Abstract
This study applied two methods to assess intra-collection duplication within 339 Nordic oat (Avena sativa L.) accessions preserved by Plant Gene Resources of Canada (PGRC). Putative duplicates, that is accessions carrying a similar accession name, were grouped into 52 duplication groups and included 230 of the 339 Nordic oat accessions. A field assessment based on visual inspection of field plots was conducted during two growing seasons to detect distinct phenotypes within each duplication group. Simultaneously, a descriptor assessment using seven characters with altogether sixteen character states was used in both years for the same purpose. The combined results of both assessments and both years indicated that among the 230 accessions in duplication groups only 118 could be identified as distinct. This would allow for a reduction of 33% of the Nordic oat accessions at PGRC. The field assessment method detected fewer (75%) distinct accessions than the descriptor assessment (84%), when considering all accessions identified as distinct as 100%. Repeatability between years was higher in the field assessment (70%) than in the descriptor assessment (64%). The field assessment requires an experienced germplasm evaluator, but allows for handling large numbers of germplasm accessions and for detecting functional, fitness related, and user-relevant diversity. Combining field assessment with descriptor assessment and more sophisticated methods on selected subgroups may be the most efficient method for determination of internal duplication in genebank collections. Bulking phenotypically similar accessions within duplication groups is preferable to eliminating duplicate accessions when collection rationalization is required, as it reduces the risk of loosing diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Canadian national seed genebank, Plant Gene Resources of Canada (PGRC), preserves with more than 27,000 accessions of the genus Avena L. the largest oat collection in the world. More than 10,000 accessions are A. sativa L., which covers the cultivated hexaploid taxa (Diederichsen 2008). Based on an agreement with the International Board for Plant Genetic Resources (IBPGR, now: Bioversity International), PGRC was assigned the task of maintaining the world base collection of this genus. Therefore, a large amount of duplication exists between the oat collection at the PGRC genebank and other ex situ collections around the world. For example, based on passport data, 81% of the PGRC oat collection is duplicated in the National Small Grain Collection of the United States Department of Agriculture (Diederichsen et al. 2001).
Fowler (2007) pointed at the large increase in genebank accessions world-wide between 1984 and 1996 from 2 million to about 6.5 million accessions, and that most of this increase is due to the fact that “…genebanks were trading samples back and forth with each other in a totally uncoordinated and basically unknown way….” He used the term “hyper-inflation” for this development as it indicates a high amount of redundant material. Van Hintum and Knüpffer (1995) made a similar statement: “Most collections in genebanks are still rather haphazard.” The historical genebanks in Russia (N.I. Vavilov Institute St. Petersburg) and Germany (Gatersleben genebank at Leibniz Institute of Plant Genetics and Crop Plant Research) had a systematic approach to germplasm acquisition (Hammer 1993). However, several of the genebanks that emerged after the 1960s may have more un-coordinated duplication within and among the collections and duplication deserves some attention.
For budget reasons, the pressure on genebank collections to rationalize their operations is growing (Hammer 2003). Therefore, the core collection principle and other strategies to optimize the composition of genebank collections have recently received increased attention (Sackville Hamilton et al. 2003; van Treuren et al. 2008). To rationalize germplasm preservation efforts globally, it will be essential to address the issues of duplication within and among genebanks systematically (van Hintum and Visser 1995; van Hintum 2000). Crop specific networks within the European context (European Cooperative Programme for the Conservation and Exchange of Crop Genetic Resources, ECP/GR) and recently on a global scale (Global Crop Diversity Trust in Rome) will have to assess duplication among and within ex situ collections in order to gain a more objective overview of the diversity preserved in genebanks.
Duplication of accessions within a genebank is not unusual, in particular in older cultivars or landrace material. Such duplication is confusing to genebank clients and a burden both for curator and genebank budget. However, not all duplicates are true duplicates. According to the terminology of van Hintum and Knüpffer (1995) duplicates can be (1) identical duplicates (genetically identical) or (2) common duplicates (derived from the same initial population). In living collections evolutionary processes can not be avoided and the term “maintenance breeding” points at the fact that reselection to original cultivar types is often necessary when breeders want to maintain their material over several generations. Only a few genebanks have voucher samples of each accession that, similar to botanical nomenclatural typus specimens, can serve as reference when conducting true to type verification of material form different regeneration cycles or origin (Hammer 1993). It may be a good starting point for each genebank to come to terms with internal duplication before approaching duplication among genebanks.
Despite the high cost, molecular methods have recently received more attention for investigating redundancy in germplasm collections (Virk et al. 1995; Zeven et al. 1998; Phippen et al. 1997; Fu 2006). Lund et al. (2003) demonstrated the use of molecular methods for investigating duplication groups in Nordic barley. Van Treuren et al. (2008) showed that diversity of relevance for plant breeders may be missed when exclusively relying on molecular assessments of diversity for managing a lettuce germplasm collection.
Oat from the Nordic countries has historically contributed significantly to oat breeding in Canada and the United States (McKenzie and Harder 1995; Stanton 1955), as well as in many European countries (Zade 1918; de Haan 1954). Therefore, this group was selected for the study. Two low-cost methods that have the capacity to be applied to large numbers of germplasm accessions were chosen for duplication assessment. The objectives were to evaluate the two methods for assessing distinctness among potential duplicates in germplasm collections and to discuss the results from a genebank perspective.
Materials and methods
All 339 accessions from the PGRC oat collection with the species identification A. sativa and with a Nordic country of origin were selected for this experiment. This resulted in 234 accessions from Sweden, 68 from Finland, 30 from Denmark and 7 from Norway. These accessions were grouped by the information recorded in the passport data as “accession name”, which contains in the PGRC database the cultivar name, the landrace name, the breeding line code, or other accession identifiers. The majority of the accessions had a cultivar name (284 accessions), followed by breeding lines (43 accessions), landraces (7 accessions) and unknown status (5 accessions). For 196 of the accessions with cultivar names, a year of release of the cultivar in one of the Nordic countries was available (Nordic Genebank 2008); the cultivars were released between 1894 (‘Ligowo’ from Sweden) and 1981 (‘Veli’ from Finland). The 339 accessions were grouped into potential duplication groups based on the recorded accession names. For example, there were five different accessions that had the accession name ‘Seger’, which is a cultivar that originated from the Swedish Plant Breeding Station at Svalöv. It goes back to a single plant selection made in 1892 and was released as a cultivar in Sweden in 1908 (Stanton 1955). Possibly, all ‘Seger’ accessions are duplicates. In addition to these, there were 15 accessions that had the accession name ‘Victory’, which is the English translation of the Swedish name. The cultivar ‘Seger’ was marketed in North America after 1908 as ‘Victory’ (Stanton 1955). The English name was also used in the Netherlands (de Haan 1954), although one accession included in this duplication group had the Dutch name ‘Zegehaver’, which translates to “victory oat”. Furthermore, there were five accessions with the name ‘Swedish Victory’, which probably refers to the same original Swedish oat cultivar. One accession had the name ‘Pobeda’ which is Russian for “victory”. As a result, all 27 accessions were placed in one duplication group that received the most authentic name, which in this case was “Seger”. Spelling variations of a name also frequently occurred. Other large duplication groups were “Guldregn I” (18 accessions), “Sol II” (12), “Guldregn II” (11) and “Örn” (8). In total, the 339 A. sativa accessions preserved by PGRC consisted of 109 accessions which were unique based on accession names, and of 54 putative duplication groups, each of them including two or more accessions, accounting for the remaining 230 accessions (Table 1).
All accessions were spring-sown at the Agriculture and Agri-Food Canada Saskatoon Research Centre experimental farm at Saskatoon, Saskatchewan, Canada (52°10′ N, 106°41′ W, altitude 501 m above sea level) on loamy, dark chernozemic soil in 2003 and 2007. Random seed samples of 7 g from each accession were sown in single rows of 3 m length without replication. After 20 rows (2003) or 24 rows (2007), single rows of the Canadian oat cultivars ‘CDC Pacer’ and ‘AC Assiniboia’ were planted in alternation, in order to detect major plot location effects on the plants’ performance during cultivation.
In 2003, the accessions were planted in the field in alphabetical order based on the accession name or based on the name of the duplication group into which they belonged. Within the duplication groups, the accessions were ordered by their accession number in the Canadian genebank. In 2007, the accessions were planted in random order, but the accessions belonging to each duplication group were planted in neighbouring plots. Within the duplication groups, the order of the accessions was randomized.
Two methods for assessing the genetic distinctness of accessions belonging into the same duplication group were applied.
Field assessment of diversity
In each growing season, the same evaluator inspected the plots at two plant development stages (heading and early maturity) and visually investigated each single row belonging to a duplication group for phenotypical differences. Any observed phenotypic differences in plant characters were recorded. Accessions that appeared similar within a duplication group were assigned the same letter code. In some duplication groups no differences were detected, i.e. all accessions were assigned the letter “a”, while in other duplications groups two, three or more plant types (“b”, “c”,…) could be distinguished. The results of the field assessments in the two years were considered separately and then combined. Any accessions that appeared differently in one of the two years or in both years received a different letter code for this combined assessment.
Descriptor assessment of diversity
At plant maturity, the following panicle descriptors were rated: panicle type, panicle erectness and panicle density. Five panicles were harvested separately from each single row and on the seeds of these panicles the following characters were rated: lemma colour, lemma hairiness, dorsal awn on lemma, kernel covering and spikelet separation (Table 2). Letter codes were assigned to each distinct combination of single character states observed within a duplication group in the same way as described above for the field assessment. The results of descriptor assessments in the two years were considered and then combined, so that any accessions that appeared differently in one of the two years or in both years received a different letter code.
Combined field and descriptor assessment
For obtaining an overall rating of genetic distinctness of accessions within a duplication group, the combined results of the field assessments and the combined results of the descriptor assessments were combined. Accessions within a duplication group that differed in one or both of the assessments received a different letter code, resulting in an overall assessment of distinctness within each duplication group.
Results
The field assessments showed that plant height, leaf colour (green vs. bluish green), lodging, panicle length, lemma length and earliness were useful characters to distinguish phenotypes by visual inspection of the plots. Additionally, panicle type (equilateral vs. unilateral), panicle density, lemma colour and oat type, which were also used in the independently conducted descriptor assessment (Table 2), were occasionally recorded as indicating phentotypic differences when doing the field assessment. None of the accessions showed intra-accession diversity for the investigated characters, however, a careful analysis of more single plants per accessions would possibly find such differences.
Both the field assessments and the descriptor assessments for phenotypic distinctness showed that within many duplication groups more than one phenotype could be distinguished. However, both assessments also indicated redundancy of material in the duplication groups (Table 1). The combined consideration of both assessment methods is the most careful estimation of distinctness this study allowed for. It resulted in the recognition of 118 accessions with distinct phenotypes among the 230 accessions belonging to duplication groups (Table 3). This indicated that 51% of the accessions were redundant when considering all duplication groups together.
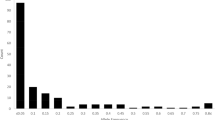
The field assessments alone detected 89 of these 118 distinct phenotypes and the descriptor assessment detected 99 distinct phenotypes. When relying on only one year of field or descriptor assessment, fewer distinct phenotypes were detected (Table 3). The field assessment in 2007 resulted in discrimination of nine more phenotypes than the assessment of 2003. Not all accessions within a duplication group that were phenotypically different in one year appeared different in the other year; in other words, not all of these accessions were consistently detected as being distinct in both years (Fig. 1). Similarly to that, in the descriptor assessment, combination of two years of descriptor assessments resulted in detection of more distinct phenotypes than only using the results of one year (Fig. 1).
Set diagrams showing absolute and relative (%) numbers of distinct accessions within 54 duplication groups of Nordic oat comprising 230 accessions in total. The years of assessment and the assessment methods are indicated. The overlapping area (intersection) of the two sets contains the number of accessions that was detected as distinct in both years or by both assessment methods. The union of two sets represents the combined assessments
Combining the field assessments and the descriptor assessments again increased the detection of distinct phenotypes. The percentage of accessions that were consistently detected as distinct in both years were used as an indicator of the repeatability of the applied assessments. Figure 1 shows these numbers. While the combined descriptor assessments detected more distinct accessions than the combined field assessments (99 vs. 89 accessions), the repeatability was lower in the descriptor assessments (64% vs. 70%). When considering both years combined, 84% of the total diversity that was detected could be identified by the descriptor assessment alone, while the field assessment alone detected only 75% of the total diversity (Table 3).
The number of accessions within a duplication group ranged from two in 21 duplication groups to 27 in the “Seger” duplication group; the number of phenotypically distinct accessions ranged from one, which meant phenotypical similarity of all accessions, to eight (Table 1). The maximum number of eight distinct phenotypes was found within the “Seger” duplication group. An association of number of accessions within a duplication group and the number of distinct phenotypes detected could be expected and was also found, but there were also duplication groups with several accessions that were phenotypically similar (Fig. 2). An example for the latter was the group “Guldregn II” with eleven accessions that could not be distinguished from each other. The other extreme represented the group “Bambu II” consisting of six accessions representing five different phenotypes.
Discussion
A morphological approach for assessing diversity as applied here may be useful when dealing with large collections and for capturing functional diversity (Love and Spaner 2007). The two approaches taken for assessing the distinctness of phenotypes in this study have a fundamental difference. The descriptor assessment is a very traditional method. Assessments are made using an a priori fixed list of descriptors.
The field assessment is based on a different principle than the descriptor assessment, because the characters used for assessing the distinctness of two or more accessions are not a priori defined. The observation skills and experience of the person conducting the assessment influence the results of the field assessment. A more experienced evaluator may detect differences among the accessions that the untrained eye may overlook. The characters that are used to distinguish among accessions are only a posteriori known and as such are part of the results. The experience factor might explain, why the assessments in 2007 resulted in more distinct accessions that the assessments in 2003, as the skills of the evaluator had increased over that time. Although well established terms, the factor of experience for skilful plant breeding has only recently received scientific attention and first steps to investigate the role and function of the so-called “breeder’s eye” were made (Timmermann 2006, 2007).
Quantitative characters such as plant height, leaf colour, lodging, panicle length and lemma length were found being useful for detecting distinct accessions in the field assessment. These characters were purposely not included in the descriptor assessment because they are very difficult to assess when absolute measurements are taken and need to be compared. Replicated field trials would be required to test for statistically significant differences and it is realistically impossible to accomplish this given the large amount of germplasm in genebanks (Yang et al. 1991). It seems more realistic to use the trained eye and perception of a person familiar with the crop to assess differences in quantitative characters in the field for the purpose of detecting differences in these traits.
A principal question is, whether combination with other methods of assessment, including more descriptors, or replication in a third year, would further increase the amount of phenotypes distinguished. The answer is certainly yes. Rodionova (1974) pointed at the possibility to classify a given oat cultivar into different botanical varieties depending on the growing season because the formation of lemma awns is influenced by environment. The results from the present study also show that replication of the field and descriptor assessment in different years is useful for obtaining better assessment results.
It is important to notice that even modern cultivars are often not genetically completely homogeneous and growing seeds of the same cultivar for some years in different environments may result in different allelic compositions of such duplicates due to genetic shift (selection) or genetic drift (random). The usefulness of intra-accession diversity in phenotypically homogeneous genebank preserved landrace accessions when re-introducing such genebank material into on-farm situations was recently demonstrated in lentil which, similar to oat, is a self-pollinating species (Horneburg and Becker 2008).
Van Hintum and Knüpffer (1995) introduced the term “common duplicates” for accessions that trace back to the same initial population but may have altered genetic identities (allelic compositions) and they distinguish them from “identical duplicates”. Sensitive methods for assessment of genetic diversity will pick these differences up. Van Treuren et al. (2001) stated that molecular variance needs to be interpreted and a decision is required “… to evaluate, whether samples display sufficient genetic variation in order to consider them distinct”. The question from a conservationist’s perspective clearly is, whether nearly similar accessions deserve to be considered and preserved as separate entries, in particular if the differences have no phenotypic impact and therefore probably no selective value.
Van Hintum et al. (2002) provided a thorough discussion regarding the splitting and lumping (bulking) of genebank accessions and reported that the Centre for Genetic Resources in The Netherlands (CGN) created so-called “umbrella varieties” of cabbage (Brassica oleracea L.) landraces that were similar in appearance. For the PGRC oat collection, a similar approach might be feasible by combining those accessions that appeared similar within a duplication group. However, in a self pollinating species the population dynamics are different. Bulking the similar phenotypes within the 52 duplication groups of all 339 Nordic oat accessions in PGRC would result in a reduction to 227 accessions, which equals a reduction by 33%.
The risk of eliminating diversity, i.e. gene erosion, when discarding potential duplicates can not be excluded and bulking may be preferable for reducing the total number of accessions. Bulking means in this context creating phenotypically homogeneous, but potentially genetically still heterogeneous bulk-accessions. The strategy could be described as “phenotypical homogenization of accessions”, which can be the result of separating accessions as suggested by Lehmann and Mansfeld (1957) or of bulking of phenotypically identical duplicates.
A decision which entry in a duplication group most authentically represents the cultivar name it carries would be very desirable. Reducing the number of entries in a duplication group to distinct phenotypes does not resolve this problem since these distinct accessions all carry the same accession name. If the accession has the status of a landrace, then it may well be that all distinct types can claim to be authentic, because the accession name is not closely associated with a single genotype. Using the terminology of van Hintum and Knüpffer (1995) this would fall in the category of “partly duplication”. This relates to the fact that genebank accessions can be heterogeneous for morphological characters: that is, the inherent accession diversity may not allow unambiguous assessments for distinguishing between two or more members of a duplication group. Landraces and older cultivars are more likely to represent such populations. The duplication group “Probsteier” pointed in this direction. The five entries of “Probsteier” had four distinct phenotypes and the three entries of “Awnless Probsteier” had three different phenotypes. All these entries may claim the same degree of authenticity. The true origin of the landrace ‘Probsteier’ is a small region of northern Germany called “Probstei”, and ‘Probsteier’ oat was widely cultivated in the Nordic and other countries (Zade 1918; de Haan 1954). ‘Awnless Probsteier’ is a selection made from the landrace at Svalöv in 1892 (Stanton 1955). Zade (1918) reported 17 oat cultivars directly or indirectly selected from the landrace ‘Probsteier’ without involving crosses, providing evidence of a tremendous amount of variation in this landrace. Grau Nersting et al. (2006) demonstrated the diversity of oat landraces historically cultivated in the Nordic countries. It is probably impossible or a mistake to declare one single accession of the PGRC collection of such a landrace-cultivar as the most authentic representative. Even if these accessions are common or partial duplicates in the terminology of van Hintum and Knüpffer (1995), they should be kept as separate genebank accessions.
In older cultivars, several phenotypes may carry the same cultivar name. Besides genetic heterogeneity in the original cultivar or landrace, such diversity may also be due to the fact that prior to variety protection, seed traders used unprotected but well known names for selling different types of seed under a fashionable cultivar name. Stanton (1955) mentioned this for oat in the United States in the first half of the 20th century.
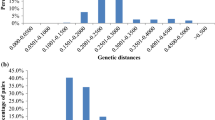
In some cases it may be easy to declare certain phenotypes within a duplication group as not authentic. Amongst the four entries of the duplication group “Bambu” one accession was hull-less. This points at a mistake: the hull-less type can not be considered a legitimate representative of ‘Bambu’, as this cultivar does not even have any hull-less oat in its pedigree (Nordic Genebank 2008). A suspicion was that more distinct phenotypes would be found in duplication groups of older cultivars, because they might not have been as genetically homogeneous to start with and they had more time to evolve differently when received from different sources. Such an association, however, was not found in the Nordic oat accessions at PGRC (Fig. 3).
This study provides clear evidence that bulking or eliminating accessions in duplication groups identified using passport data only is not appropriate, as distinct accessions would be merged or lost, respectively. Van Hintum and Knüpffer (1995) came to the same conclusion. The field assessment method might be a very useful approach for a primary assessment of duplication, in particular if the budgets are limited and rationalisation pressure is high. The field assessment for distinctness by a trained evaluator should allow differences in quantitative or qualitative characters to be detected that enhance or decrease the fitness of an accession in comparison to putative duplicate accessions. These characters are particularly important when bulking accessions, as they would enhance or decrease the proportion of these particular genotypes within the newly created accessions during subsequent regeneration of these composed populations. To make decisions regarding genebank accession management based on functional diversity, and in particular based on characters that impact the fitness of a distinct genotype, is definitely meaningful. The descriptor assessment can be used simultaneously with the field assessment for refining the results. The descriptor assessment method requires more resources than the field assessment method. The descriptor assessment is more objective in the traditional understanding of science. The combination with molecular methods as applied by Zeven et al. (1998), albeit on a smaller number of accessions, is certainly useful. The field assessment method might be criticized as being subjective, but relying on an experienced germplasm evaluator and his or her decisions may be as practical and efficient as relying on experienced plant breeders, who are steadily making similar decisions.
References
De Haan H (1954) Oat breeding in the Netherlands. Euphytica 3:81–180. doi:10.1007/BF00029954
Diederichsen A (2008) Assessments of genetic diversity within a world collection of cultivated hexaploid oat (Avena sativa L.) based on qualitative morphological characters. Genet Resour Crop Evol 55:419–440. doi:10.1007/s10722-007-9249-y
Diederichsen A, Timmermans E, Williams DJ, Richards KW (2001) Holdings of Avena germplasm at Plant Gene Resources of Canada and status of the collection. Oat Newsl 47:35–42.
Fowler C (2007) Global dimensions of conserving crop diversity. Seed Savers 2006 Harvest Edition. Seed Savers Exchange Inc, Decorah, pp 33–42
Fu YB (2006) Redundancy and distinctness in flax germplasm as revealed by RAPD dissimilarity. Plant Genet Resour 4:117–124. doi:10.1079/PGR2005106
Grau Nersting L, Andersen SB, von Bothmer R, Gullord M, Bagger Jørgensen R (2006) Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 150:327–337. doi:10.1007/s10681-006-9116-5
Hammer K (1993) The 50th anniversary of the Gatersleben genebank. Plant Genet Resour Newsl 91/92:1–8
Hammer K (2003) A paradigm shift in the discipline of plant genetic resources. Genet Resour Crop Evol 50:3–10. doi:10.1023/A:1022944910256
Horneburg B, Becker HC (2008) Crop adaptation in on-farm management by natural and conscious selection: a case study with lentil. Crop Sci 48:203–212. doi:10.2135/cropsci2007.03.0170
Lehmann CO, Mansfeld R (1957) Zur Technik der Sortimentserhaltung. Kulturpflanze 5:108–138. (On the technique for collection-maintenance.) doi:10.1007/BF02095492
Love B, Spaner D (2007) Agrobiodiversity: its value, measurement, and conservation in the context of sustainable agriculture. J Sustain Agric 31:53–82. doi:10.1300/J064v31n02_05
Lund B, Ortiz R, Skovgaard IM, Waugh R, Andersen SB (2003) Analysis of potential duplicates in barley gene bank collections using re-sampling of microsatellite data. Theor Appl Genet 106:1129–1138
McKenzie RIH, Harder DE (1995) Oat. In: Slinkard AE, Knott DR (eds) Harvest of gold: the history of field crop breeding in Canada. University Extension Press, Saskatoon, pp 98–112
Nordic Genebank (2008) SESTO Gene bank documentation system. http://tor.ngb.se/sesto/index.php?scp=ngb&thm=sesto&r=437596376. Accessed 18 September 2008
Phippen WB, Kresovich S, Candelas FG, McFerson JR (1997) Molecular characterization can quantify partition variation among genebank holdings: a case study with phenotypically similar accessions of Brassica oleracea var. capitata (cabbage) ‘Golden Acre’. Theor Appl Genet 94:227–234. doi:10.1007/s001220050404
Rodionova NA (1974) Opredelenie podlinnosti vidov i sortov ovsa po morfoligičskim priznakam zerna. [Determination of authenticity of oat species and cultivars by morphological characters of the grain]. Trudy po prikladnoj botanike, genetike i selekcii 51(2):54–61
Sackville Hamilton R, Engels JMM, van Hintum ThJL (2003) Rationalization of genebank management. In: Engels JMM, Visser L (eds) A guide to effective management of germplasm collections. IPGRI, Rome, pp 80–92
Stanton TR (1955) Oat identification and classification. Technical Bulletin No. 1100. United States Department of Agriculture, Washington DC
Timmermann M (2006) The breeder’s eye – theoretical aspects about the breeder’s decision-making. In: Øestergård H, Fontaine L (eds) Proceedings of the COST SUSVAR workshop on cereal crop diversity: implications for production and products, 13–14 June 2006, La Besse, France. ITAB, France, pp 118–123
Timmermann M (2007) Phänomenologie der Natur: eine methodologische Erweiterung der quantifizierenden Naturwissenschaften [Phenomenology of nature: methodological amplification of the quantifying natural sciences]. In: Zikeli S, Claupein W, Dabbert S, Kaufmann B, Müller T, Valle Zárate A (eds) Proceedings of Zwischen Tradition und Globalisierung—9. Wissenschaftstagung Ökologischer Landbau, Universität Hohenheim, Stuttgart, Deutschland, 20–23 March 2007, vol 2, pp 787–790. Köster, Hohenheim (Also available at: http://orgprints.org/9453/. Accessed 18 September 2008)
Van Hintum ThJL (2000) Duplication within and between germplasm collections III. A quantitative model. Genet Resour Crop Evol 47:507–513. doi:10.1023/A:1008703031415
Van Hintum ThJL, Knüpffer H (1995) Duplication within and between germplasm collections I. Identifying duplication on the basis of passport data. Genet Resour Crop Evol 42:127–133. doi:10.1007/BF02539516
Van Hintum ThJL, Visser DL (1995) Duplication within and between germplasm collections II. Duplication in four European barley collections. Genet Resour Crop Evol 42:135–145. doi:10.1007/BF02539517
Van Hintum ThJL, Sackville Hamilton NR, Engels JMM, van Treuren R (2002) Accession management strategies: splitting and lumping. In: Engels JMM, Ramanatha Rao V, Brown AHD, Jackson MT (eds) Managing plant genetic diversity. International Plant Genetic Resources Institute, Rome
Van Treuren R, van Hintum ThJL, van de Wiel CCM (2008) Marker-assisted optimization of an expert-based strategy for the acquisition of modern lettuce varieties to improve a genebank collection. Genet Resour Crop Evol 55:319–330. doi:10.1007/s10722-007-9237-2
Van Treuren R, van Soest LJM, van Hintum ThJL (2001) Marker-assisted rationalization of genetic resources collections: a case study in flax using AFLPs. Theor Appl Genet 103:144–152. doi:10.1007/s001220100537
Virk PS, Newbury HJ, Jackson MT, Ford-Lloyd BV (1995) The identification of duplicate accessions within a rice germplasm collection using RAPD analysis. Theor Appl Genet 90:1049–1055. doi:10.1007/BF00222920
Yang RC, Jana S, Clarke JM (1991) Phenotypic diversity and association of some potentially drought-responsive characters in durum wheat. Crop Sci 31:1484–1491
Zade A (1918) Der Hafer – eine Monographie auf wissenschaftlicher und praktischer Grundlage [Oats – a monograph on scientific and practical grounds]. Gustav Fischer, Jena
Zeven AC, Dehmer KJ, Gladis T, Hammer K, Lux H (1998) Are the duplicates of perennial kale (Brassica oleracea L. var. ramosa DC.) true duplicates as determined by RAPD analysis? Genet Resour Crop Evol 45:105–111
Acknowledgement
The technical assistance of Mr. D.J. Williams for setting up the field plots and careful descriptor evaluation of the germplasm is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diederichsen, A. Duplication assessments in Nordic Avena sativa accessions at the Canadian national genebank. Genet Resour Crop Evol 56, 587–597 (2009). https://doi.org/10.1007/s10722-008-9388-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-008-9388-9