Abstract
Ribosomal DNA (rDNA) spacer length polymorphism was studied in foxtail millet (Setaria italica ssp. italica) landraces from Pakistan and Afghanistan and in its wild ancestor (S. italica ssp. viridis) from Pakistan by PCR-based methods. Sequence polymorphism was also investigated for accessions selected based on the observed length polymorphism. The PCR-based length polymorphism and sequence polymorphism of rDNA intergenic spacer (IGS) clearly demonstrated genetic differentiation between cultivated and wild forms in the region. Genetic differentiation was observed between different areas to some extent in the cultivated form, and between different regions in the wild form of northern Pakistan. Based on the results, we discuss the genetic differentiation of foxtail millet and wild ancestor in this region and possible utility of rDNA markers to trace the dispersal of this crop in the region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foxtail millet, Setaria italica (L.) P. Beauv. ssp. italica is one of the oldest cereals in Eurasia. It has been used in various ways peculiar to different areas of Eurasia (Sakamoto 1987a); it is thought to have played an important role in the Old World’s early agriculture. The geographical origin of foxtail millet remains a controversial issue. Cytological and genetic studies has indicated that the wild ancestor of this crop is S. italica ssp. viridis (L.) Thell. (Kihara and Kishimoto 1942; Li et al. 1945; Le Thierry d’Ennequin et al. 2000; Wang et al. 1995). The geographical origin of domesticated ssp. italica cannot be determined by the distribution of ssp. viridis, which is commonly found in different areas in Europe and Asia and colonizing in the New World. Various hypotheses have advanced monophyletic and polyphyletic origins such as a single in East Asia including China and Japan (Vavilov 1926) and polyphyletic origins in China and Europe (Harlan 1975). Recently, Li et al. (1995) concluded that foxtail millet was domesticated independently in China and Europe and landraces in Afghanistan and Lebanon had been domesticated separately in recent times because they had primitive characteristics such as numerous tillers with small panicles. In contrast to the hypotheses of multiple origins, Sakamoto (1987a) suggested that foxtail millet originated somewhere in Central Asia, Afghanistan, Pakistan and northwestern India, because strains with less restricted compatibility (Kawase and Sakamoto 1987) and with primitive morphological traits were found there. This hypothesis was unique in treating China as a secondary center of origin of foxtail millet and treating Afghanistan and Pakistan as key regions for foxtail millet origin. Scheibe (1943) described that S. italica ssp. viridis was cultivated mainly as a fodder crop but also for grains in Hindukush. This fact indicates that foxtail millet may have been domesticated in this area.
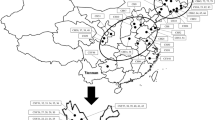
Kyoto University Expedition Teams were sent to Afghanistan in 1977 and 1978 and to Pakistan in 1987, for collecting genetic resources in these regions, respectively. Foxtail millet landraces and its wild ancestor, S. italica ssp. viridis, were collected directly from the fields (Sakamoto 1987b; Kawase and Sakamoto 1989). Landraces from northern Pakistan were classified into three groups, Chitral, Baltistan and Dir based on Ochiai et al. 1994 (Table 1 and Fig. 1). The three groups are different from each other in quantitative characteristics and qualitative characteristics and also in geographical distribution in northern Pakistan (Ochiai et al. 1994). Landraces of Chitral Group and Afghan landraces commonly have primitive characteristics such as many tillers with small panicles, short plant height, and long bristle on the panicles that are similar to ssp. viridis. Kawase et al. (1997) also characterized these landraces based on intraspecific hybrid pollen sterility but it was difficult to elucidate the phylogenetic relationships of these landrace groups with landraces from other regions by this method.
The exploration route and collection sites (black circles) in the northern mountainous area of Pakistan. Broken circles indicate geographical distribution of foxtail millet landraces groups, the Baltistan Group, the Chitral Group and the Dir Group (Ochiai et al. 1994). Circle graphs indicate frequencies of S type (20-bp deletion) and L type of rDNA type I
Ribosomal DNA (rDNA) is arranged in tandem arrays and contains genes for 25 S, 18 S and 5.8 S rRNAs. The intergenic spacer (IGS) of rDNA is highly variable in length, even within a species or among cultivars, because it contains subrepeats which vary in repeat number (Rogers and Bendich 1987). We also analyzed rDNA RFLP in foxtail millet, cloned a repeat unit (Fukunaga et al. 1997), determined the sequence of the intergenic spacer, and identified the polymorphic region (Fukunaga et al. 2005). In the recent paper, we classified landraces at the IGS sequence level (Fukunaga et al. 2006). We found two major types designated as types I and–II. Type I is about 300 bp shorter a ribosomal DNA repeat unit than types II and is distributed broadly in temperate zone; while type II is predominantly in the subtropical and tropical zones. Type I was further classified into seven subtypes, Ia to Ig. Type II was classified further into four subtypes, IIa-IId (Fukunaga et al. 2006). Among subtypes of type I, types Ia and Ib are distributed broadly from East Asia to Europe but type Ic and Id are restrictedly distributed in East Asia and Afhganistan-northwestern Pakistan (Chitral group according to Ochiai et al. 1994), respectively. We also found types Ie to Ig restrictedly in the wild ancestor. It is interesting that type Id has 20-bp deletion in subrepeat 3 (see Fig. 3) and has the limited distribution in the morphologically primitive landraces in Afghanistan and northwestern Pakistan (Chitral Group). It implies that these landraces originated independently as suggested by Li et al. (1995) based on morphological characters.
To test the hypothesis of independent origins of primitive morphological characters of foxtail millet in this region, we analyzed several accessions of foxtail millet and also ssp. viridis collected in this region. In this paper, we focused on length polymorphisms of the IGS such as types I and II and 20 bp deletion in type I, and we also analyzed the sequence polymorphism of selected samples.
Materials and methods
Plant materials
As shown in Table 1 and Figs. 1 and 2, we used 77 samples of ssp. italica and 40 of ssp. viridis collected from northern Pakistan, and 17 ssp. italica accessions from Afghanistan. All of the landraces have been maintained by selfing and DNA of ssp. italica was extracted by bulked seedlings. For ssp. viridis, DNA was extracted from 20 grains from each individual directly collected in natural habitats.
Detection of length polymorphism
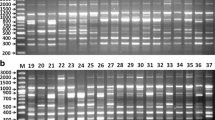
Polymerase chain reaction (PCR) was done with a primer combination of IGS 8 and IGS 4 (Fig. 3) using PCR conditions described by Fukunaga et al. 2005 and Fukunaga et al. 2006. PCR products were resolved on an 1.5% agarose gel and determined the length polymorphism as types I and II as Fukunaga et al. 2005, 2006 and. Further, PCR products were digested with Rsa I according to manufacturer’s manual and run on 2.5% agarose gel in order to detect 20 bp deletion in subrepeat 3 (type Id in Fukunaga et al. 2006). We designated the accessions having the 20 bp deletion (Figs. 3 and 4) as S (short)-type and those lacking the 20-bp deletion as L (long)-type of the type I previously reported.
Structure of ribosomal DNA IGS. Black boxes indicate rRNA genes. Grey boxes and a white box indicate subrepeats and C repeat, respectively (See Fukunaga et al. 2006). 20-bp deletion which is found specifically in type-Id was indicated. Horizontal arrows indicate the primers and vertical arrows indicate restriction enzyme Rsa I sites
Sequencing
We picked up 22 landraces from Pakistan (6 S-type accessions and 16 L-type ones), 9 landraces from Afghanistan (5 L-type accessions and 4 S-type accessions) and 17 ssp. viridis accessions from Pakistan (16 L-type accessions and one heterogeneous accession of L and S types) judging from the length polymorphism data and sequenced their rDNA. We also used six ssp. viridis samples of type II-rDNA for sequencing analysis. Direct sequencing was adopted based on the previous paper (Fukunaga et al. 2006) except one ssp. viridis accessions that is a heterogeneous one of L and S types. For this heterogeneous accession, we ligated the PCR products with pGEM vector and transformed into JM109 E. coli cell. We picked up three clones, and determined the sequences of the inserts.
All sequences obtained were classified into subtypes according to Fukunaga et al. 2006, and when a sequence does not fit to any of known types we designated it as a new type.
Results and discussion
Length polymorphism
Types I and II were readily determined on 1.5% agarose gel as previously described (Fukunaga et al. 2005, 2006). In type I accessions, a 20-bp deletion (S-type) was successfully distinguished from L-type by digestion with RsaI followed by electrophoresis on 2.5% agarose gel (Fig. 4). As shown in Figs. 1 and 2, all of the 17 Afghan landraces and the 77 accessions of northern Pakistani landraces were of type I whereas 26 of the 39 accessions of ssp. viridis were of type I and 13 were of type II. The S-type was found in a high frequency (94%) in the Chitral Group, followed by the Dir Group (54%) and Afghan landraces (24%) and the Baltistan Group (24%).
Polymorphism at sequence level
We genotyped an intergenic spacer region of rDNA in 22 Pakistani landraces with type I accession (6 S-type and 16 L-type), 9 Afghan landraces (5 S-type and 4 L-type) and 17 ssp. viridis accessions (15 type I and 2 type II). The sequences of S-type lanrdraces from Pakistan to Afghanistan are identical to that of subtype Id, while L type is classified into subtype Ib. In ssp. viridis accessions, most of the type I accessions had subtype Ig as in the previous study, one had type Ib and one had a new type, type Ih (Table 2, DDBJ AB543494). Type Ih was different at one nucleotide from type Ia, in which G at positon 131 of type Ia had been changed to A in this type. As all of these sequence peaks were clear, a single rDNA type was predominant in a tandem array in each accession. One accession (accession No. 87-9-28-5-2. see Fig. 2) from Pengal, Gilgit Agency, had both of S-type and L-type. We cloned a PCR product into a plasmid and picked up three clones: two clones had type Id sequence and one had type Ib one by sequencing. This accession is determined to be a heterozygote of these types or has admixture of these types in a tandem repeat.
Randomly chosen six type II accessions were sequenced, one of which was identical to type IIe (renamed from type III; Fukunaga et al. 2006) and different from the rest. Remaining five accessions were identical to each other at nucleotide sequence level but it is not same as any known subtypes of type II. We designated their genotype as subtype IIf (DDBJ AB543493).
Genetic differentiation between landraces and wild ancestor and within and between areas
As shown in Figs. 1 and 2, the difference in rDNA types between foxtail millet, ssp. italica and its wild ancestor, ssp. viridis in this region was very clear. All of ssp. italica accessions from Pakistan and Afghanistan had type I rDNA, whereas the wild ancestor had both of types I (67%) and II (33%).
The 54% of type I, ssp. italica was determined as S-type but only one accession of ssp. viridis was S-type. At the sequence level, L-type of ssp. italica was also different from ssp. viridis (Table 2). All of ssp. italica including Afghan landraces had subtype Ib whereas most of ssp. viridis had subtype Ig but one had Ih and one Ib. Ig was different from type Ib not only in two nucleotide sequences in subrepeats but also C repeats (Fukunaga et al. 2006), where Ib had a CCCCCCC (C7) sequence but Ig had C6 sequence (Fukunaga et al. 2006). In northern Pakistan, differentiation between foxtail millet and its wild ancestor was distinct as for the distribution of type II, deletion of 20 bp in type I and also nucleotide sequence in L-type of type I.
Our previous results implied that only morphologically primitive landraces have subtype Id rDNA and we expected that type Id would be specifically found in such landraces as Afghan landraces and Chitral Group (Fukunaga et al. 2006). Subtype Id-like type, however, was dominant in Chitral Group but also found in Dir and Baltistan Groups. Afghan landraces, all of which showed primitive morphological characters, showed both of L-type (74%) and S-type (26%).
In this analysis we also investigated local population of ssp. viridis. So far, genetic structure of ssp. viridis population was analyzed in a large scale in Eurasia by isozymes (Jusuf and Pernes 1985, Wang et al. 1995) and AFLP (Le Thierry d’Ennequin et al. 2000) but no clear differentiation pattern was observed. In this analysis, we analyzed the population structure of local populations of ssp. viridis and found clear genetic differentiation of populations from Gilgit Agency-Baltistan and those from the NWFP as shown in Fig. 2. All populations have type I rDNA from Gilgit Agency—Baltistan, whereas 50% of individuals from NWFP had type II rDNA. This distribution pattern of different ssp. viridis rDNA types was reflected by history of colonizing or adaptation of this weed to different places in Pakistan. Further analysis of ssp. viridis from various regions will reveal the expansion pattern of ssp. viridis in this area.
Origin of foxtail millet landraces from Pakistan
As there was one viridis accession with type Ib and another accession with type Ib/Id that were commonly found in foxtail millet, it is possible to suppose that they are ancestral forms of foxtail millet from which foxtail millet was domesticated in this region (Ochiai et al. 1994; Ochiai 1996). It is also possible to suppose the hetergeneous type Ib/Id in ssp. viridis arose by gene-flow from foxtail millet into ssp. viridis as these accessions were collected in the places near foxtail millet cultivation (Fig. 3). Kawase and Sakamoto (1989) observed intermediate types between foxtail millet and sympatric ssp. viridis. Therefore, it is not strongly suggested that Pakistan is one of the original places of foxtail millet, as a large difference between ssp. italica and ssp. viridis in the region was found in this analysis. Id-type of ssp. viridis was rarely found in Pakistan although Scheibe (1943) described that ssp. viridis had been cultivated for the grains in Hindukush. As it is still not easy to explain that change from other subtypes of type I into type Id occurred in a simple mutation step, type Id foxtail millet may have been domesticated independently. Further analysis of both of foxtail millet and ssp. viridis was needed and the 20-bp deletion determined in this paper might be a useful clue to elucidate crop evolution of this millet in the analysis.
This study indicates that history of foxtail millet in this region is complex as expected by intraspecific hybrid pollen sterility (Kawase et al. 1997), which is not always consistent with morphological variation (Ochiai et al. 1994). If type- Id foxtail millet is found in more east or west in further analysis, it will be a good marker to trace foxtail millet dispersal. It will be important to analyzing foxtail millet landraces in the neighboring areas such as Iran (Hammer and Khoshbakht 2007) and Central Asia in detail.
References
Fukunaga K, Domon E, Kawase M (1997) Ribosomal DNA variation in foxtail millet, Setaria italica (L.) P. Beauv. and a survey of variation from Europe and Asia. Theor Appl Genet 97:751–756
Fukunaga K, Ichitani K, Taura S, Sato M, Kawase M (2005) Ribosomal DNA intergenic spacer sequence in foxtail millet, Setaria italica (L.) P. Beauv. and its characterization and application to typing of foxtail millet landraces. Hereditas 142:38–44
Fukunaga K, Ichiatani K, Kawase M (2006) Phylogenetic analysis of rDNA intergenic spacer subrepeats and its implication for domestication history of foxtail millet, Setaria italica. Theor Appl Genet 113:261–269
Hammer K, Khoshbakht K (2007) Foxtail millet (Setaria italica (L.) P. Beauv.) in Mazandaran/Northern Iran. Genet Resour Crop Evol 54:907–911
Harlan JR (1975) Crops and man. Soc Amer Agron Crop Sci Soc America, Madison, WI
Jusuf M, Pernes J (1985) Genetic variability of foxtail millet (Setaria italica (L.) P. Beauv.): electrophoretic study of five isoenzyme systems. Theor Appl Genet 71:385–391
Kawase M, Sakamoto S (1987) Geographical distribution of landrace groups classiffed intraspecific pollen sterility in foxtail millet Setaria italica (L.) P. Beauv. Jpn J Breed 37:1–9
Kawase M, Sakamoto S (1989) Field observation and collection of foxtail millet and common millet in northern mountains areas of Pakistan. In: Sakamoto S (ed) A preliminary report of the studies on millet cultication and its agro-pastoral culture complex in the Indian Subcontinent, II (1987). Research team for the studies on millet cultivation and its agro-pastroral culture complex in the Indian Subcontinent. Kyoto University, Kyoto, pp 51–68
Kawase M, Ochiai Y, Fukunaga K (1997) Characterization of foxtail millet Setaria italica (L.) P. Beauv. in Pakistan based on intraspecific hybrid pollen sterility. Breed Sci 47:45–49
Kihara H, Kishimoto E (1942) Bastarde zwischen Setaria italica und S. viridis (in Japanese with German summary). Bot Mag 20:63–67
Le Thierry d’Ennequin M, Panaud O, Toupance B, Sarr A (2000) Assessment of geneticrelationships between Setaria italica and its wild relatives S. viridis using AFLP marker. Theor Appl Genet 100:1061–1066
Li HW, Li CH, Pao WK (1945) Cytological and genetic studies of interspecific cross of the cultivated foxtail millet, Setaria italica P. Beauv. and the green foxtail millet, S. viridis L. J Amer Soc Agron 37:32–54
Li Y, Wu SZ, Cao YS (1995) Cluster analysis of an international collection of foxtail millet (Setaria italica (L.) P. Beauv). Genet Resour Crop Evol 45:279–285
Ochiai Y (1996) Variation in tillering and geographical distribution of foxtail millet (Setaria italica P. Beauv.). Breed Sci 46:143–146
Ochiai Y, Kawase M, Sakamoto S (1994) Variation and distribution of foxtail millet (Setaria italica P. Beauv.) in the mountainous areas of northern Pakistan. Breed Sci 44:413–418
Rogers BO, Bendich AJ (1987) Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Sakamoto S (1987a) Origin and dispersal of common millet and foxtail millet. Japan Agric Res Quart 21:84–89
Sakamoto S (1987b) Origin and phylogenetic differentiation of cereals in the Southwest Eurasia. In: Sakamoto S (ed) Domesticated plants and animals of the southwest Eurasian agro-pastroral culture complex. Kyoto University, Kyoto, pp 1–45
Scheibe A (1943) Die Hirsen im Hindukusch Ein Beitrag zur Kenntnis von Kulturpflanzen in geographischer Rückzugsposition. Z. Pflanzenzüchtung 25:392–436
Vavilov NI (1926) Studies on the origin of cultivated plants. Inst Appl Bot Plant Breed, Leningrad
Wang RL, Wendell J, Dekker J (1995) Weedy adaptation in Setaria spp.: I. Isozyme analysis of the genetic diversity and population genetic structure in S. viridis. Am J Bot 82:308–317
Acknowledgments
This work is supported by a Grand-in-Aid for Young Scientists (B) of Japanese Society of Promotion of Science (JSPS) to KF.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fukunaga, K., Ichitani, K. & Kawase, M. rDNA polymorphism of foxtail millet (Setaria italica ssp. italica) landraces in northern Pakistan and Afghanistan and in its wild ancestor (S. italica ssp. viridis). Genet Resour Crop Evol 58, 825–830 (2011). https://doi.org/10.1007/s10722-010-9619-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9619-8