Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis in many cancer cells but not in normal ones. Recombinant TRAIL and agonistic antibodies to its cognate receptors are currently being studied as promising anticancer drugs. However, preclinical and clinical studies have shown that many types of human cancers are resistant to TRAIL agonists. We previously reported that a deficiency of fucosylation, which is one of the most common oligosaccharide modifications, leads to resistance to TRAIL-induced apoptosis. In contrast, DNA methylation is associated with silencing of various tumor suppressor genes and resistance of cancer cells to anticancer drugs. The aim of this study is to clarify the involvement of DNA methylation in the regulation of cellular fucosylation and the susceptibility to TRAIL-induced apoptosis. When nineteen cancer cell lines with relatively low fucosylation levels were treated with a novel methyltransferase inhibitor, zebularine, an increase in the fucosylation level was observed in many cancer cell lines. The expression of fucosylation-related genes, such as the FX, GDP-fucose transporter, and Fut4 genes, was significantly increased after the treatment with zebularine. Moreover, a synergistic effect of zebularine on TRAIL-induced apoptosis was observed in several cancer cell lines, in which fucosylation was increased by treatment with zebularine. This synergistic effect was independent of the expression of TRAIL receptors and caspase-8. These results indicate that cellular fucosylation is regulated through DNA methylation in many cancer cells. Moreover, zebularine might be useful as a combination drug with TRAIL-based therapies in patients with TRAIL-resistant cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oligosaccharides are one of the most important factors in the posttranslational modification of proteins [1, 2]. It is a well-known fact that oligosaccharide structure change during malignant transformation [3]. Therefore, oligosaccharides have been used as bio-markers for various types of tumors and have the potential to become targets for effective treatment of them. Fucosylation, which comprises the transfer of a fucose residue to an oligosaccharide or protein, is one of the most important oligosaccharide changes in cancer [4]. A representative example is fucosylated alpha-fetoprotein (AFP). While AFP is a well-known tumor marker for hepatocellular carcinoma (HCC) [5], its level is also increased in chronic liver diseases, such as chronic hepatitis and liver cirrhosis. In contrast, fucosylated AFP, AFP-L3, is a very specific marker for HCC, and has been clinically used since 1996 in Japan [6, 7] and 2005 in the United States [8].
Fucosylation is catalyzed by fucosyltransferases, guanosin-5′-diphosphate (GDP)-fucose synthetic enzymes, and GDP-fucose transporter (GDP-Fuc Tr). Thirteen fucosyltransferases have been identified in the human genome, and are localized in the endoplasmic reticulum and Golgi apparatus. GDP-fucose is synthesized in the cytosol via two pathways, namely salvage and de novo pathways (Fig. 1). The salvage pathway synthesizes GDP-fucose from free fucose derived from an extracellular or lysosomal source. The de novo pathway transforms GDP-mannose to GDP-fucose via three reactions catalyzed by GDP-mannose-4,6-dehydratase (GMDS) [9, 10] and GDP-4-keto-6-deoxy-mannose-3,5-epimerase-4-reductase, known as FX [11]. The salvage pathway is responsible for only about 10% of the cellular pool of GDP-fucose. Most cellular GDP-fucose is synthesized via the de novo pathway. FX knockout mice show a defect of this pathway, followed by a virtually complete deficiency of cellular global fucosylation [12]. GDP-fucose synthesized in the cytosol is transported to the Golgi apparatus through GDP-Fuc Tr to serve as a substrate for fucosyltransferases [13, 14]. We have shown that GDP-fucose is a more important regulatory factor for fucosylation in HCC than fucosyltransferases. The level of GDP-fucose, and expression of the FX and GDP-Fuc Tr genes are significantly increased in HCC tissues compared with adjacent chronic hepatitic or normal liver tissues [15, 16]. We previously found that interleukin 6 secreted from pancreatic cancer cells increased the expression of several fucosylation-related genes, which resulted in the production of fucosylated haptoglobin [17, 18]. However, the details of the mechanisms by which the expression of fucosylation-related genes is regulated remain unknown.
Fucose metabolism and its regulatory factors. GDP-fucose is mainly synthesized through de novo pathway including the enzymatic reactions by GMDS and FX. GDP-fucose synthesized in cytosol is transported into the Golgi apparatus by GDP-fucose transporter. Thus, GMDS, FX, and GDP-fucose transporter are key molecules in the supply of cellular GDP-fucose to all fucosyltransferases. Finally, fucose residue is attached to acceptor oligosaccharides or proteins from GDP-fucose by each fucosyltransferase. Fucosyltransferases are divided into four groups; α1-2, α1-3/4, α1-6, and O-fucosyltransferases
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a type II transmembrane protein that belongs to the tumor necrosis factor superfamily [19]. TRAIL on immune cells including NK cells is important because it mediates host immune surveillance against tumors [20]. Binding of TRAIL to TRAIL receptors results in receptor oligomerization, and the recruitment of Fas-associated death domain (FADD) and caspase-8. The formation of this multi-protein complex, designated as the death-inducing signaling complex (DISC), allows autoactivation of the recruited caspase-8 and initiation of apoptosis. TRAIL can initiate apoptosis in a wide variety of tumor cells, but not normal ones. Thus, the TRAIL pathway is an attractive therapeutic target for the treatment of cancer. In fact, optimized soluble recombinant human TRAIL or agonistic antibodies targeting TRAIL receptors as proapoptotic antitumor therapeutic agents are currently undergoing phase 1 or 2 clinical evaluation in patients with several types of tumors [21, 22]. However, despite TRAIL’s promising anticancer activity, the early clinical trials showed that many types of tumor cells are resistant to TRAIL-based therapies [23]. To overcome the resistance capacity of many cancer cells as to single-agent treatment involving recombinant TRAIL or agonistic TRAIL receptor antibodies, attempts are being made to estimate the therapeutic efficiency of co-administration of TRAIL-targeting drugs and various conventional chemotherapeutics through preclinical or clinical experiments. In previous papers, we reported that a deficiency of fucosylation due to a GMDS mutation leads to the acquisition of resistance to TRAIL [24]. This report suggested that examination of the fucosylation levels in tumor tissues might be promising for predicting the efficiencies of TRAIL-based therapies. Moreover, the combination of TRAIL-based therapies with the agent, which can up-regulate the fucosylation level, might have a synergistic therapeutic effect.
Epigenetic alterations in DNA methylation are a hallmark of human cancer [25, 26]. This modification only accounts for a cytosine that precedes a guanosine in the DNA sequence (the CpG dinucleotide). Around half of the known human genes contain CpG-rich sites within their promoter regions (CpG islands). Several glycogenes are known to be regulated through DNA methylation. An increased level of sialyl Lewis A, which is a cancer-associated carbohydrate determinant, results from incomplete synthesis of disialyl Lewis A due to silencing of βGlcNAc: α2,6 sialyltransferase through DNA methylation [27]. Furthermore, loss of expression of Sda antigen due to DNA methylation of Sda-β1,4 N-acetylgalactosaminyltransferase is also involved in the synthesis of sialyl Lewis A in gastrointestinal cancer [28]. However, it remains unclear whether or not cellular global fucosylation is regulated through DNA methylation.
In the present study, we investigated whether or not cellular fucosylation is epigenetically regulated through DNA methylation in several types of cancer cells. Moreover, we examined the synergistic effect of a novel methyltransferase inhibitor, zebularine, on TRAIL-induced apoptosis.
Materials and methods
Cells and reagents
Human bladder cancer cell line T24, human prostate cancer cell line PC3, human breast cancer cell line MDA-MB-231, human ovarian cancer cell lines ES2, TOV21G, and KOC7C, human acute T cell leukemia cell line Jurkat, human chronic myeloid leukemia cell line K562, human histiocytic lymphoma cell line U937, human acute lymphocytic leukemia cell line Molt4, human acute myeloid leukemia cell line HL60, human hepatocellular carcinoma cell lines Hep3B, HLF and HLE, and human colon cancer cell lines HT29, HCA7, SW837 and SW620 were obtained from the American Type Culture Collection (Manassas, VA), and human choriocarcinoma cell lines JEG3, HTR8/SV40, CC1 and CC3 were kindly provided by Dr E. Yamamoto (Nagoya University). These cells were cultured in RPMI1640 (Sigma, Saint Louis, MO) supplemented with 100 U/ml penicillin (Meiji, Tokyo, Japan), 100 μg/ml streptomycin (Wako, Osaka, Japan), and 10% fetal calf serum. Human recombinant TRAIL, Zebularine and z-VAD-IETD were purchased from BIOMOL (Plymouth Meeting, PA), Wako and R&D systems (Minneapolis, MN), respectively. A stock solution of zebularine was prepared by dissolving it in DMSO and stored at −30°C until used. Rabbit anti-human cleaved poly (ADP-ribose) polymerase (PARP) polyclonal and β-actin antibodies and mouse anti-human caspase-8 antibody were purchased from Cell Signaling (Beverly, MA) and ALEXIS (Plymouth Meeting, PA), respectively. Anti-rabbit and mouse IgG antibodies were purchased from Cell Signaling and Promega (Madison, WI), respectively.
Treatment with zebularine
Adhesive cells were plated at a density of 1.125 or 2.25 × 105 cells in 6 cm dishes 24 h prior to treatment with zebularine. At 48 h after the first treatment, the medium was replaced with fresh medium containing of 100 μM zebularine. Protein lysates were prepared 24 h or 48 h later. In the case of floating cells, the cells were plated at a density of 0.9 or 1.8 × 106 cells in 6 cm dishes, and simultaneously treated with zebularine. Protein lysates were prepared 48 h or 72 h later. An equal concentration of DMSO was used as a control.
Western blotting
The cells were harvested from culture dishes. After precipitation by centrifugation at 2,000 rpm for 5 min at 4°C, they were resuspended in TNE buffer (10 mM Tris-HCl [pH 7.8], 1% NP40, 0.15 M NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA]) including a protease inhibitor cooktail (Roche, Basel, Switzerland), and then placed on ice for 30 min to allow solubilization. After samples were centrifuged at 15,000 rpm for 15 min at 4°C, the supernatants were collected. The cell lysates were quantitated using a Bicinchoninic acid kit (BCA kit, Pierce, Rockford, IL). Cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, followed by transfer to a polyvinylidine difluoride (PVDF) membrane (Millipore, Woburn, MA). After blocking with phosphate-buffered saline (PBS) containing 5% skim milk for 1 h at room temperature (RT), the membrane was incubated with diluted first antibodies overnight at 4°C. After washing of the membrane with Tris-buffered saline (TBS) (136 mM NaCl, 2.6 mM KCl, 24 mM Tris, pH 7.4) containing 0.05% Tween 20 (TBST), it was incubated with diluted second antibodies at RT. It was again washed and then developed with an enhanced chemiluminescence system (ECL kit., GE Healthcare BioSciences, Chalfont St. Giles, UK) according to the manufacturer’s protocol.
Lectin blotting
Duplicate samples were subjected to SDS-PAGE under reducing conditions. One gel was subjected to Coomassie Brilliant Blue (CBB) staining and the other was subjected to transfer to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany) for lectin blot analysis with Aleuria aurantia lectin (AAL). AAL interacts with α1-2, α1-3 , α1-4, and α1-6 fucosylated oligosaccharides. After blocking with PBS containing 3% BSA overnight at 4°C, the membrane was incubated in diluted biotinylated AAL (Seikagaku Corp., Tokyo, Japan) for 20 min at RT. It was then washed three times with TBST and incubated with diluted avidin-peroxidase conjugates (ABC kit., Vector Res. Corp., Burlingame, CA) for 20 min at RT. The development was performed using Immobilon™ Western (Millipore).
Real-time RT-PCR analysis
The cells were treated with Trizol (Invitrogen), and then collected in 2 ml microtubes. After 15 min, 200 μl of chloroform was added, followed by vortexing for 15 s. After standing at RT for 10 min, samples were centrifuged at 15,000 rpm for 15 min at 4°C. An equal amount of 2-propanol was added to each supernatant, followed by mixing well and standing for 15 min. Samples were centrifuged at 15,000 rpm at 4°C for 15 min and the pellets were washed with 0.5 ml of 80% ethanol twice. The pellets were dried and dissolved in 50 μl of nuclease-free H2O (Ambion, Austin, TX). The concentrations of all RNA samples were determined spectrophotometrically and the samples were stored at −80°C until used.
Using a PrimeScript® RT reagent kit (TAKARA BIO, Shiga, Japan), each RNA in 1×PrimeScript® Buffer, Prime Script® RT Enzyme Mix I, 25 pmol Oligo dT Primer, and 50 pmol Random 6 mers was adjusted to a volume of 10 μl with RNase-free H2O according to the manufacturer’s protocol. The reverse-transcription reaction was performed for 15 min at 37°C and terminated for 5 s at 85°C. Each synthesized cDNA was diluted ten-fold. Two μl of each diluted cDNA was adjusted to a 20 μl solution containing 1×SYBR Premix Ex Taq II and 0.4 μM forward and reverse primers. Real time PCR analysis was carried out using a Chromo4 Real-Time PCR Detector (BioRad Laboratories, Tokyo, Japan). After initial polymerase activation and a denaturation step of 10 s at 95°C, the samples underwent 40 amplification cycles, each comprising 5 s at 95°C and 20 s at 66°C. Fluorescence was measured at the end of each cycle to monitor the progress of amplification. A melting curve was recorded by heating slowly at 0.5°C/sec from 66°C to 95°C and holding for 3 s. Fluorescence was measured continuously during the slow temperature increase to monitor the dissociation of synthesized double strand PCR products. The primer sequences for the genes used in this study are summarized in Table 1. Hypoxanthine guanine phosphribosyl transferase (HPRT) was used as an internal control. The results were normalized as relative values using HPRT.
Measurement of caspase-8 activation
Caspase-8 activity was measured using a FLICE/Caspase-8 Colorimetric Assay Kit (MBL, Nagoya, Japan). Briefly, the cells were harvested from culture dishes. After precipitation by centrifugation at 2,000 rpm for 5 min at 4°C, they were resuspended in cell lysis buffer, and then placed on ice for 10 min to allow solubilization. Samples were then centrifuged at 15,000 rpm for 1 min at 4°C, the supernatants being collected. The cell lysates were quantitated using a BCA kit. Equal amounts of protein extracts were loaded onto a 96-well microplate, followed by incubation at 37°C for 3 h with 2×Reaction buffer, 1 M DTT, and caspase-8 substrate (Ac-IETD-pNA). Free pNA, which is a caspase-8-dependent cleaved product, was calorimetrically quantified using a microplate reader at 405 nm.
FACS analysis
To determine the sub-G1 DNA content, cells were treated with various concentrations of recombinant TRAIL for 24 h after treatment with zebularine. Both adherent and floating cells were collected by centrifugation at 2,000 rpm for 5 min at 4°C, and then washed twice with PBS. Fixation was performed in ice-cold 50% ethanol for 40 min. After centrifugation at 2,000 rpm for 5 min at 4°C, the pellets were resuspended in 200 μl of 1 mg/ml RNase/PBS and warmed at 37°C for 20 min to degrade cellular RNA. Then, 100 μg/ml propidium iodide (Sigma)/PBS was added and cellular DNA was stained for 10 min at 4°C. The cells were sorted according to their DNA content with FACS Calibur operated with CellQuest software and analyzed with Modfit software (Becton Dickinson, San Diego, CA). Cell debris and fixation artifacts were gated out. The cells containing subG1 DNA were calculated as a percentage of the total cells. The results are represented as the means of three experiments.
The cells treated with zebularine were detached from culture dishes using 1.0 mM EDTA/PBS. The harvested cells were incubated with mouse anti-human DR4 or DR5 monoclonal antibodies (R&D, Minneapolis, MN) diluted with 0.1% bovine serum albumin (BSA)/PBS for 20 min at 4°C. After the cells had been washed with 0.1% BSA/PBS, they were incubated with Alexa fluor 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR) for 20 min at 4°C. Then the cells were washed. Flow cytometric analysis was performed with FACS Calibur.
Results
Fucosylation is regulated through DNA methylation in many cancer cells
It is a well-known fact that regulation of cellular fucosylation differs among the many types of cancer cell lines. To determine the involvement of DNA methylation in the regulation of cellular fucosylation, we searched for cancer cell lines with relatively low fucosylation levels. When we performed lectin blotting with AAL, which binds to fucosylated oligosaccharides, using many cancer cell lines, 19 cancer cell lines other than U937, HLE, and HLF were selected as ones with relatively low fucosylation levels (Fig. 2a). To examine the possibility that DNA methylation contributes to the low fucosylation levels in these cancer cell lines, each cancer cell line was treated with a DNA methyltransferase inhibitor, zebularine (Fig. 2b and c). As shown in Fig. 2b, the fucosylation levels in MDA-MB-231, PC3, T24, Hep3B, TOV21G, JEG3, HCA7, and HL60 cells were increased in a concentration-dependent manner after the treatment with zebularine for 72 h. The treatment with zebularine for 96 h resulted in more significant increases of the fucosylation levels in HT29, K562, Molt4, and Jurcat cells (Fig. 2c). Other data from zebularine treatment for 96 h are shown in supplementary figure 1. Other cell lines, ES2, SW620, SW834, CC1, CC3, and KOC7C, showed no change of fucosylation in several conditions of zebularine treatment. These results indicated that cellular fucosylation is decreased through the DNA methylation in many cancer cell lines.
Alteration of the fucosylation level on treatment with zebularine in several cancer cell lines. a Lectin blotting involving AAL showed the expression patterns of fucosylated glycoproteins in 22 cancer cell lines. PC: positive control, NC: negative control. b, c When 19 cell lines were treated with a methyltransferase inhibitor, zebularine, for 72 (b) or 96 h (c), changes of cellular fucosylation were examined by AAL letin blotting. The described number (0, 50, 100) indicates the concentration of zebularine. Treatment with 100 μM zebuarine for 72 h lead to an excessive cell death in ES2 and HTR8/SV40 cells. Thus, proteins from these cells couldn’t be prepared. Upper and lower panel showed the results from Coomassie brilliant blue (CBB) staining and AAL letin blotting, respectively. CBB staining indicated that equal amounts of protein were loaded in the lanes
The expression levels of several kinds of fucosylation-related genes are regulated through DNA methylation
Cellular fucosylation is regulated in a cell type-specific manner in which many fucosylation-related genes might be involved (Fig. 1). To determine whether fucosylation-related genes were induced by the treatment with zebularine, we performed real-time PCR analyses. As shown in Fig. 3, the expression of the FX, GDP-Fuc Tr, and Fut4 genes was significantly increased in MDA-MB-231 cells after the treatment with zebularine. The expression of these three genes was also increased in K562 cells after the treatment with zebularine (Supplementary figure 2). These results indicate that the expression of several fucosylation-related genes is regulated through DNA methylation in cancer cell lines. Moreover, the enhancement of expression of the FX, GDP-Fuc Tr, and Fut4 genes on treatment with zebularine might be responsible for the increase in cellular fucosylation in MDA-MB-231.
Alteration of expression of fucosylation-related genes after treatment with zebularine. Real-time RT-PCR analyses of the GMDS, FX, GDP-Fuc Tr, FUT3, FUT4, FUT5, FUT6, FUT7, and FUT8 genes were performed using total RNA prepared from MDA-MB-231 cells treated with zebularine at indicated concentration. The results are represented as the means of three experiments. Bars indicate SD
Zebularine treatment results in sensitization to TRAIL-induced cell death
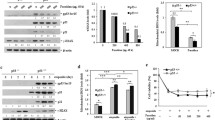
The remodeling of cell surface glycoproteins, such as growth factor receptors and adhesion molecules, through fucosylation is associated with their biological activities. We previously reported that a deficiency of cellular fucosylation due to mutation of the GMDS gene resulted in resistance to TRAIL-induced apoptosis. We investigated whether or not DNA demethylation on treatment with zebularine leads to greater susceptibility to TRAIL-induced apoptosis. When MDA-MB-231 cells were pretreated with zebularine, the expression of cleaved PARP, which serves as a marker of cells undergoing apoptosis, was significantly increased after additional treatment with recombinant TRAIL (Fig. 4a). Furthermore, pretreatment with zebularine resulted in a marked increase in the subG1 population, which is a hallmark of cell death, in TRAIL-treated MDA-MB-231 cells (Fig. 4b). These results indicate that zebularine sensitized MDA-MB-231 cells to TRAIL-induced apoptosis. In addition, the significant increase of TRAIL-induced expression of cleaved PARP by pretreatment with zebularine was also observed in HL60, PC3, and T24 cells (Fig. 4c). In HT29 cells, we slightly observed the synergistic effect of zebularine on TRAIL-induced apoptosis. Fucosylation level in these cells was significantly increased by zebularine treatment (Fig. 2b). However, this phenomenon was not observed in SW620, SW834, and KOC7C cells in which fucosylation didn’t change by treatment with zebularine. These results suggest the involvement of fucosylation in the synergistic effect of zebularine on TRAIL-induced apoptosis.
The effect of zebularine on the susceptibility to recombinant TRAIL. a After treatment with 100 μM zebularine for 72 h followed by treatment with the presented concentration of TRAIL for 2.5 h, the expression level of cleaved PARP were examined. Western blotting of β-actin indicated that equal amounts of protein were loaded in each lane. b The population of subG1 cells after treatment with 100 μM zebularine for 72 h followed by treatment with the presented concentration of TRAIL for 24 h was determined with a flow cytometer. Caspase-8 activity and SubG1 population data are represented as the ratio to those of the cells untreated with TRAIL. Bars indicate SD. c After HL60, PC3, T24, HT29, SW620, SW834, and KOC7C cells were pretreated with zebularine and then treated with TRAIL, expression of cleaved PARP was examined by Western blotting
Zebularine affected TRAIL-induced apoptosis at the upstream of caspase-8 in TRAIL signaling pathway
To demonstrate how zebularine affected TRAIL-induced apoptosis, the activation of caspase-8, which is the most upstream molecule in TRAIL signaling pathway, by treatment with zebularine and TRAIL was examined. As shown in Fig. 5a, TRAIL-induced activation of caspase-8 was increased by pretreatment with zebularine. In addition, when MDA-MB-231 cells were pretreated with a caspase-8 inhibitor, z-VAD-IETD, increased expression of cleaved PARP by treatment with zebularine and TRAIL was suppressed (Fig. 5b). These results indicate that zebularine affected TRAIL-induced apoptosis at the upstream of caspase-8 in TRAIL signaling pathway, and that the increase of caspase-8 activity contributed to synergistic effect of zebularine on TRAIL-induced apoptosis.
Zebularine affected TRAIL-induced apoptosis at the upstream of caspase-8 in TRAIL signaling pathway. a After treatment with 100 μM zebularine for 72 h followed by treatment with the presented concentration of TRAIL for 2.5 h, caspase-8 activity was examined. b After treatment with 50 μM caspase-8 inhibitor for 1 h, MDA-MB-231 cells were treated with TRAIL in the presence 25 μM the inhibitor for 2.5 h. Prepared protein lysates were analyzed by Western blotting using anti-cleaved PARP and β-actin antibodies
Synergistic effect of zebularine on TRAIL-induced apoptosis is independent of expression of TRAIL receptors and caspase-8
Although treatment with zeularine is considered to lead to the DNA demethylation of many genes and their re-expression, the expression level of caspase-8 was not affected by the treatment in MDA-MB-231 cells (Fig. 6a). In contrast, the treatment with zebularine increased the expression of caspase-8 in HCA7 cells, indicating that zebularine had the ability to increase the expression of caspase-8 in cells other than MDA-MB-231 ones. The increase in caspase-8 activity on the addition of TRAIL in MDA-MB-231 cells pretreated with zebularine indicated that zebularine affected factors upstream of caspase-8 in the TRAIL-induced apoptotic signaling cascade. Next, the expression of proapoptotic TRAIL receptors, DR4 and DR5, on the cell surface was investigated by flow cytometry. The expression levels of both receptors were similar in MDA-MB-231 cells treated with and without zebularine (Fig. 6b). These results indicated that the expression levels of caspase-8 and TRAIL receptors were not involved in the increases in apoptosis induced by co-treatment with zebularine and TRAIL in MDA-MB-231 cells.
The expression of DR4, DR5 and caspase-8 did not change on treatment with zebularine. a, b At 72 h after treatment with 100 μM zebularine in MDA-MB-231 cells, the expression of caspase-8 and TRAIL receptors was determined by Western blotting and flow cytometry, respectively. The solid or dotted line indicates the cells treated with or without zabularine, respectively. Negative control staining indicated with gray line was done without each antibody
Discussion
Abnormal hypermethylation of the promoters of numerous tumor suppressor genes or cancer-related genes is commonly found in primary neoplasms and various cancer cell lines. Because epigenetic processes are potentially reversible in contrast to genetic mutations, inhibitors of DNA methylation have been developed for cancer therapy. 5-Azacytidine and its deoxy analog, 5-aza-2′-deoxycitidine, are two of the most well-known DNA methylation inhibitors. However, both drugs are quite toxic in vitro and in vivo, and they are unstable in aqueous solution, making them difficult to administer both experimentally and clinically. Under these circumstances, zebularine was developed as a novel DNA methylation inhibitor. Due to its great stability in acidic and neutral solutions, and minimal toxicity both in vitro and in vivo, zebularine is expected to become a more effective drug for cancer [29].
Oligosaccharide structures dramatically change through a genetic or epigenetic mechanism during carcinogenesis. In this study, we demonstrated that cellular fucosylation was regulated through DNA methylation in many cancer cell lines. Among many kinds of fucosylation-related genes, expression of the FX, GDP-Fuc Tr, and Fut4 genes was increased after the treatment with zebularine in MDA-MB-231 cell lines. Although it remains unknown whether the promoter regions of the FX, GDP-Fuc Tr, and Fut4 genes can be directly methylated, databases for searches for CpG islands (MethPrimer: http://www.urogene.org/methprimer/index1.html and CpG Island Searcher: http://cpgislands.usc.edu/) and Methyl Primer Express® Software v1.0 (Applied Biosystems, Foster City, CA) showed the existence of several CpG islands in 5′-upstream sequence of these genes. When methylation status of one of candidates of CpG island in 5′-upstream of genomic region coding mRNA of FX (NM_003313) or GDP-Fuc Tr (NM_018389) was preliminary examined by methylation-specific PCR analysis, hypermethylation of these regions were not observed (Supplementary figure 4). However, in order to confirm whether expression of these genes were directly regulated by DNA methylation, further investigations, such as identification of correct promoter region of these genes and examination of other CpG islands, would be required. In addition, methylation status of the Fut4 gene also needs to be analyzed in detail.
DR4 and caspase-8 were previously reported to be frequently inactivated through epigenetic silencing in many cancer cells resistant to TRAIL-induced apoptosis. Treatment with 5-aza-2′-deoxycitidine reversed hypermethylation of the DR4 [30–32] and caspase-8 [33–37] genes, resulting in restoration of these molecules and activation of TRAIL-induced apoptosis. However, no alteration of the expression of the DR4, DR5, and caspase-8 genes was observed in MDA-MB-231 cells treated with or without zebularine. The difference in effects of methyltransferase inhibitors on the expression of the DR4 and caspase-8 genes might be caused by the substantial diversity of the molecular activities of the inhibitors [38] or cell type specificity.
As shown in Fig. 5b, z-VAD-IETD suppressed synergistic effect of zebularine on TRAIL-induced apoptosis. On the other hand, it couldn’t affect zebularine-induced apoptosis, indicating that zebularine induced apotposis in caspase-8 independent manner. This result suggested that zebularine induced apoptosis through the activation of intrinsic pathway as well as its synergistic effect on TRAIL-induced extrinsic pathway.
Among many kinds of apoptosis-related molecules, including caspases and Bcl-2 family members, involved in the TRAIL-induced apoptotic signaling cascade, caspase-8 is one of the most upstream molecules. In our previous experiments, the restoration of fucosylation in a fucosylation-deficient colon cancer cell line, HCT116, resulted in a significant increase in caspase-8 activation when the cells were treated with TRAIL (unpublished data). While the mechanism by which fucosylation regulates TRAIL-induced activation of caspase-8 remains unknown, the increase in caspase-8 activity on treatment with TRAIL in MDA-MB-231 cells pretreated with zebularine might be dependent on an increase in cellular fucosylation. In addition, Fig. 4c suggests the involvement of fucosylation in synergistic effect of zebularine on TRAIL-induced apoptosis. To confirm direct involvement of fucosylation, we established FX-, GDP-fuc Tr-, and its double-knockdown MDA-MB-231 cells. However, increased fucosylation by treatment with zebularine was not decreased in these knockdown cells (Supplementary figure 3). The remained activities might be enough to maintain cellular fucosylation or Fut4 might also play an important role in increased fucosylation by treatment with zebularine in MDA-MB-231 cells.
In our recent immunohistochemical analysis, impaired cellular fucosylation was observed in certain kinds of colon cancer tissues (manuscript in preparation). These cancers might escape from NK cell-mediated tumor surveillance through the acquisition of resistance to TRAIL, followed by tumor progression and metastasis [21]. Although various mechanisms by which fucosylation is decreased in these cancer tissues have been considered, the present study suggests that DNA hypermethylation of fucosylation-related genes leads to impaired fucosylation, followed by further development of cancer. Further studies should be performed to determine the correlation between cellular fucosylation and the DNA methylation status of fucosylation-related genes in many human cancer tissues.
Abbreviations
- TRAIL:
-
Tumor necrosis factor-related apoptosis-inducing ligand
- AAL:
-
Aleuria aurantia lectin
- GMDS:
-
GDP-mannose-4,6-dehydratase
- AFP:
-
alpha-fetoprotein
- GDP-fucose:
-
guanosin 5′-diphosphate-fucose
- GDP-Fuc Tr:
-
GDP-fucose transporter
References
Ohtsubo, K., Marth, J.D.: Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006)
Haltiwanger, R.S., Lowe, J.B.: Role of glycosylation in development. Annu. Rev. Biochem. 73, 491–537 (2004)
Hakomori, S.: Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv. Cancer Res. 52, 257–331 (1989)
Miyoshi, E., Moriwaki, K., Nakagawa, T.: Biological function of fucosylation in cancer biology. J. Biochem. 143, 725–729 (2008)
Alpert, M.E., Uriel, J., de Nechaud, B.: Alpha-1 fetoglobulin in the diagnosis of human hepatoma. N. Engl. J. Med. 278, 984–986 (1968)
Aoyagi, Y., Isemura, M., Suzuki, Y., Sekine, C., Soga, K., Ozaki, T., Ichida, F.: Fucosylated alpha-fetoprotein as marker of early hepatocellular carcinoma. Lancet 2, 1353–1354 (1985)
Aoyagi, Y., Isemura, M., Suzuki, Y., Sekine, C., Soga, K., Ozaki, T., Ichida, F.: Change in fucosylation of alpha-fetoprotein on malignant transformation of liver cells. Lancet 1, 210 (1986)
Food and Drug Administration, HHS: Medical devices; immunology and microbiology devices; classification of AFP-L3% immunological test systems. Final rule. Fed. Regist. 70, 57748–57750 (2005)
Ohyama, C., Smith, P.L., Angata, K., Fukuda, M.N., Lowe, J.B., Fukuda, M.: Molecular cloning and expression of GDP-D-mannose-4, 6-dehydratase, a key enzyme for fucose metabolism defective in Lec13 cells. J. Biol. Chem. 273, 14582–14587 (1998)
Sullivan, F.X., Kumar, R., Kriz, R., Stahl, M., Xu, G.Y., Rouse, J., Chang, X.J., Boodhoo, A., Potvin, B., Cumming, D.A.: Molecular cloning of human GDP-mannose 4, 6-dehydratase and reconstitution of GDP-fucose biosynthesis in vitro. J. Biol. Chem. 273, 8193–8202 (1998)
Tonetti, M., Sturla, L., Bisso, A., Benatti, U., De Flora, A.: Synthesis of GDP-L-fucose by the human FX protein. J. Biol. Chem. 271, 27274–27279 (1996)
Smith, P.L., Myers, J.T., Rogers, C.E., Zhou, L., Petryniak, B., Becker, D.J., Homeister, J.W., Lowe, J.B.: Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus. J. Cell Biol. 158, 801–815 (2002)
Lübke, T., Marquardt, T., Etzioni, A., Hartmann, E., von Figura, K., Körner, C.: Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 28, 73–76 (2001)
Lühn, K., Wild, M.K., Eckhardt, M., Gerardy-Schahn, R., Vestweber, D.: The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 28, 69–72 (2001)
Noda, K., Miyoshi, E., Gu, J., Gao, C.X., Nakahara, S., Kitada, T., Honke, K., Suzuki, K., Yoshihara, H., Yoshikawa, K., Kawano, K., Tonetti, M., Kasahara, A., Hori, M., Hayashi, N., Taniguchi, N.: Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 63, 6282–6289 (2003)
Moriwaki, K., Noda, K., Nakagawa, T., Asahi, M., Yoshihara, H., Taniguchi, N., Hayashi, N., Miyoshi, E.: A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology 17, 1311–1320 (2007)
Okuyama, N., Ide, Y., Nakano, M., Nakagawa, T., Yamanaka, K., Moriwaki, K., Murata, K., Ohigashi, H., Yokoyama, S., Eguchi, H., Ishikawa, O., Ito, T., Kato, M., Kasahara, A., Kawano, S., Gu, J., Taniguchi, N., Miyoshi, E.: Fucosylated haptoglobin is a novel marker for pancreatic cancer: a detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int. J. Cancer 118, 2803–2808 (2006)
Narisada, M., Kawamoto, S., Kuwamoto, K., Moriwaki, K., Nakagawa, T., Matsumoto, H., Asahi, M., Koyama, N., Miyoshi, E.: Identification of an inducible factor secreted by pancreatic cancer cell lines that stimulates the production of fucosylated haptoglobin in hepatoma cells. Biochem. Biophys. Res. Commun. 377, 792–796 (2008)
Wiley, S.R., Schooley, K., Smolak, P.J., Din, W.S., Huang, C.P., Nicholl, J.K., Sutherland, G.R., Smith, T.D., Rauch, C., Smith, C.A., et al.: Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3, 673–682 (1995)
Takeda, K., Hayakawa, Y., Smyth, M.J., Kayagaki, N., Yamaguchi, N., Kakuta, S., Iwakura, Y., Yagita, H., Okumura, K.: Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 7, 94–100 (2001)
Ashkenazi, A., Herbst, R.S.: To kill a tumor cell: the potential of proapoptotic receptor agonists. J. Clin. Invest. 118, 1979–1990 (2008)
Ashkenazi, A.: Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat. Rev. Drug Discov. 7, 1001–1012 (2008)
Johnstone, R.W., Frew, A.J., Smyth, M.J.: The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer 8, 782–798 (2008)
Moriwaki, K., Noda, K., Furukawa, Y., Ohshima, K., Uchiyama, A., Nakagawa, T., Taniguchi, N., Daigo, Y., Nakamura, Y., Hayashi, N., Miyoshi, E.: Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology 137, 188–198 (2009)
Herman, J.G., Baylin, S.B.: Gene silencing in cancer in association with promoter hypermethylation. N Engl. J. Med. 349, 2042–2054 (2003)
Jones, P.A., Baylin, S.B.: The epigenomics of cancer. Cell 128, 683–692 (2007)
Kannagi, R., Yin, J., Miyazaki, K., Izawa, M.: Current relevance of incomplete synthesis and neo-synthesis for cancer-associated alteration of carbohydrate determinants–Hakomori’s concepts revisited. Biochim. Biophys. Acta 1780, 525–531 (2008)
Kawamura, Y.I., Toyota, M., Kawashima, R., Hagiwara, T., Suzuki, H., Imai, K., Shinomura, Y., Tokino, T., Kannagi, R., Dohi, T.: DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 135, 142–151 (2008)
Cheng, J.C., Yoo, C.B., Weisenberger, D.J., Chuang, J., Wozniak, C., Liang, G., Marquez, V.E., Greer, S., Orntoft, T.F., Thykjaer, T., Jones, P.A.: Preferential response of cancer cells to zebularine. Cancer Cell 6, 151–158 (2004)
Elias, A., Siegelin, M.D., Steinmüller, A., von Deimling, A., Lass, U., Korn, B., Mueller, W.: Epigenetic silencing of death receptor 4 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in gliomas. Clin. Cancer Res. 15, 5457–5465 (2009)
Bae, S.I., Cheriyath, V., Jacobs, B.S., Reu, F.J., Borden, E.C.: Reversal of methylation silencing of Apo2L/TRAIL receptor 1 (DR4) expression overcomes resistance of SK-MEL-3 and SK-MEL-28 melanoma cells to interferons (IFNs) or Apo2L/TRAIL. Oncogene 27, 490–498 (2008)
Horak, P., Pils, D., Haller, G., Pribill, I., Roessler, M., Tomek, S., Horvat, R., Zeillinger, R., Zielinski, C., Krainer, M.: Contribution of epigenetic silencing of tumor necrosis factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL resistance and ovarian cancer. Mol. Cancer Res. 3, 335–343 (2005)
Fulda, S., Debatin, K.M.: 5-Aza-2′-deoxycytidine and IFN-gamma cooperate to sensitize for TRAIL-induced apoptosis by upregulating caspase-8. Oncogene 25, 5125–5133 (2006)
Hopkins-Donaldson, S., Ziegler, A., Kurtz, S., Bigosch, C., Kandioler, D., Ludwig, C., Zangemeister-Wittke, U., Stahel, R.: Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 10, 356–364 (2003)
Fulda, S., Küfer, M.U., Meyer, E., van Valen, F., Dockhorn-Dworniczak, B., Debatin, K.M.: Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene 20, 5865–5877 (2001)
Eggert, A., Grotzer, M.A., Zuzak, T.J., Wiewrodt, B.R., Ho, R., Ikegaki, N., Brodeur, G.M.: Resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Cancer Res. 61, 1314–1319 (2001)
Grotzer, M.A., Eggert, A., Zuzak, T.J., Janss, A.J., Marwaha, S., Wiewrodt, B.R., Ikegaki, N., Brodeur, G.M., Phillips, P.C.: Resistance to TRAIL-induced apoptosis in primitive neuroectodermal brain tumor cells correlates with a loss of caspase-8 expression. Oncogene 19, 4604–4610 (2000)
Stresemann, C., Brueckner, B., Musch, T., Stopper, H., Lyko, F.: Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 66, 2794–2800 (2006)
Acknowledgments
This study was performed by a grant from the New Energy and Industrial Technology Development Organization (NEDO) as a part of the developing technology project on implementing sugar chain functions in Japan, a Grant-in-Aid for Scientific Research (A), No. 21249038, from the Japan Society for the Promotion of Science, a Grant-in-Aid for Cancer Research and Scientific Research on Priority Areas, No. 20014011, from the Ministry of Education, Science, and the Global COE program of Osaka University funded by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 855 kb)
Rights and permissions
About this article
Cite this article
Moriwaki, K., Narisada, M., Imai, T. et al. The effect of epigenetic regulation of fucosylation on TRAIL-induced apoptosis. Glycoconj J 27, 649–659 (2010). https://doi.org/10.1007/s10719-010-9310-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-010-9310-5