Abstract
Family 18 chitinases hydrolyze chitin through a substrate-assisted catalytic mechanism and are to a variable extent able to catalyze transglycosylation reactions. Previously Aspergillus fumigatus AfChiB1 was found to be able to catalyze transglycosylation reactions. Structural analysis reveals that AfChiB1 consists of an eight-stranded β/α-barrel. Like other members of the family 18 hydrolases, AfChiB1 has conserved substrate binding site and catalytic acid, while a suitable nucleophile is missing. In this study, Trp137, Asp246, and Met243, which are close to the glycosidic cleavage site, were mutated to glutamate individually. As a result, the W137E remained its hydrolytic activity and was completely devoid of transglycosyl activity, while the D246E reduced its chitinolytic activity and increased its transglycosyl activity. And the M243E showed a remarkable reduction of chitinolytic activity and complete loss of transglycosyl activity. These results suggested that the transglycosyl reaction catalyzed by the AfChiB1 is due to lacking of nucleophile.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Family 18 chitinases (CAZY GH 18 at http://afmb.cnrs-mrs.fr/CAZY) hydrolyze chitin, a linear polymer of (β1–4) linked N-acetylglucosamine (NAG). They are found in organisms ranging from bacteria to mammals [4, 6, 7–9, 12, 16–17, 25, 27], where they have diverse functions ranging from accessing chitin as an energy resource to pathogen defense [6–8]. Inhibitors of these enzymes are of considerable interest as research tools, but also may have chemotherapeutic potential against pathogenic fungi, insects, and nematodes. Due to their important roles, chitinases have been studied extensively.
It has been described that family 18 chitinases are to a variable extent able to catalyse transglycosyl reactions [13, 23]. Nowadays, family 18 chitinases are proposed to function via a retaining mechanism, which involves a catalytic acid and a nucleophile [1]. However, a suitable nucleophile is missing in these proteins. Based on the crystal structure of hevamine, a substrate-assisted catalytic mechanism was proposed [19]. With the accumulation of structural information, the catalytic mechanism for these glycoside hydrolases has been well established [14, 19–22, 24]. The family 18 chitinases employ an unusual reaction mechanism, in which the acid protonating the glycosidic bond is a conserved glutamate and the nucleophile is the oxygen of the N-acetyl group on the -1 sugar (the sugar on the non-reducing end of the glycosidic bond), forming an oxazolinium ion intermediate. However, little effort has been made to probe the mechanism of transglycosyl activity associated to the family 18 chitinases.
The opportunistic fungal pathogen Aspergillus fumigatus is known to produce a wide range of chitinolytic enzymes [5, 29]. One of its secreted chitinases, AfChiB1 consists of 395 amino acids and exhibits a high homology with other members of the family 18 glycosyl hydrolases [28]. Previously, a direct evidence has been obtained to show that (NAG)2 can be transferred to the non-reducing end of PNP-(NAG)2 during hydrolysis of PNP-(NAG)2 [29]. Based on the resolved structure of the AfChiB1 (PDB: molecule 1WNO and 1W9P), Glu177 is identified as a catalytic acid [10, 15]. However, like other members of family 18 chitinases, a suitable nucleophile is absent in its catalytic center, suggesting a substrate-assisted catalytic mechanism. In this study, we introduced a glutamate residue into the site spatially closed to Glu177 or cleavage site. Our results showed that replacement of Trp137 with glutamate could completely remove transglycosyl activity associated to AfChiB1, while other mutations displayed an altered binding affinity, hydrolytic/transglycosyl activity and stability.
2 Methods
2.1 Chemicals and reagents
Colloidal chitin was prepared according to Vyas and Deshpande [26]. Restriction enzymes, Taq DNA polymerase, and T4 DNA ligase were from Takara Biotech. Co. Ltd. (Dalian, China). IPTG, X-gal, DEPC, and glycol chitosan were from Sigma Chemical Co. (St. Louise, USA). All other chemicals were from commercial sources and of the highest quality. NAG, (NAG)2, (NAG)3, (NAG)4, (NAG)5, (NAG)6, PNP-(NAG)2, and PNP-NAG were from Northstar BioProducts™ Associates of Cape Cod, Inc.
2.2 Microorganism and plasmids
Aspergillus fumigatus YJ-407 isolated from soil (China General Microbiological Culture Collection Center, CGMCC 0386) was maintained on potato glucose (2%) agar slant. E. coli strain DH5α was from Bethesda Research Laboratories. E. coli strain BL21(DE3) pLysS and expression vector pET-15b were from Novagen.
2.3 Expression and purification of chitinases in E. coli
The PCR was carried out using pGEM-chi as template [28]. The primers used were: PE1, 5′-CTCCATGGCTAGCTCCGGTTATCGCT-3′; and PE2, 5′-GCGGATCCTTAGGTTTGCATGCCATTGCGC-3′, and the sites of NcoI and BamHI was introduced to 5’ terminal of PE1 and PE2, respectively. The PCR product was digested with NcoI and BamHI and subsequently cloned into the sites between NcoI and BamHI of pET-15b. The resulting plasmid was confirmed by DNA sequencing and designated as pET-chi.
The site-directed mutagenesis was carried out using GeneTailor™ (Invitrogen) by following the manufacturer’s instruction. The PCR was preformed with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) for 20 cycles (94°C for 2 min, 94°C for 30 s, 56°C for 1 min, 68°C for 7 min) and further incubated at 68°C for 10 min. pET-chi was used template. The primer pairs used for generating the D246E were 5′-TGGAATCTCATGGCCTACGAATATGCTGGCA-3′ and 5′-TCGTAGGCCATGAGATTCCAGAAGTCCA AT-3′, for the M243E were 5′-AATTGGACTTCTGGAATCTCGAAGCCTACGATTA-3′ and 5′-GAGATTC CAGAAGTCCAATTGCTGATCCAT-3′, and for the W137E were 5′-TTCTCTTGTCCATTGGCGGGGAA ACCTACTCTCC-3′ and 5′-CCCGC CAATGGACAAGAGAACCTTGAGGTT-3′, respectively.
E. coli BL21(DE3)pLysS strain harboring recombinant plasmid was inoculated in LB containing ampicillin and incubated at 37°C. When the cell density reached 0.5 (OD600), IPTG was added to a final concentration of 1 mM. After induction at 37°C for 3 h, the cells were harvested and resuspended in 0.2 M sodium acetate buffer (pH 5.0) and then disrupted by sonication. The cellular debris was removed by centrifugation. The soluble protein in supernatant was precipitated by the addition of solid ammonium sulfate. The precipitated proteins of 30–60%s were collected and dissolved in 10 ml of 50 mM Tris–HCl (pH 8.0). After dialysis against 50 mM Tris–HCl buffer (pH 8.0), the enzyme solution was applied to a DEAE-Sephadex Fast Flow column pre-equilibrated with 50 mM Tris–HCl buffer (pH 8.0). The enzyme fractions eluted between 0.10–0.15 M of NaCl were pooled, dialyzed against distilled water and lyophilized. All purification steps were carried out with AKTÄ FPLC (Amersham Pharmacia Biotech) at 4°C.
2.4 Enzyme and protein assay
The enzyme reaction was initiated by the addition of the chitinase solution to the assay mixture containing 0.4 ml 0.5% colloidal chitin and 50 mM sodium acetate buffer, pH 5.0 in a total volume of 1 ml. After incubation at 60°C for 15 min, the reaction mixture was heated in a boiling water bath for 3 min. The liberated reducing sugar was measured by Schales and Schales’ procedure [18] modified by Imoto and Yagishita [11]. One unit of chitinase activity is defined as the amount that produces the reducing sugars corresponding to 1 μmol of NAG per min under the standard assay condition. The protein was measured by using the method described by Bradford [3]. BSA was used as standard.
2.5 Enzymatic hydrolysis of PNP-(NAG)2
For analyzing the products released from PNP-(NAG)2, the reaction mixture contained 8 nmol of substrate and 1 μg of purified chitinase in 20 μl of 50 mM sodium acetate buffer, pH 5.0. After incubating at 37°C for predetermined time, the reaction mixture was heated at 100°C for 3 min to stop the reaction. After centrifugation, 10 μl of supernatant were analyzed with HPAEC-PAD using PA-1 column and the products were eluted with 18 mM NaOH (1 ml/min). Standard PNP-(NAG)2, PNP-NAG, (NAG)2, and NAG were used to calibrate the retention time.
2.6 Enzymatic hydrolysis of chitooligosaccharide
For analysing the products released from chitooligosaccharides, the reaction mixture contained 5 μg chitooligosaccharides and 1 μg purified chitinase in 15 μl 50 mm sodium acetate buffer pH 5.0. After incubating at 37°C for predetermined time, the reaction mixture was heated at 100°C for 3 min to stop the reaction. The entire incubation mixture was applied onto a TLC plate. The plate was developed by n-butanol: acetic acid: water (2:1:1, v/v/v).
2.7 Computer analysis of protein
Analysis of 3-D structure of the proteins was carried out using DeepView/ Swiss-PdbViewer (Version 3.7) (http://www.expasy.org/spdbv/).
3 Results
3.1 Construction of the mutant enzymes
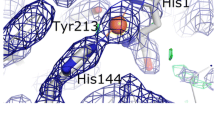
As revealed by crystal structure of the AfChiB1 [10, 15], the AfChiB1 contains a (βα)8 fold. Asp175 and Glu177 form part of the family 18 chitinase DxE motif (http://afmb.cnrs-mrs.fr/CAZY), with Glu177 as the catalytic acid. The two catalytic residues and the conserved Asp173 constitute a proton transfer system, which promotes protonation of the glycosidic oxygen in the substrate during the hydrolysis process (Fig. 1a). Asp175, Glu177, Tyr245, Asp246, and Arg301 make direct hydrogen bonds to the substrate, while Trp137, Ala217, Met243, and Phe251 are involved in hydrophobic contacts with the substrate/reaction intermediate by stacking with the substrate. Among these residues crucial for substrate binding and catalysis, Asp-246 is on the opposite side of the active site Glu177, while Met-243 and Trp-137 are close to the catalytic site (Fig. 1b). To introduce a glutamate residue near the cleavage site, we constructed the mutant enzyme M243E, W137E and D246E as described in Methods.
Stereo structure of the catalytic site of the AfChiB1. Docking model of the AfChiB1 in complex with (NAG)3 (a) and stereo figure of the AfChiB1 (PDB molecule: 1WNO) active site (b) are shown. Glu177, Asp175 and Asp173 constitute a proton transfer system, which promotes protonation of the glycosidic oxygen in the substrate during the hydrolysis process. The substrate and side chain of amino acids are shown as sticks. The backbone is shown as a ribbon drawing
As shown in Fig. 2, the mutant enzymes W137E, M243E, and D246E were expressed in E. coli and purified to homogeneity as described in the “Methods” section. The specific activities of purified wild-type, D246E, M243E, and W137E for swollen chitin were 11.01, 7.94, 6.19, and 6.94 U/mg, respectively (Table 1), suggesting a reduction of chitinolyitc activity of the mutant enzymes.
Expression and purification of the mutant enzymes. The wild-type and mutant enzymes were induced (a) and purified (b) as described in the “Methods” section. Gel, 12%, was used to separate the proteins by SDS-PAGE
3.2 Properties of the mutant enzymes
As summarized in Table 1, the mutant enzymes showed a reduction of hydrolytic rate toward swollen chitin, and the Km values of the wild-type, D246E, M243E, and W137E were determined as 0.44, 0.20, 0.30, and 0.27 mg/ml, respectively, suggesting an increase of binding affinity of the mutant enzymes. Since previously we showed that AfChiB1 could degrade chitosan [29], we also tested the binding affinity of the mutant enzymes with chitosan. The K m values of the wild-type, D246E, M243E, and W137E were 2.94, 5.18, 3.13, and 3.85 mg/ml, respectively. It is interesting to note that M243E showed a significant loss of activity toward chitosan, although its binding affinity was almost the same as the wild-type (Table 1).
As shown in Fig. 3a, the wild-type enzyme showed an optimum temperature of 60°C, the D246E was similar to the wild-type. While both the M243E and W137E exhibited their highest activities at 70°C. Moreover, over 70% activity of the W137E could be detected at temperature from 30°C to 80°C. The W137E shared similar optimum pH with the wild-type (pH 5–7), the D246E had the highest activity around pH 6.0, and the M243E showed an optimum pH of 4–5 (Fig. 3b). When the mutant enzymes were incubated at 40, 55, 60, and 65°C, the W137E was found to be more stable than the wild-type. The M243E became less stable and the D246E was significantly less stable as compared with the wild-type (Fig. 4).
Optimum temperature (a) and pH (b) of the mutant enzymes. a 10 μg of purified enzyme were incubated with 0.5% swollen chitin at 30–90°C for 15 min in a standard reaction mixture. b 10 μg of purified enzyme were incubated with 0.5% swollen chitin at 60°C for 15 min in 100 mM buffer with different pHs (HAc-NaAc for pH 3.6–5.8 and KH2PO4–NaOH for pH 6.2–8.0). The released NAG was assayed as described in the “Methods” section. The same experiment was repeated at least three times, a representative data is shown
Effect of temperature on the stability of the mutant enzymes. The purified enzyme in a concentration of 1 mg per ml 50 mM sodium acetate buffer, pH 5.0 was separately incubated at 45°C (filled circles), 55°C (filled triangles), 60°C (empty circles), and 65°C (empty triangles) for various times. An aliquot of the enzyme solution was removed from each incubation mixture at intervals and assayed for the activity as described in the “Methods” section
3.3 Transglycosyl activities of the mutant enzymes
In our previous report, we have detected a final product mixture of (NAG)2, NAG, and PNP-NAG during the hydrolysis of PNP-(NAG)2 [29]. The production of PNP-NAG can not be explained simply by the action of hydrolytic activity as the AfChiB1 is unable to release NAG from the non-reducing end. Thus, the only plausible explanation is that PNP-NAG is derived from the transglycosylation, in which (NAG)2 in the enzyme-substrate complex was transferred to PNP-(NAG)2 to form PNP-(NAG)4, which is then hydrolysed immediately to yield PNP-NAG and a short-lived (NAG)3 that is finally broken down to NAG and (NAG)2. Therefore, by detecting the PNP-NAG produced from PNP-(NAG)2, we were able to evaluate the transglycosyl activities of the mutant enzymes.
When 0.4 nmol/μl of PNP-(NAG)2 was used as substrate, the wild-type enzyme could utilize 96% of the substrate to produce a mixture of (NAG)2, PNP-NAG and NAG within 30 min, in which (NAG)2 was the dominant product. In addition, a trace amount of short-lived (NAG)3 was also detected (Fig. 5a and Table 2). The ratio of PNP-NAG/(NAG)2 was 0.22. Within 45 min all substrate was converted into a final mixture of (NAG)2, PNP-NAG and NAG, and the ratio of PNP-NAG/(NAG)2 was 0.20 (Table 2). As compared with the wild-type enzyme, the W137E utilized 99% of substrate within 45 min to produce only (NAG)2, suggesting a similar hydrolytic rate to the wild-type and a complete loss of transglycosyl activity (Fig. 5d). When the M243E was incubated with PNP-(NAG)2, no PNP-NAG was detected in the product mixture (Fig. 5c), and only 8% of substrate was hydrolyzed within 45 min (Table 2), suggesting a significant decrease of hydrolytic activity and complete loss of transglycosyl activity. Interestingly, although the D246E could only catalyze 74% of the substrate within 45 min, the PNP-NAG/(NAG)2 value was calculated as 0.67. After 60 min about 80% of substrate was utilized and gave a PNP-NAG/(NAG)2 value of 0.7 (Fig. 5b and Table 2). These data demonstrated that mutation of Asp246 to Glu246 led to 26% loss of the hydrolytic activity and 3.5-fold increase of the transglycosyl activity when PNP-(NAG)2 was used as substrate.
Hydrolysis of PNP-(NAG)2 by the mutant enzymes. One microgram of purified wild-type (a), D246E (b), M243E (c) or W137E (d) was incubated with 8 nmol of PNP-(NAG)2 in 20 μl 50 mM sodium acetate buffer, pH 5.0 at 37°C for predetermined time. The reaction was stopped by heating at 100°C for 3 min. The products were analyzed with HPAEC-PAD using PA-1 column (18 mM NaOH, 1 ml/min). Standard PNP-(NAG)2, PNP-NAG, (NAG)3, (NAG)2, and NAG were used to calibrate the retention time
To investigate chitinolytic/transglycosyl activities toward natural substrate, chitooligosaccharides were used as substrate. As shown in Fig. 6, when (NAG)5 or (NAG)4 were used, (NAG)6 could be detected in reaction mixture of the wild-type AfChiB1 within 2 h, indicating a transglycosyl activity. Prolonged incubation overnight led to a final mixture of (NAG)2 and NAG. As compared with the wild-type, the W137E quickly broke down (NAG)4, (NAG)5 or (NAG)6 into (NAG)2 and (NAG)3 within 2 h, suggesting a complete loss of transglycosyl activity of the W137E. When the D246E was incubated with (NAG)4, both (NAG)5 and (NAG)6 were detected in the product mixture after 16 h, which suggested an increase of transglycosyl activity of the D246E. When the M243E was incubated with (NAG)6, it was not surprised to find (NAG)2 and (NAG)3 in product mixture after 16 h, which is consistent with the reduced hydrolytic activity toward PNP-(NAG)2. However, when (NAG)4 was used, minor (NAG)5 and (NAG)6 were also detected in product mixture within 2 h. This result indicated that M243E still remained some of its transglycosyl activity.
TLC analysis of hydrolysis of chitooligosaccharides by the mutant enzymes. Two micrograms of purified enzyme were incubated with 25 μg of chitooligosaccharide in 15 μl of 50 mM sodium acetate (pH 5.0) at 37°C for various times. After incubation, the reaction mixture was analysed by TLC using n-butanol/acetic acid/water (2:2:1, v/v/v) as solvent. S standard, G1 NAG, G2 (NAG)2, G3 (NAG)3, G4 (NAG)4, G5 (NAG)5, G6 (NAG)6
Interestingly, when (NAG)3 was used as substrate, the wild-type enzyme could produce a final mixture of (NAG)2 and NAG, the M243E showed a similar activity toward (NAG)3, while the W137E displayed a reduced activity toward (NAG)3. Since the difference between endo- and exo-chitinase is that endo-chitinase requires at least tetramer and exo-chitinase requires at least trimer as substrate [29], our observations suggest that the M243E had an exo-chitinase activity similar to the wild-type and the W137E showed a reduced exo-chitinase activity. Thus, the reduced hydrolytic activity associated with the M243E might be due to a reduction of endo-chitinase activity, while the slightly decreased hydrolytic activity associated with the W137 might be contributed by a reduction of exo-chitinase activity. As for the D246E, (NAG)3 was converted into (NAG)5, (NAG)4, (NAG)2, and NAG.
4 Discussion
Family 18 chitinases are able to catalyse transglycosyl reaction in addition to their chitinolytic activities [2, 13, 23, 29]. With the accumulation of structural information, the family 18 chitinases are proposed to employ an unusual hydrolytic mechanism, in which the acid protonating the glycosidic bond is a conserved glutamate and the nucleophile is the oxygen of the N-acetyl group on the -1 sugar [14, 19–22, 24]. However, little effort has been made to probe the mechanism of transglycosyl activity associated to the family 18 chitinases.
Considering family 18 chitinases usually have only one catalytic acid, we hypothesized that the transglycosyl reaction could be due to the lacking of a nucleophile in their active center. To obtain supporting evidence, we mutated Trp137, Asp246, and Met243 to glutamate individually, which are close to the catalytic acid Glu177 and may serve as a potential site for a proper nucleophile.
Our results demonstrated that W137E was completely devoid of transglycosyl activity. This result might be explained by simulation of the W137E in complex with (NAG)3: Asp137 in the wild-type stacks against a sugar at the +1 subsite and is thus central to selecting an incoming sugar rather than water to hydrolyse the glycosyl-enzyme intermediate (Fig. 7a), which leads to transglycosyl reaction. Glu137 can not form hydrophobic stack with the +1 subsite, and also the oxygen of Glu137 is closer to Glu177 than the oxygen of the N-acetyl group on the -1 sugar, their distances are calculated as 2.96Å and 3.52Å, respectively (Fig. 7b). Thus, Glu137 may serve as a suitable nucleophile to mediate water attack to release sugar at the −1 and −2 subsites. In addition, the increase of thermostability associated with the W137E may be due to the introduced hydrogen bond between Glu137 and Glu177 (Fig. 7b).
Computer modeling of the W137E (b), M243E (d), and D246E (f) in complex with (NAG)3. The stereo structures of the wild-type AfChiB1 in complex with (NAG)3 are shown in left panel (a, c, and e). The amino acid side chains and substrate are shown as sticks. The backbone is shown as a ribbon drawing. The hydrogen bond between AfChiB1 residue and substrate is shown as green dashed line. The interactions between Glu177 and N-acetyl group of -1 sugar/Glu137 are shown as orange dashed line. The distance (Å) between two oxygen molecules is labeled next to the line
As compared with the wild-type, the M243E only remained 8–11% of hydrolytic activity. Meanwhile, the M243E was devoid of the transglycosyl activity toward PNP-(NAG)2 (Fig. 5c) and remained a transglycosyl activity against (NAG)4 (Fig. 6). Docking model reveals that Glu243 forms hydrogen bond with backbone oxygen of −1 sugar (Fig. 7d) instead of a hydrophobic stack formed between Met243 and +1 subsite (Fig. 7c). It is likely that the increased binding affinity at −1 subsite allows the retaining of (NAG)2 at −1 and −2 subsite and the loss of hydrophobic stack at +1 subsite leads to a weak binding of incoming sugar, especially for triose. In case of tetraose, one more sugar residue at reducing end would enhance the interaction with the protein.
Interestingly, although the D246E showed a slight decrease of chitinolytic activity against PNP-(NAG)2, it produced more transglycosyl product, PNP-NAG. We assessed the changes in transglycosyl activity of the mutant by comparing the ratio of PNP-NAG/(NAG)2, which represents the rate of the transglycosyl reaction occurred during the hydrolysis. As summarized in Table 2, the transglycosylation rate of the D246E was 5.7-fold at early stage of hydrolysis (within 15 min) and dropped to 3.5-fold of the wild-type after 45 min. The PNP-NAG/(NAG)2 value of the D246E remained to be 3.5-fold of the wild-type during a prolonged hydrolysis. Computer modeling suggests that Asp246 in the wild-type AfChiB1 forms a hydrogen bond with hydroxyl group at C6 position on -1 NAG (Fig. 7e), while Glu246 in the D246E mutant appeared to interact with +1 sugar by forming hydrogen bond (Fig. 7f), which may increase the binding affinity of +1 sugar. This increase of binding affinity at +1 subsite and a reduction of binding at -1 subsite allow an easier binding of another incoming sugar and a reduced stability of the glycosyl-enzyme intermediate, which thus leads to an increased transglycosyl activity associated with the D246E.
Abbreviations
- NAG:
-
N-acetylglucosamine
- (NAG)2 :
-
N,N′-di-N-acetylchitobiose
- (NAG)3 :
-
N,N′,N″-tri-N-acetylchitotriose
- PNP-(NAG)2 :
-
p-nitrophenyl-N,N′-di-N-acetyl- β-chitobioside
- PNP-(NAG)4 :
-
p-nitrophenyl-N,N′,N″,N′″,N″″-tetra-N-acetylchitotetrose
- PNP:
-
p-nitrophenol
References
Armand, S., Tomita, H., Heyraud, A., Gey, C., Watanabe, T., Henrissat, B.: Stereochemical course of the hydrolysis reaction catalyzed by chitinases A1 and D from Bacillus circulans WL-12. FEBS Lett. 343, 177–180 (1994). doi:10.1016/0014-5793(94)80314-5
Aguilera, B., Ghauharali-van der Vlugt, K., Helmond, M.T., Out, J.M., Donker-Koopman, W.E., Groener, J.E., Boot, R.G., Renkema, G.H., van der Marel, G.A., van Boom, J.H., Overkleeft, H.S., Aerts, J.M.: Transglycosidase activity of chitotriosidase: improved enzymatic assay for the human macrophage chitinase. J. Biol. Chem. 278, 40911–40916 (2003). doi:10.1074/jbc.M301804200
Bradford, M.: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248–254 (1976). doi:10.1016/0003-2697(76)90527-3
Brurberg, M.B., Nes, I.F., Eijsink, V.G.H.: Comparative studies of chitinases A and B from Serratia marcescens. Microbiol. 142, 1581–1589 (1996)
Escort, G.M., Hearn, V.M., Adams, D.J.: Inducible chitinolytic system of Aspergillus fumigatus. Microbiol. 144, 1575–1585 (1998)
Flach, J., Pilet, P.E., Jolles, P.: What’s new in chitinase research? Experientia Basel 48, 701–716 (1992). doi:10.1007/BF02124285
Gooday, G.W.: Physiology of microbial degradation of chitin and chitosan. Biodegradation 1, 177–190 (1990). doi:10.1007/BF00058835
Gooday, G.W.: The ever-widening diversity of chitinase. Carbohydrates Eur. 19, 18–22 (1997)
Hawtin, R.E., Arnold, K., Ayres, M.D., Zanotto, P.M.D., Howard, S.C., Gooday, G.W., Chappell, L.H., Kitts, P.A., King, L.A., Possee, R.D.: Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology 212, 673–685 (1995). doi:10.1006/viro.1995.1525
Hu, H., Wang, G., Yang, H., Zhou, J., Mo, L., Yang, K., Jin, C., Jin, C., Rao, Z.: Crystallization and preliminary crystallographic analysis of a native chitinase from the fungal pathogen Aspergillus fumigatus YJ-407. Acta Crystallogr. D Biol. Crystallogr. 60(Pt 5), 939–940 (2004). doi:10.1107/S0907444904005190
Imoto, T., Yagishita, K.: A simple activity measurement of lysozyme. Agric. Biol. Chem. 35, 1154–1156 (1971)
Kuranda, M.J., Robbins, P.W.: Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266, 19758–19767 (1991)
Nanjo, F., Sakai, I., Ishikawa, M., Isobe, K., Usui, T.: Properties and transglycosylation reaction of a chitinase from Nocardia orientalis. Agric. Biol. Chem. 53, 2189–2195 (1989)
Rao, F.V., Houston, D.R., Boot, R.G., Aerts, J.M.F.G., Sakuda, S., van Aalten, D.M.F.: Crystal structures of allosamidin derivatives in complex with human macrophage chitinase. J. Biol. Chem. 278, 20110–20116 (2003). doi:10.1074/jbc.M300362200
Rao, F.C., Houston, D.R., Boot, R.G., Aerts, J.M.F.G., Hodkinson, M., Adams, D.J., Shimi, K., Omura, S., van Aalten, D.M.F.: Specificity and affinity of natural product cyclopentapeptide inhibitors against A. fumigatus, human, and bacterial chitinase. Chem. Biol. 12, 65–76 (2005). doi:10.1016/j.chembiol.2004.10.013
Renkema, G.H., Boot, R.G., Muijsers, A.O., Donker-Koopman, W.E., Aerts, J.M.F.G.: Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J. Biol. Chem. 270, 2198–2202 (1995). doi:10.1074/jbc.270.44.26252
Sakuda, S., Isogai, A., Matsumoto, S., Suzuki, A.: Search for microbial insect growth regulators. II. Allosamidin, a novel insect chitinase inhibitor. J. Antibiot. (Tokyo) 40, 296–300 (1987)
Schales, O., Schales, S.S.: A simple method for the determination of glucose in blood. Arch. Biochem. 8, 285–292 (1945)
Terwisscha van Scheltinga, A.C., Kalk, K.H., Beitema, J.J., Dijkstra, B.W.: Crystal structures of hevamine, a plant defense protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure 2, 1181–1189 (1994). doi:10.1016/S0969-2126(94)00120-0
Terwisscha van Scheltinga, A.C., Armand, S., Kalk, K.H., Isogai, A., Henrissat, B., Dijkstra, B.W.: Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis. Biochem. 34, 15619–15623 (1995). doi:10.1021/bi00048a003
Terwisscha van Scheltinga, A.C., Hennig, M., Dijkstra, B.W.: The 1.8 Å resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J. Mol. Biol. 262, 243–257 (1996). doi:10.1006/jmbi.1996.0510
Tews, I., Terwisscha van Scheltinga, A.C., Perrakis, A., Wilson, K.S., Dijkstra, B.W.: Substrate-assisted catalysis unifies two families of chitinolytic enzyme. J. Am. Chem. Soc. 119, 7954–7959 (1997). doi:10.1021/ja970674i
Usui, T., Hayachi, Y., Nanjo, F., Sakai, K., Ishido, Y.: Transglycosylation reaction of a chitinase purified from Nocardia orientalis. Biochim. Biophys. Acta 923, 302–309 (1987)
van Aalten, D.M.F., Kornmander, D., Synstad, B., Gaseidnes, S., Peter, M.G., Eijsink, V.G.H.: Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. USA 98, 8979–8984 (2001). doi:10.1073/pnas.151103798
Vinetz, J.M., Dave, S.K., Specht, C.A., Brameld, K.A., Xu, B., Hayward, R., Fidock, D.A.: The chitinase PfCHT1 from the human malaria parasite Plasmodium falciparum lacks proenzyme and chitin-binding domains and displays unique substrate preferences. Proc. Natl. Acad. Sci. USA 96, 14061–14066 (1999). doi:10.1073/pnas.96.24.14061
Vyas, P., Deshpande, M.V.: Chitinase produced by Myrothecium verrucaria and its significance for fungal mycelia degradation. J. Gen. Appl. Microbiol. 35, 343–350 (1989). doi:10.2323/jgam.35.343
Wu, Y., Egerton, G., Underwood, A.P., Sakuda, S., Bianco, A.E.: Expression and secretion of a larval-specific chitinase (family 18 glycosyl hydrolase) by the infective stages of the parasitic nematode, Onchocerca volvulus. J. Biol. Chem. 276, 42557–42564 (2001). doi:10.1074/jbc.M103479200
Wang, Y., Wang, J., Hu, H., Yang, X., Yang, S., Yu, X., Jin, C.: Cloning and expression of chitinase gene from Aspergillus fumigatus YJ-407. Chin. J. Biotechnol. 20, 843–850 (2004)
Xia, G., Jin, C., Zhou, J., Yang, S., Zhang, S., Jin, C.: A novel chitinase having a unique mode of action from Aspergillus fumigatus YJ-407. Eur. J. Biochem. 268, 4079–4085 (2001). doi:10.1046/j.1432-1327.2001.02323.x
Acknowledgements
This work was supported by the National Basic Research Program of China (2004CB719606) to C. Jin.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enzymes: exochitinases (EC 3.2.1.14)
Rights and permissions
About this article
Cite this article
Lü, Y., Yang, H., Hu, H. et al. Mutation of Trp137 to glutamate completely removes transglycosyl activity associated with the Aspergillus fumigatus AfChiB1. Glycoconj J 26, 525–534 (2009). https://doi.org/10.1007/s10719-008-9203-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-008-9203-z