Given that turnips in continental Asia (especially central Asia) show large genetic variations, almost all Japanese turnips are considered to be derived from central Asia. Among turnips from central Asia, those containing the chloroplast of haplogroup II and the nuclear genome of cluster 2 came to Japan. Those lacking seed mucilage and possessing leaf hairs migrated to eastern Japan and those possessing seed coat mucilage and lacking leaf hairs migrated to western Japan. There were two accessions that belonged to cluster 1, and of these one accession ‘Shougatsu-kabu’ grows spontaneously in field and was considered to be primitive. This suggests that turnips migrated multiple times from the Asia Continent to Japan. We examined a limited number of accessions in central Asia, including only one accession from Afghanistan. More extensive surveys of turnips in central Asia, in particular Afghanistan and neighboring countries, may reinforce our conclusion.
References
Allender CJ, Allainguillaume J, Lynn J, King GJ (2007) Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. (n = 9) and wild relatives. Theor Appl Genet 114:609–618
Aoba T (1958) Classification of the European-type varieties of turnip in Japan (in Japanese with English abstract). Bull Yamagata Univ Agric Sci 2:403–415
Aoba T (1961) Studies on the classification and geographical distribution of local varieties of vegetables in Japan. III. On the relationship and geographical distribution of the local varieties of turnip in the north-western part of Central Japan (in Japanese with English abstract). J Japan Soc Hortic Sci 22:318–324
Aoba T (1981a) Vegetables (in Japanese). Hosei University Press, Tokyo
Aoba T (1981b) Seeds of Brassica, especially morphology of seed coat (in Japanese). Agric Hortic 56:1239–1244
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Chung SM, Staub JE (2003) The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theor Appl Genet 107:757–767
De Candolle A (1886) Origin of cultivated plants (English translation), 2nd edn. Hafner Publishing, New York
Evanno G, Regnault S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Flannery ML, Mitchell FJG, Coyne S, Kavanagh TA, Burke JI, Salamin N, Dowding P, Hodkinson TR (2006) Plastid genome characterization in Brassica and Brassicaceae using a new set of nine SSRs. Theor Appl Genet 113:1221–1231
Gomez-Campo C, Prakash S (1999) Origin and domestication. In: Gomez-Campo C (ed) Biology of Brassica coenospecies. Elsevier, Amsterdam, pp 33–58
Hammer K, Gladis Th, Laghetti G, Pignone D (2013) The wild and the grown—remarks on Brassica. Int J AgriSci 3:453–480
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 117:309–334
Langella O (2007) POPULATIONS (version 1.2.30). (http://bioinformatics.org/~tryphon/populations/)
Louarn S, Torp AM, Holme IB, Andersen SB, Jensen BD (2007) Database derived microsatellite markers (SSRs) for cultivar differentiation in Brassica oleracea. Genet Resour Crop Evol 54:1717–1725
Mizushima U, Tsunoda S (1967) A plant exploration in Brassica and allied genera. Tohoku J Agric Res 17:249–276
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol 19:153–170
Nishi S (1980) Differentiation of Brassica crops in Asia and the breeding of ‘HAKURAN’, a newly synthesized leafy vegetable. In: Tsunoda S, Hinata K, Gomez-Campo C (eds) Brassica crops and wild allies, biology and breeding. Japan Science Society Press, Tokyo, pp 109–120
Peakall R, Smouse PE (2006) GENALEX 6, genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pino Del Carpio D, Basnet RK, De Vos RCH, Maliepaard C, Visser R, Bonnema G (2011) The patterns of population differentiation in a Brassica rapa core collection. Theor Appl Genet 122:1105–1118
Prakash S, Hinata K (1980) Taxonomy, cytogenetics and origin of crop Brassica, a review. Opera Bot 55:1–57
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trend Ecol Evol 16:142–147
Rakow G (2004) Species origin and economic importance of Brassica. In: Pua EC, Douglas CJ (eds) Biotechnology in agriculture and forestry, vol 54. Brassica. Springer, Heidelberg, pp 3–11
Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Galvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, pp 1–10
Saitou N, Nei M (1987) The neighbor-joining method, a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schebalina MA, Sazonova LV (1985) Root crops (Brassica—turnip, rutabaga, radish). In: Dorofeev VF (ed) Flora of the cultivated plants (USSR), vol 18. Agropromizdat, Leningrad
Shibutani S, Okamura T (1954) On the classification of turnips in Japan with regard to the types of epidermal layers of the seed (in Japanese with English abstract). J Japan Soc Hortic Sci 22:235–238
Sinskaia EN (1928) The oleiferous plants and root crops of the family crusiferae. Bull Appl Bot Genet Plant Breed 19:555–607
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto M (2006) SSR-based comparative genomics between Brassica rapa and Arabidopsis thaliana, the genetic origin of clubroot resistance. Genetics 173:309–319
Takuno S, Kawahara T, Ohnishi O (2007) Phylogenetic relationships among cultivated types of Brassica rapa L. em. Metzg. as revealed by AFLP analysis. Genet Resour Crop Evol 54:279–285
Wolfe AD, Liston A (1998) Contributions of PCR-based methods to plant systematics and evolutionary biology. In: Soltis DE, Soltis PS, Doyle JJ (eds) Plant molecular systematics II. Kluwer, Boston, pp 43–86
Yamane K, Lu N, Ohnishi O (2009) Multiple origins and high genetic diversity of cultivated radish inferred from polymorphism in chloroplast simple sequence repeats. Breed Sci 59:55–65
Yazawa S, Ueyama H, Namiki T (1986) Simple discrimination method of seed coat type of Brassica campestris L. (in Japanese). Agric Hortic 61:556–558
Zhao J, Wang X, Deng B, Lou P, Wu J, Sun R, Xu Z, Vromans J, Koornneef M, Bonnema G (2005) Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet 110:1301–1314
Acknowledgments
The authors gratefully thank the gene banks mentioned in Table 1 for providing the seeds of turnip accessions, and Dr. H. Iwata of The University of Tokyo for providing valuable advice concerning statistical analysis. This work was supported by a Grant-in Aid for Scientific Research (KAKENHI) Grant No. 12J01146 from the JSPS (Japanese Society for the Promotion of Science).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Takahashi, Y., Yokoi, S. & Takahata, Y. Genetic divergence of turnip (Brassica rapa L. em. Metzg. subsp. rapa) inferred from simple sequence repeats in chloroplast and nuclear genomes and morphology. Genet Resour Crop Evol 63, 869–879 (2016). https://doi.org/10.1007/s10722-015-0290-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-015-0290-y