Abstract
Twenty-one RAPD and twelve ISSR primers were used for assessment of genetic diversity and establishing phenetic relationships among 35 genotypes of six currently cultivated Indian Momordica species and five genotypes of two Luffa species. A total of 436 RAPD and 230 ISSR scorable fragments were produced of which 99.8% fragments showed polymorphism among the species and varieties of Momordica and Luffa. The level of polymorphism detected by the 33 random primers was higher among the species (99.8%) of Momordica than that estimated among the varieties (61.3%). The varieties belonging to dioecious Momordica species (75.6%) showed a higher level of polymorphism as compared to monoecious species (50.3%). A significant level (68.6%) of polymorphism however was detected by the two marker types among the Indian varieties of monoecious M. charantia species. A wider range of molecular diversity (16–95%) detected by both RAPD and ISSR markers reflected presence of high level of genetic variation among the species and Indian varieties of Momordica and Luffa. The level of inter-specific diversity was maximum (90%) between annual monoecious M. charantia and perennial dioecious M. cochinchinensis whereas the extent of intra-specific diversity was highest particularly in dioecious species (51%) as compared to monoecious species like M. charantia (38%). Wider divergence of the taxon of controversial identity, M. cymbalaria from the other Indian cultivated Momordica species and their evolutionary closeness with Luffa species was evident. The clustering pattern obtained among the 40 genotypes belonging to different Momordica and Luffa species corresponded well with their morphological, cytological and taxonomic classification, which was further supported by high boot-strap values and PCA analysis. Species and genotype-specific fragments detected by the random markers would be useful in introgression breeding for genetic improvement of Momordica cultivated in India. A smaller set of 28 informative random markers screened in this study could precisely differentiate the Momordica genotypes from each other and thus would be of use in many marker-based genotyping applications in Momordica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Momordica derives its name from Latin ‘mordeo’ (momordi = to bite) in allusion to the jagged seeds and comprises 59 species. Momordica charantia (common name: bitter gourd, karela or balsam pear or bitter melon) is a vegetable with many culinary uses especially in Asia and Africa. It is also grown as an ornamental and has been used for centuries in ancient traditional Indian, Chinese, and African pharmacopoeia. Among the cucurbits, M. charantia and M. dioica are considered prized vegetables because of their high nutritive values especially ascorbic acid and iron (Chakravarty1959; Behera et al. 2008a) and medicinal properties. In India, the Momordica species except bitter gourd (M. charantia), are being gathered from wild and eaten besides being used in genetic improvement of the species.
Taxonomic confusion exists in Momordica spp. and details of their floral biology, system of evolution and inheritance are poorly understood. The botanical names and common names are often used incorrectly or interchangeably (Joseph et al. 2007). Different taxonomic classification approaches have resulted in controversies about the number of species that exist and the phylogenetic relationships among these species. According to the latest revision of Momordica spp. of Indian occurrence (Joseph 2005), there are six well identified species of which four are dioecious and two are monoecious. The monoecious taxa are M. charantia L. (2n = 22) and M. balsamina L. (2n = 22). The dioecious taxa are M. dioica Roxb. ex Willd. (2n = 28), M. sahyadrica Joseph et Antony (2n = 28), M. cochinchinensis (Lour.) Spreng. (2n = 28) and M. subangulata Blume subsp. renigera (G. Don) W.J.J de Wilde (2n = 56). However, M. cymbalaria (Hook., Fenzl ex Naud.), which is expected to be under Momordica has not been included under the class of Momordica of Indian occurrence.
Though a number of varieties belonging to different Momordica species have been developed in India, no information is available on their genetic base. Advancements in DNA technology have resulted in an array of tools for DNA polymorphism assays. DNA based molecular markers are useful tools that provide a relatively unbiased estimation of genetic diversity and establish genetic relationship more precisely than morphological and biochemical markers (Soller and Beckmann 1983). Among these, PCR based random molecular markers such as Random Amplified Polymorphic DNA (RAPDs) and Inter Simple Sequence Repeats (ISSRs) are more commonly used in species in which there is a lack of DNA sequence information.
Understanding the extent of natural variation and phylogenetic relationship at molecular level is essential to develop new strategies for genetic improvement of Momordica. Earlier, molecular markers like RAPD have been used to assess genetic diversity among the species of bitter gourd (Dey et al. 2006), spine gourd (Rasul et al. 2007), cucumber (Horejsi and Staub 1999), pumpkin (Gwanama et al. 2000), watermelon (Lee et al. 1996) and ash gourd (Sureja et al. 2006). Besides, ISSR markers have been employed in genetic diversity analysis of different cucurbits (Dje et al. 2006; Levi et al. 2004; Ritschel et al. 2004) and in phenetic studies among related Momordica species namely, Citrullus, Cucumis and Praecitrullus fistulosus (Levi et al. 2005). Phylogenetic relationship among the different monoecious and dioecious Momordica species has also been studied using plastid and mitochondrial DNA based markers (Schaefer and Renner 2009).
No systematic effort has yet been made to understand the existing diversity pattern and phenetic relationships among the cultivated varieties included under Indian Momordica spp., using molecular markers. The present study was undertaken for assessment of genetic diversity and establishing phenetic relationships among different genotypes of six currently cultivated Indian Momordica spp. and a taxon of controversial identity, Momordica cymbalaria using RAPD and ISSR markers.
Materials and methods
Plant materials
Thirty-five genotypes of different Momordica species including 11 of M. charantia, seven each of M. subangulata subsp. renigera and M. dioica, four of M. cymbalaria, three of M. sahyadrica, two of M. cochinchinensis and one of M. balsamina were included in the present study. Five genotypes of Luffa (three of L. acutangula and two of L. cylindrica) were also included for comparison. The details of genotypes used and general morphology of the species are provided in Table 1. In M. charantia, M. balsamina, L. acutangula, L. cylindrica and M. sahyadrica, selfed (three generations) seeds were used to raise the plant for DNA extraction. Leaf samples of M. cochinchinensis, M. subangulata subsp. renigera and M. dioica were collected from the strains maintained at Central Horticultural Experiment Station, Bhubaneswar. Tubers of two genotypes of M. sahyadrica and four genotypes of M. cymbalaria were collected from the natural habitat in Kerala and Tamil Nadu, respectively. The collected tubers were grown at IARI, New Delhi for collection of leaf sample.
DNA isolation and PCR amplification
Total genomic DNA was isolated from bulked young leaves of field grown plants of each genotype using modified CTAB method (Doyle and Doyle1987) and purified. The quality and quantity of DNA isolated from these leaf samples was determined by agarose gel electrophoresis using a known quantity of λ DNA as standard. The genomic DNA was subjected to PCR amplification using 25 random decamer primers (RAPD) and 15 ISSR primers (15–23 decameric oligonucleotides) selected based on previous study on Momordica charantia accessions (Behera et al. 2008b). PCR reaction was performed in a 25 μl volume containing 1 μl of 10× Taq buffer A (10 mM Tris–HCl, pH 8.3 with 15 mM MgCl2), 0.5 μl of 10 mM dNTPs, 0.2 μl of 3 unit of Taq DNA polymerase (Bangalore Genei Pvt. Ltd, Bangalore, India), 2 μl (25 ng) of template genomic DNA and 2 μl (5 pM) each of RAPD/ISSR primers. PCR reactions were run on a Biometra T Gradient thermocycler (Biometra, USA) using the cycling temperature profiles described earlier by Jain et al. (1994) for RAPD analysis. Cycling conditions used for ISSR PCR amplification were: initial denaturation at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 55°C for 2 min, 72°C for 2 min and a final extension step at 72°C for 5 min. The amplified products were resolved on 2.5% agarose gel at 70–80 V for 3.5–4 h, using 0.5× TBE (Tris-Boric acid-EDTA) buffer, visualized under UV light after staining with ethidium bromide and photographed using gel documentation system. The size (bp) of the amplicons was determined by comparing their mobility with 100 bp DNA ladder as size standard.
Data analysis
Only clear and reproducible DNA fragments were scored as 1–0 binary data matrix for the presence and absence of a band, respectively. Cluster analysis among the 40 genotypes of Momordica and Luffa species was based on Jaccard similarity coefficient (Jaccard 1908) using the unweighted pair group method (UPGMA) and SAHN clustering algorithm. Principal components were derived for each genotype using eigen vectors and eigen values extracted from a correlation matrix among the markers that was obtained from a standardized data matrix. All the above analyses were carried out using NTSYS-pc (Version 2.02e, Applied Biostatistics) program. Boot-strap analysis (1,000 iterations) of the binary data was performed using the WINBOOT programme (Yap and Nilsson 1996) to determine the confidence limits of the UPGMA based dendrograms and boot-strap of 50% majority rule consensus tree was constructed. Average discrimination coefficients (D) of each RAPD and ISSR primers were estimated for all the 40 genotype-pairs with band differences ranging from one to five using the PowerMarker (Liu and Muse 2005) software tool.
Results
Level of DNA polymorphism in Momordica spp.
A total of 25 RAPD and 15 ISSR primers were used to amplify two genotypes of Momordica species to optimize PCR conditions namely, annealing temperature, template DNA and primer concentration. Twenty-one RAPD and twelve ISSR primers produced reproducible and clear scorable amplification products and thus were selected in the present study for genetic diversity analysis. Various gel concentrations (2–2.5%) were also tried depending on the size of amplified products for better resolution. Both RAPD and ISSR showed clear polymorphic fragment patterns among the species and varieties of Momordica. A total of 436 and 230 scorable fragments with mean of 20.8 and 19.2 were produced by 21 RAPD and 12 ISSR primers, respectively (Table 2). Out of 666 fragments, one produced by the RAPD primer OPW-2 was found monomorphic in all the genotypes of Momordica and Luffa and the remaining were polymorphic (99.8%). The polymorphic primers produced distinct banding patterns in all the Momordica and Luffa species examined (Figs. 1, 2). The size of the amplified products ranged from 250 to 4,000 bp with RAPD and 250–3,000 bp in case of ISSR. The level of polymorphism varied from species to species as well as with the markers used (Table 3). The PIC ranged from 0.15 (OPC 17) to 0.36 (OPW 13) with an average of 0.28 for RAPD and 0.22 (UBC 841 and UBC 890) to 0.34 (UBC 808) with an average of 0.27 for ISSR (Table 2). The level of polymorphism among the varieties of Momordica was lower (61.3%) than the level among the species (99.8%). The intra-specific polymorphism detected by ISSR was maximum in M. cochinchinensis (93.2%) followed by M. dioica (91.2%), M. subangulata (80%) and M. cymbalaria (72.3%), whereas RAPD revealed maximum level of polymorphism in M. cochinchinensis (92.6%) followed by M. subangulata (89.5%) and M. dioica (83.2%). The RAPD and ISSR primers showed minimum intra-specific polymorphism in L. cylindrica. The extent of polymorphism detected by these two marker types was 68.6% among the eleven varieties of M. charantia. A total of 228 scorable fragments were produced in this species by 21 RAPD (Fig. 1) and 12 ISSR (Fig. 2) primers with an average of 6.9 fragments per primer (Table 2). Number of fragments amplified per primer ranged from three (OPC 17, OPW 2 and OPW 20) to fourteen (OPW 18) in RAPD and three (UBC 809) to twelve (UBC 848) in ISSR. The percentage of polymorphic fragments detected by these two marker types varied from primer to primer (Fig. 1) that ranged from 20 to 100%. One hundred fifty-seven (68.9%) of 228 bands showed polymorphism among the varieties of M. charantia and more than 60% of the polymorphic bands were produced by 1/3rd of the primers used. The PIC of the RAPD and ISSR primers in the varieties of M. charantia ranged from 0.08 to 0.43 with an average of 0.20. In the seven varieties of M. dioica and M. subangulata, 231 (85.9%) of the 269 amplified fragments and 229 (86.1%) of 266 fragments showed polymorphism with the mean PIC value of 0.32 and 0.31, respectively.
The proportion of species-specific fragments was low across all the Momordica species, whereas the percentage of genotype specific fragments within each Momordica species was significantly higher (Table 3). The RAPD primers detected highest number of species as well as genotype-specific fragments as compared to ISSR primers (Table 2). Interestingly, a combined profile of nineteen RAPD and nine ISSR markers could identify almost all the genotypes of M. dioica, M. subangulata and M. cochinchinensis as well as seven of the 11 genotypes in M. charantia, and thus were found informative with regard to number of genotypes identified. Overall, based on discrimination coefficient (D), three of the RAPD primers (OPF8, OPW3 and OPW19) and two of the ISSR primers (UBC841 and UBC 890) were identified as most informative among randomly selected 55 genotype-pairs at one to five band levels.
Assessment of molecular genetic diversity and phenetic relationship
The genetic similarity was computed for all combinations of 40 genotypes belonging to seven Momordica spp. and two Luffa spp. based on 12 ISSR and 21 RAPD primers (Tables 4, 5). The range of pair-wise similarity was broad (0.05 to 0.84) with an average of 0.45. The similarity between Luffa and Momordica ranged from 0.05 (L. cylindrica and M. cochinchinensis) to 0.12 (L. cylindrica and M. cymbalaria). Among the Momordica species, maximum similarity was observed between M. dioica and M. sahyadrica (0.34) followed between M. subangulata subsp. renigera and M. dioica (0.28) and between M. charantia and M. balsamina (0.26), and minimum between M. charantia and M. cochinchinensis (0.1). The varieties within the L. cylindrica showed the maximum genetic similarity (0.80) followed by the varieties of M. sahyadrica (0.72), L. acutangula (0.70), M. cymbalaria (0.64), M. charantia (0.62), M. dioica (0.47) and M. subangulata (0.42) and M. cochinchinensis (0.33).
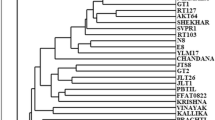
The RAPD and ISSR markers used in the study clearly discriminated all the 40 genotypes from each other and resulted in definitive grouping among different species and varieties of Momordica and Luffa (Fig. 3) that corresponded well with their known phenetic relationships as well as morphological, cytological and taxonomic classifications. The dendrogram generated based on both RAPD and ISSR markers clustered all the 40 genotypes under Momordica and Luffa spp. together into two major groups. Group I contained six Momordica species and group II consisting of one Momordica species (M. cymbalaria) and two Luffa species (L. cylindrica and L. acutangula). The cultivated M. charantia and the wild M. balsamina, which are monoecious in nature, clustered together in sub-group Ia. The dioecious species M. dioica, M. sahyadrica, M. subangulata subsp. renigera and M. cochinchinensis formed the sub-group Ib. The monoecious M. cymbalaria clustered with the two monoecious Luffa spp. in group II. The eleven genotypes of M. charantia were grouped (Table 5) into two major clusters (Fig. 3). A single accession (CHA 2) being divergent from other varieties of M. charantia clustered separately with an average similarity of 0.57. The genotypes, DBTG 501 and Nakhra showed highest similarity (0.84), while CHA 2 and WBBG 29 showed the lowest similarity (0.51). All the major clusters and sub-clusters were supported by high boot-strap values except those for the M. charantia and M. dioica species, which showed low level of inter-varietal polymorphism. Majority rule consensus tree constructed using the boot-strap of the binary data in two marker types showed a high degree of correspondence (Fig. 3). The results of PCA analysis were consistent with the pattern of clustering among the genotypes used (Fig. 4).
Discussion
The random molecular markers like RAPD and ISSR are particularly useful for studying polymorphism and genetic diversity pattern in plant species where no genomic information is available. In the present study, these two marker systems were used for assessing molecular diversity and establishing phenetic relationship among different Momordica species and varieties cultivated in India. The amplification success rate of both RAPD and ISSR markers in Momordica and Luffa species was 82.5%. Overall, 99.8% markers used in this study detected polymorphism among 35 genotypes of Momordica and five genotypes of Luffa. This is higher than the level of polymorphism (74.5%) reported earlier with the RAPD and ISSR markers in Momordica and its related genera (Singh et al. 2007; Behera et al. 2008b). Expectedly, the level of polymorphism detected by these markers among the Momordica species (99.8%) was much higher than that observed among the varieties (61.3%) within a species.
The RAPD and ISSR markers used in this study were equally informative in terms of detecting inter-varietal polymorphism. Each of these markers differentiated all the varieties belonging to Momordica and Luffa except for one RAPD primer OPW-2. The level of inter-varietal polymorphism detected by random markers was higher than that (74.5%) reported earlier among varieties of M. charantia (55.6%) using the RAPD and ISSR markers (Dey et al. 2006; Singh et al. 2007; Behera et al. 2008b). The average level of intra-specific polymorphism was higher in the dioecious species namely, M. cochinchinensis (92.8%), M. subangulata subsp. renigera (86.1%) and M. dioica (85.9%) as compared to monoecious species of Momordica (50.3%). This is in contrast to earlier studies of Rasul et al. (2007) where very low level of polymorphism was observed among the varieties of dioecious M. dioica using RAPD markers. The four dioecious Momordica species used in this study are perennial and highly heterozygous in nature, which are being maintained at Central Horticultural Experiment Station through multiplication of perenniating structure (tuberous roots). Thus a significant level of polymorphism (75.6%) detected by RAPD and ISSR markers among the varieties belonging to dioecious Momordica species is expected. Interestingly, both the markers also detected a significant level (68.8%) of polymorphism even among the Indian varieties of monoecious M. charantia. The genotypic difference among the varieties of M. charantia was possibly due to their wide geographic distribution, and considerable ecological and morphological variation with respect to fruit shape, size and colour (Dey et al. 2006).
Seventy-six species-specific fragments were detected by 33 RAPD and ISSR primers having potential applications in introgression breeding of Momordica. These markers can be utilized for broadening the genetic base of Momordica varieties cultivated in India by inter-specific hybridization followed by marker-assisted monitoring of introgression. A combination of nineteen RAPD and nine ISSR markers screened in this study could precisely differentiate the Momordica genotypes from each other and thus would be of use in purity testing, maintenance breeding and other genotyping applications in Momordica.
A wide range of molecular diversity (16–95%) detected by both RAPD and ISSR markers reflected presence of high level of genetic variation among the species and Indian varieties of Momordica and Luffa. High level of differentiation of Momordica (n = 11, 14) and Luffa (n = 13) from each other, which is expected in view of the variation observed in morphological characteristics and basic chromosome number. Within Momordica, high degree of diversity between annual monoecious M. charantia and perennial dioecious M. cochinchinensis is supported by earlier studies using the plastid, mitochondrial and nuclear DNA markers (Schaefer and Renner 2009). M. dioica and M. sahyadrica clustered together in a group with least inter-specific diversity (66%) which was supported by formation of interfertile hybrid between these two species (Bharathi et al. 2011). Further, M. subangulata subsp. renigera, presumed to be an amphidiploid, clustered together with M. dioica which seems reasonable since M. dioica is one of the putative parent of the amphidiploid M. subangulata as has been deduced recently through morphological and cytological studies (Bharathi et al. 2010). The close relationship between M. charantia and M. balsamina is expected because of their monoecious (2n = 22) and annual nature and similar morphological features. Occurrence of a high bivalent frequency with normal meiotic cycle in the hybrid progeny of M. charantia and M. balsamina (Singh 1990) further supported our results.
Momordica cymbalaria, being highly divergent from the other Momordica species, clustered distinctly with Luffa species in a separate group. Although M. cymbalaria has been treated as one of the species of Momordica in earlier reports (De Wilde and Duyfjes 2002), our recent morphological and cytological studies (Bharathi et al. 2011) clearly suggested its distinctness from all other Momordica spp. of Indian occurrence. M. cymbalaria differs from other Indian Momordica species in the basic chromosome number, and the morphology of fruit, seed and flower. Higher divergence of M. cymbalaria from Indian Momordica species and their close genetic similarity with African species of Momordica like M. humilis and M. boivinii have been reported by Schaefer and Renner (2009) using the plastid, mitochondrial and nuclear DNA markers. It is possible that M. cymbalaria originated along with other African species from a progenitor species different from the Momordica species of Indian origin. It however, most likely got introduced into India from Africa, but has remained isolated following introduction. Further studies are required to clearly establish origin, evolution and the taxonomic status of M. cymbalaria using a larger set of M. cymbalaria genotypes, African species and more number of informative markers.
The level of intra-specific diversity was highest (67%) in M. cochinchinensis followed by M. subangulata (58%), M. dioica (53%) and M. sahyadrica (28%). The two genotypes CHSG11 and CHSG27 in M. dioica and one genotype IC553771 in M. subangulata having significant variability in morphological features clustered separately from other genotypes belonging to these two Momordica species. The two genotypes SUB2 and SUB3 in case of M. subangulata and like-wise two genotypes CHSG1 and CHSG26 in M. dioica were found closely related, which is supported by high level of similarity in morphological, taxonomical and cytological characteristics (Bharathi et al. 2010). An intermediate level (38%) of diversity was observed among 11 varieties of M. charantia possibly due to their significant level of morphological, ecological and geographical variation. The wild free living M. charantia var. muricata (CHA 2) having bifid tendrils clustered separately from domesticated M. charantia var. charantia, which is supported by its significant morphological dissimilarity with cultivated M. charantia varieties particularly for fruit traits.
The present study revealed high-level of DNA polymorphism as revealed by RAPD and ISSR markers among the Momordica species. The pattern of clustering of the genotypes used corresponded with the known taxonomic relationships. M. cymbalaria, being distinctly different from other Momordica species, appeared to have a different mode of origin and evolution. Species and variety specific markers identified in the study would be useful in introgression and maintenance breeding programmes of Momordica.
References
Behera TK, Singh AK, Staub JE (2008a) Comparative analysis of genetic diversity in Indian bitter gourd (Momordica charantia L.) using RAPD and ISSR markers for developing crop improvement strategies. Sci Hort 115:209–217
Behera TK, Staub JE, Behera S, Simon PW (2008b) Bitter gourd and human health. Med Arom Plant Sci Biotech 1:224–226
Bharathi LK, Vinod, Munshi AD, Behera TK, Chandrashekaran S, Kattukunnel JJ, Das AB, Vishalnath (2010) Cytomorphological evidence for segmental allopolyploid origin of Teasle gourd (Momordica subangulata subsp. renigera). Euphytica 176:79–85
Bharathi LK, Munshi AD, Vinod, Chandrashekaran S, Behera TK, Das AB, John JK, Vishalnath (2011) Cyto-taxonomical analysis of Momordica L. (Cucurbitaceae) species of Indian occurrence. J Genet 90:21–30
Chakravarty HL (1959) Monograph of Indian Cucurbitaceae. Rec Bot Surv India 17:81
De Wilde WJJO, Duyfjes BEE (2002) Synopsis of Momordica (Cucurbitaceae) in South East Asia and Malaysia. Bot Zhurn 57:132–148
Dey SS, Singh AK, Chandel D, Behera TK (2006) Genetic diversity of bitter gourd (Momordica charantia L.) genotypes revealed by RAPD markers and agronomic traits. Sci Hort 109:21–28
Dje Y, Tahi GC, Zoro BIA, Malice M, Baudoin JP, Bertin P (2006) Optimization of ISSR marker for African edible-seeded Cucurbitaceae species genetic diversity analysis. Afr J Biotech 5:83–87
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure from small tissues of fresh leaf tissue. Phytochem Bull 19:11–15
Gwanama C, Labuschagne MT, Botha AM (2000) Analysis of genetic variation in Cucurbita moschata by random amplified polymorphic DNA (RAPD) markers. Euphytica 113:19–24
Horejsi T, Staub JE (1999) Genetic variation in cucumber (Cucumis sativus L.) as assessed by random amplified polymorphic DNA. Genet Resour Crop Evol 46:337–350
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bul Soc Vaudoise Sci Nat 44:223–270
Jain A, Bhatia S, Banga SS, Prakash S, Laksmikumaran M (1994) Potential use of random amplified polymorphic DNA (RAPD) technique to study the genetic diversity in Indian mustard Brassica juncea and its relationship to heterosis. Theor Appl Genet 88:116–122
Joseph JK (2005) Studies on ecogeography and genetic diversity of the genus Momordica L. in India. Ph.D. Thesis, Mahatma Gandhi University, Kottayam
Joseph JK, Antony VT, Roy YC (2007) On the occurrence, distribution and taxonomy of Momordica subangulata Blume ssp. renigera (G. Don) de Wilde in India. Genet Resour Crop Evol 54:1327–1332
Lee SJ, Shin JS, Park KW, Hong YP (1996) Detection of genetic diversity using RAPD-PCR and sugar analysis in watermelon [Citrullus lanatus (Thumb) Mansf.] germplasm. Theor Appl Genet 92:719–725
Levi A, Thomas CE, Newman M, Reddy OUK, Zhang X, Xi Y (2004) ISSR and AFLP markers differ among American watermelon cultivars with limited genetic diversity. J Am Soc Hort Sci 129:553–558
Levi A, Thomas CE, Simmons AM, Thies JA (2005) Analysis based on RAPD and ISSR markers reveals closer similarities among Citrullus and Cucumis species than with Praecitrullus fistulosus (Stocks) Pangalo. Genet Resour Crop Evol 52:465–472
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Rasul MG, Hiramatsu M, Okubo H (2007) Genetic relatedness (diversity) and cultivar identification by randomly amplified polymorphic DNA (RAPD) markers in teasle gourd (Momordica dioica Roxb.). Sci Hort 111:271–279
Ritschel PS, Lins TCDL, Tristan RL, Buso GSC, Buso JA, Ferreira ME (2004) Development of microsatellite markers from an enriched genomic library for genetic analysis of melon (Cucumis melo L.). BMC Plant Biol 4:9–23
Schaefer H, Renner SS (2009) A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Mol Phylogenet Evol 54:553–560
Singh AK (1990) Cytogenetics and evolution in the Cucurbitaceae. In: Bates DM, Robinson RW, Jeffrey C (eds) Biology and utilization of Cucurbitaceae. Comstock Publishing, Associates Cornell University Press, Ithaca, pp 10–28
Singh AK, Behera TK, Chandel D, Sharma P, Singh NK (2007) Assessing genetic relationships among bitter gourd (Momordica charantia L.) accessions using inter simple sequence repeat (ISSR) markers. J Hort Sci Biotechnol 82:217–222
Soller M, Beckmann JS (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Sureja AK, Sirohi PS, Behera TK, Mohapatra T (2006) Molecular diversity and its relationship with hybrid performance and heterosis in ash gourd [Benincasa hispida (Thumb.) Cogn.]. J Hort Sci Biotchnol 81:33–38
Yap IV, Nilsson RJ (1996) Winboot: a program for performing boot strap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI discussion paper series no. 14, International Rice Research Institute, Manila
Acknowledgments
The authors thank Director, NBPGR, New Delhi for providing the seeds of M. balsamina and M. sahyadrica, and Dr. S. Anbu, Dean, Horticulture College and Research Institute, Periyakulam, Tamil Nadu for providing the tubers/seeds of M. cymbalaria genotypes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bharathi, L.K., Parida, S.K., Munshi, A.D. et al. Molecular diversity and phenetic relationship of Momordica spp. of Indian occurrence. Genet Resour Crop Evol 59, 937–948 (2012). https://doi.org/10.1007/s10722-011-9735-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-011-9735-0