Abstract
Random amplified polymorphic DNA markers were used to study sub-structure and genetic differentiation amongst 31 populations (seven cultivated and 24 wild populations) belonging to 14 Asiatic Vigna species. Ten pre-selected RAPD primers generated 152 polymorphic amplification products. Estimates of polymorphism indices were higher for the wild taxa in comparison to the cultivated forms. FST values between populations ranged from 0.111 to 0.801 and Nei’s genetic diversity values between and within species varied from 0.26 to 0.70 and 0.04 to 0.56 respectively. The high FST and FCT values indicated strong subdivision of populations and high differentiation among species. Analysis of molecular variance was performed by grouping the populations conforming to specific species. AMOVA was also performed separately to better resolve the differentiation of species within mungo–radiata complex. Molecular phylogenetic relationships amongst the species of radiata–mungo complex; namely, black gram (V. mungo (L.) Hepper), green gram (V. radiata (L.) Wilczek), V. radiata var. sublobata, V. radiata var. setulosa, V. mungo var. silvestris and V. hainiana, were studied through cluster analyses. Two distinct groups were recognized within the complex, with population samples of V. hainiana forming one cluster. Further, V. hainiana appeared to be equidistant to both V. radiata and V. mungo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Vigna Savi has approximately 150 species that are grouped into seven subgenera. Asiatic grams are assigned to subgenus Ceratotropis (Piper.) Verdc. with 16–17 recognized species (Verdcourt 1970; Maréchal et al. 1978; Tateishi 1996). Of these eight species, namely, V. radiata, V. mungo, V. angularis, V. aconitifolia, V. umbellata, V. trilobata, V. trinervia and V. reflexo-pilosa var. glabra are used as human and animal food. Thus, Asiatic Vigna constitutes an economically important group of cultivated and wild species, of which rich diversity occurs in India (Arora 1985; Babu et al. 1985).
The taxonomy of Vigna species is complex and needs detailed analysis, as the estimation of genetic diversity and delineation of several important taxa remain uncertain till date. Few studies have addressed genetic variation in most important Asiatic pulse yielding Vigna species, namely V. radiata and V. mungo, in relation to the wild forms which have been designated as distinct species or alternatively as botanical varieties of the cultivated species by different workers. For a long period, V. radiata var. sublobata had been considered the common ancestor of green gram and black gram. However, Lukoki et al. (1980) and Chandel et al. (1984) recognized divergence of both species before their domestication, and reported that V. radiata and V. mungo were domesticated from two very different taxa, namely, V. radiata var. sublobata and V. mungo var. silvestris. Babu et al. (1985) did not recognize such varietal status for wild forms; instead they described all these wild forms as one species, namely, V. sublobata on the basis of morphological similarities observed in the natural populations occurring in Indian subcontinent and also, described a new species, V. hainiana, to delineate more primitive and wild forms with greater diversity for morphological traits in comparison to the members of the V. sublobata species.
Presence of gaps in the available information has necessitated the use of molecular markers for the analysis of the population substructure, genetic diversity and phylogenetic relationships amongst selected species of the genus Vigna. The present study was formulated with the following objectives: (a) to study population substructure and genetic diversity in selected Asiatic Vigna species (b) to investigate status and distinctiveness of V. radiata var. sublobata, V. radiata var. setulosa, V. mungo var. silvestris and V. hainiana (c) to investigate relationships of the wild forms with V. radiata and V. mungo. These objectives are addressed in the selected Vigna species through RAPD analysis.
Materials and methods
Plant material and DNA extraction
A total of 31 selected populations of 14 Vigna species were analyzed in the study (Table 1). Total genomic DNA was extracted from 15-day-old individual etiolated seedlings using CTAB extraction procedure (Saghai-Maroof et al. 1984). DNA was treated with Bovine Pancreatic RNase A and extracted once with phenol:chloroform:isoamylalcohol (25:24:1 v/v) and twice with chloroform:isoamylalcohol (24:1 v/v). After precipitation with chilled absolute ethanol, two washes of 70% ethanol were given. The concentration of DNA samples was estimated using DNA flourometer (Hoefer scientific, San Francisco, USA) employing Hoechst 33258 as DNA intercalating dye (Brunk et al. 1979). A diluted stock of 20 ng/μl DNA was used for setting up PCR amplification reactions.
Primer selection
Forty-two deca-nucleotide RAPD primers from kits OPA, OPB, OPC, OPD and OPH of Operon Technologies (USA) were screened for repeatability and scorability using two samples each of V. radiata and V. mungo. Out of these 42 primers, 10 primers yielding repeatable, good and polymorphic amplification products were selected. The nucleotide sequences of these primers and characteristics of amplification products recorded are listed in Table 2. The PCR amplification reaction mixture contained 2.5 mM MgCl2, 50 mM KCl, 10 mM tris HCl (pH 9), 0.1% Triton X-100, 0.2 mM dNTPs, 1 unit Taq DNA polymerase (Bangalore Genei, Bangalore, India), 0.4 μM primer and 20 ng genomic DNA in a reaction volume of 25 μl. Amplification was performed using GeneAmp 9600 Thermal Cycler (Perkin Elmer Cetus, Norwak, CT, USA) with the following temperature profile: a pre-denaturation step of 3 min at 94°C, amplification reactions were cycled 40 times at 94°C for 1 min, 32°C for 1 min and 72°C for 1 min, a final extension step was allowed for 10 min at 72°C. The amplification products were separated in 1.8% agarose gel by electrophoresis in 1× TAE buffer, pH 8.0. The 1 kb DNA marker ladder (MBI Fermentas, Germany) was used as molecular weight standard. After electrophoresis, the gels were stained with ethidium bromide, viewed under UV light and photographed with Polaroid 667 film.
Data scoring and analysis
All gel patterns were scored for presence and absence of RAPD fragments, and data was entered into a binary data matrix as discrete variables (1 for presence and 0 for absence). Genetic similarity (GS) between each pair of species was estimated using the method of Jaccard (1908): GS = nxy/nt − nz, where nxy is the number of bands common to accession A and B; nt the total number of bands present in all samples and nz the number of bands not present in both A and B but found in other samples. The similarity indices between the samples of mungo–radiata complex were also computed using the same formula. The amount of genetic variation within each population and species was quantified by determining number of polymorphic loci, percent polymorphic loci (p), observed number of alleles per locus (na) and effective number of alleles per locus (ne). Genetic diversity statistics were calculated after Nei (1987). Expected heterozygosity was calculated for all populations using the procedure of Lynch and Milligan (1994). At each polymorphic locus, the total allelic diversity was represented by Ht, which was partitioned into mean allelic diversity within populations (Hs) and the allelic diversity among populations (Dst). The proportion of total allelic diversity or genetic differentiation among populations (Gst) was calculated as the ratio Dst/Ht. FST values representing differentiation between populations were also calculated using Arlequin Version 3.01 software (Excoffier et al. 1992).
The Shannon–Weaver information index (Shannon and Weaver 1949) was calculated to measure the extent of diversity in each sample. The Wright’s F-statistics, FCT, FSC and FST were also computed (Wright 1965). These are hierarchically related descriptors of the distribution of genetic variation within and among populations. The statistical analyses were performed using POPGEN version 1.31 (http://www.ualberta.ca/~fyeh/fyeh).
An analysis of molecular variance (AMOVA) was also performed to partition the total genetic variation into that occurring within population, among population within groups and among groups using Arlequin Version 3.01 software (Excoffier et al. 1992). It was performed by grouping the populations conforming to species to two level of analysis, firstly, by including all 31 populations belonging to 14 Vigna species, secondly, by including only those species that belong to mungo–radiata complex. These two types of AMOVA were performed to resolve the differentiation of species within mungo–radiata complex.
The Jaccard’s similarity matrix was utilized to construct a dendrogram using UPGMA procedure. Principal components analysis (PCA) was also performed to study the differences among the populations within a cluster. These analyses were conducted using NTSYS-pc software (Rohlf 1992).
Results
The 10 RAPD primers used (Table 2) generated 152 amplification products in the 152 individuals of 31 populations belonging to 14 Vigna species. The number of amplified fragments varied from 13 to 19 per primer. The pair-wise similarity values between species of mungo–radiata complex ranged from 0.081 to 0.910 with the average value of 0.339. The FST values (Table 3) between populations ranged from 0.084 (between accessions of V. hainiana i.e. BB-6-2K and TCR26) to 0.801 (between accessions of V. mungo and V. radiata var. sublobata TCR11).
The wild Vigna forms showed higher intra-specific variability in comparison to the cultivars (Tables 4, 5). Diversity parameters in the populations were indicators of the diversity prevalent in each of the species analyzed (Table 4). The number of polymorphic loci ranged from 15 (BBL69, V. radiata var. setulosa) to 99 (BB18-2K, V. hainiana) and the percent polymorphic loci varied from 9.87% (BBL69, V. radiata var. setulosa) to 65.13% (BB18-2K, V. hainiana). The estimates for observed and effective number of alleles per locus ranged from 1.09 (TCR66 of V. radiata var. sublobata) to 1.65 (BB18-2K of V. hainiana) and 1.06 (TCR66 of V. radiata var. sublobata) to 1.41 (BB18-2K of V. hainiana) respectively. The Nei’s genetic diversity (h) estimates ranged from 0.04 (TCR66 of V. radiata var. sublobata) to 0.24 (BB18-2K, V. hainiana). The values for expected heterozygosity ranged from 0.07 (BBL69 of V. radiata var. setulosa) to 0.31 (BBL18-2K of V. hainiana). The Shannon-Weaver information index values varied from 0.09 (TCR66 of V. radiata var. sublobata) to 0.56 (BB18-2K, V. hainiana).
Inter-species variability was analyzed by computing the parameters, number of polymorphic loci (P), percent polymorphic loci, observed number of alleles per locus (na), effective number of alleles per locus (ne), Shannon-Weaver information Index (I), total diversity (Ht), diversity within accessions (Hs) and diversity between accessions, Dst (Table 5). Maximum number of polymorphic loci (136) and percent polymorphic loci (89.47) were observed for V. hainiana while, minimum number of polymorphic loci (30) and percent polymorphic loci (19.74) were observed for V. radiata var. setulosa. Observed number of alleles per locus (na) and effective number of alleles per locus (ne) ranged from 1.19 (V. radiata var. setulosa) to 1.71 (V. hainiana) and 1.13 (V. radiata var. setulosa) to 1.45 (V. hainiana) respectively. Shannon–Weaver information index (I) ranged from 0.1 (V. radiata var. setulosa and V. reflexo-pilosa var. glabra) to 0.4 (V. hainiana). Total diversity, which is partitioned into diversity within accessions and diversity between accessions varied from 0.07 (V. radiata var. setulosa and V. reflexo-pilosa var. glabra) to 0.26 (V. hainiana). The estimated within accession diversity ranged from 0.01 (V. trilobata) to 0.19 (V. angularis), whereas, between accession diversity ranged from 0.04 (V. mungo) to 0.12 (V. trilobata). The gene-flow among populations within species varied from 0 to 1.63 (V. mungo).
Wright Fixation indices (FSC, FST and FCT) were calculated to evaluate population subdivision and population substructure (Table 6). The computations for these indices were performed for all the populations and separately for mungo–radiata complex. For mungo–radiata relatives, FCT, FSC and FST estimates were 0.87, 0.43 and 0.56 respectively. Similarly for all Vigna species combined the fixation indices FCT, FSC and FST were 0.56, 0.42 and 0.25 respectively.
AMOVA (Table 7) indicated that 43.71% of the total variation was accounted by within population variation in comparison to 31.60% by among population variation within the species, and among species variation accounted for 24.71% of total variation. Similarly, for mungo–radiata complex, of the total genetic diversity, 40.42% was attributed to individuals within populations, 30.95% to populations within species and 28.63% to amongst species.
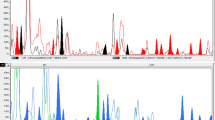
The UPGMA tree constructed on the basis of Jaccard’s similarity matrix by incorporating V. unguiculata as an outgroup (Fig. 1) classified the populations representing six species of mungo–radiata complex into two groups (I and II). Out of six species, group I included all populations belonging to V. radiata, V. mungo, V. radiata var. sublobata, V. radiata var. setulosa and V. mungo var. silvestris, whereas, group II incorporated all the populations belonging to V. hainiana. The PCA analysis (Fig. 2) was performed for mungo–radiata complex. The six species segregated into six groups, with all populations of a species forming a distinct single group. The first three most informative principal components accounted for 14.82%, 10.82% and 9.20% of the total variation and 22 components are required to explain the total variation.
Discussion
The knowledge of population substructure, genetic diversity and phylogenetic relationships between wild forms and cultigens of Vigna is essential for devising strategies for efficient genetic diversity maintenance and utilization. The environmental challenges faced by wild and cultivated species, today, underline the innate importance of measurement and assessment of genetic variability. The loss of genetic diversity is the major threat for the maintenance and adaptive potential of species. Moreover, accurate identification of genetic relatedness between cultivars and varieties is a pre-requisite for plant breeding programmes. In the present study, extensive genetic variability, high differentiation among species and strong subdivision among populations of the species were observed. High polymorphism observed for amplicons generated with each primer can be attributed to the fact that 14 distinct and diverse species of Vigna were analyzed in the present study. In fact, earlier only 3% bands were reported as common in 23 accessions belonging to subgenus Ceratotropis (Kaga et al. 1996).
Of the species analyzed, higher polymorphism was observed for the wild species in comparison to the cultivated ones except for the wild species where sample size is very small due to their limited occurrence in the wild habitats (Tables 4, 5). This is in contrast to the high genetic similarity reported earlier among the cultivars of V. radiata using RAPDs (Lakhanpaul et al. 2000). Perusal of the estimates of genetic diversity statistics among the populations representing various species indicated high levels of population differentiation. Averaged over all loci, the total genetic diversity is moderate and high level of variability is found between species. However, total diversity (Ht), within accession diversity (Hs) and between accession diversity (Dst) observed were higher than earlier studies based on isozyme electrophoresis, where values for total diversity is less than 0.2 for V. vexillata (Sonnante et al. 1997) and V. luteola and V. marina (Sonnante et al. 1998) and 0.3 for V. unguiculata (Pasquet 1999). In these reports, the within accession diversity reported was less than 0.01 (Sonnante et al. 1997; Pasquet 1999). Higher diversity and differentiation reported in the present study could be due to presence of greater diversity in wild populations of the species and higher polymorphism observed for the RAPD markers in comparison to isozymes and also due to the fact that larger number of polymorphic markers was available with RAPDs (Williams et al. 1990). On the other hand, protein/isozymes profiles reveal only a fraction of the genetic changes in the coding regions of the genome.
Considerable gene-flow was detected between the populations of V. mungo, V. mungo var. silvestris, V. radiata, V. radiata var. sublobata, V. hainiana and V. umbellata. This indicates occurrence of inter-crossing between wild populations of these species, as these populations occur in contiguous areas and mostly their geographic areas of occurrence overlap (Table 1). Minimum values for FST were observed between populations of V. hainiana indicating that these populations are least differentiated, whereas, within a group maximum values of FST was recorded for populations of V. radiata var. sublobata indicating that this group comprises divergent populations.
Fixation index is defined as increased homozygosity resulting from inbreeding. Wright’s F statistics could be negative or positive indicating a deficiency or excess of homozygotes in relation to a random mating population. F equals to zero, when the population is in Hardy–Weinberg equilibrium, while when all genotypes are homozygous, F equals 1. In the present study, high estimates of fixation index indicated a predominantly autogamous behavior of the wild species. Hartl (1987) suggested that the range 0–0.05 indicates little differentiation, 0.05–0.15 moderate, 0.15–0.25 large differentiation and above 0.25 indicates very large differentiation. The high values for Fct observed here (which is equivalent to Wright’s Fst), indicates strong subdivision of populations representing a species and high genetic differentiation among the species. A comparison of the Fct values for the mungo–radiata complex (0.87) and all the Vigna species together (0.25) indicates existence of very large genetic differentiation among the designated species and lower taxonomic levels in this complex. The partitioning of molecular variance with AMOVA revealed that substantial variation exists among the species and populations indicating that greater number of population samples should be sampled and studied to delineate the species identities and boundaries in this species complex. It could be seen that the partitioning of variation is similar in magnitude in both mungo–radiata complex as well as when all 14 Vigna species are compared together.
UPGMA tree based on RAPD profiles indicated that V. mungo, V. radiata, V. mungo var. silvestris, V. radiata var. sublobata and V. radiata var. setulosa are distinct taxonomic groups. However, as these species were placed in the same cluster in the dendrogram, their origin from a common ancestor is evident. In addition, cultivated forms are grouped close to their proposed wild forms. Though, Kaga et al. (1996) reported similar grouping in their study of subgenus Ceratotropis, more variation between cultivated and wild forms was found in V. radiata than in V. mungo, in the present study. Therefore, wild forms of V. radiata have differentiated greatly since domestication and some of them retain only partial affinity with the related species V. mungo (Miyazaki 1982). Clustering of all the accessions of V. hainiana together suggests it to be a distinct taxa and its close and equidistant grouping also appeared to support the view that V. hainiana might be the common putative progenitor of wild relatives of both V. mungo and V. radiata (Babu et al. 1985). However, elaborate analysis of diverse populations is required to further confirm whether V. hainiana could be considered as the putative progenitor of green gram and black gram and whether the closest wild relative of green gram, namely, V. radiata var. sublobata and V. radiata var. setulosa also have their origin from V. hainiana.
To conclude, the evidences suggest that: (1) the wild species, V. radiata var. sublobata and V. mungo var. silvestris have high similarity to their respective cultivated forms. (2) V. hainiana, V. radiata var. sublobata, V. radiata var. setulosa and V. mungo var. silvestris are distinct taxa. (3) V. hainiana is equidistant to both V. radiata and V. mungo. (4) V. hainiana is more primitive compared to V. radiata var. sublobata, V. radiata var. setulosa and V. mungo var. silvestris. Hence, V. hainiana could be the common pivotal progenitor species of both V. radiata var. sublobata and V. mungo var. silvestris from which the cultivated mung (V. radiata) and urd (V. mungo) have evolved.
References
Arora RK (1985) Diversity and collection of wild Vigna species in India. FAO/IBPGR. Plant Genet Resour Newsl 63:26–35
Babu CR, Sharma SK, Johri B (1985) Taxonomic revision of Indian Phaseolus, Vigna, Macroptilium, Dysolobium. Bull Bot Surv India 27:1–28
Brunk CF, Jones KC, James TW (1979) Assay for nanogram quantities of DNA in cellular homogenates. Anal Biochem 92:497–500. doi:10.1016/0003-2697(79)90690-0
Chandel KPS, Lester RN, Starling RJ (1984) The wild ancestor of urid and mungbeans (V. mungo (L.) Hepper and Vigna radiata (L.) Wilczek). Bot J Linn Soc 89:85–96. doi:10.1111/j.1095-8339.1984.tb01002.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction sites. Genetics 131:479–491
Hartl DL (1987) A primer of population genetics, 2nd edn. Sinauer Associates Inc. Publishers, Sunderland
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kaga A, Tomooka N, Egawa Y, Hosaka K, Kamijima O (1996) Species relationships in the subgenus Ceratotropis (genus Vigna) as revealed by RAPD analysis. Euphytica 88(1):17–24. doi:10.1007/BF00029261
Lakhanpaul S, Chadha S, Bhat KV (2000) Random Amplified Polymorphic DNA (RAPD) analysis in Indian mung bean (Vigna radiata (L.) Wilczek) cultivars. Genetica 109:227–234. doi:10.1023/A:1017511918528
Lukoki L, Maréchal R, Otoul E (1980) The wild ancestors of cultivated beans V. radiata and V. mungo. Bull Jard Bot Nat Belg 28:23–30
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Maréchal R, Mascherpa JM, Stainier F (1978) Etude taxonomique d’ un group complexe d’ especes des generes Phaseolus et Vigna (Papillionaceae) sur la base de donnees morphologiques, et polliniques traitees par l’ analyse informatique. Boissera 28:1–273
Miyazaki S (1982) Classification and phylogenetic relationships of the Vigna radiata-mungo-sublobata complex. Bull Nat Inst Agr Sci D 33:1–61
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Pasquet RS (1999) Genetic relationships among species of V. unguiculata (L.) Walp. based on allozyme variation. Theor Appl Genet 98(1–2):1104–1119. doi:10.1007/s001220051174
Rohlf FJ (1992) NTSYS-pc Numerical taxonomy and multivariate taxonomy system, version 1.8
Saghai-Maroof MA, Soliman KM, Jorgenson RA, Allard AW (1984) Ribosomal spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA 81:8014–8018. doi:10.1073/pnas.81.24.8014
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Sonnante G, Spinosa A, Marangi A, Pignone D (1997) Isozymes and RAPD analysis of the genetic diversity within and between V. luteola and V. marina. Ann Bot (Lond) 80:741–746. doi:10.1006/anbo.1997.0511
Sonnante G, Spinosa A, Pignone D (1998) Assessment of genetic variation in a working collection of Vigna vexillata (L.) Rich by isozyme and RAPD analyses. Genet Resour Crop Evol 45:347–354
Tateishi Y (1996) Systematics of the species of Vigna subgenus Ceratotropis. In: Srinives P, Kitbamroong C, Miyazaki S (eds) Mungbean germplasm: collection, evaluation and utilization for breeding program. JIRCAS, Japan, pp 9–24
Verdcourt B (1970) Studies in the Leguminosae-Papiliondideae for the “Flora of Tropical East Africa” IV. Kew Bull 24:507–569. doi:10.2307/4102859
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535. doi:10.1093/nar/18.22.6531
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420. doi:10.2307/2406450
Acknowledgements
The facilities provided by Director, National Bureau of Plant Genetic Resources for conducting this work is gratefully acknowledged. RV gratefully acknowledges receipt of research fellowship for her Ph.D. work from Council of Scientific and Industrial Research, Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vir, R., Bhat, K.V. & Lakhanpaul, S. Analysis of population substructure, genetic differentiation and phylogenetic relationships among selected Asiatic Vigna Savi species. Genet Resour Crop Evol 56, 783–795 (2009). https://doi.org/10.1007/s10722-008-9401-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-008-9401-3