Abstract
The XA21 protein has broad spectrum resistance against Xanthomonas oryzae pv. oryzae. Although Xa21-mediated immunity is well characterized, little is known about the origin and evolutionary history of this gene in grasses. Therefore, we analyzed all Xa21 gene homologs in eight whole-genome sequenced rice lines, as well as in four gramineous genomes, rice, Brachypodium, sorghum and maize; using Arabidopsis Xa21 homologs as outgroups, 17, 7, 7 and 3 Xa21 homologs were detected in these four grasses, respectively. Synteny and phylogenetic analysis showed that frequent gene translocation, duplication and/or loss, have occurred at Xa21 homologous loci, suggesting that they have undergone or are undergoing rapid generation of copy number variations. Within the rice species, the high level of nucleotide diversity between Xa21-like orthologs showed a strong association with the presence/absence haplotypes, suggesting that the genetic structure of rice lines plays an important role in the variations between these Xa21-like orthologs. Strongly positive selection was detected in the core region of the leucine-rich repeat domains of the Xa21 subclade among the rice lines, indicating that the rapid gene diversification of Xa21 homologs may be a strategy for a given species to adapt to the changing spectrum of species-specific pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial blight disease, caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the most serious rice diseases, especially in Asia and Africa. To date, over 29 genes have been reported with resistance to blight disease (Leung 2008) and some were isolated. Among these genes, Xa21 encodes a receptor-like kinase (RLK) with leucine rich repeats (LRRs), a transmembrane domain and a cytoplasmic serine/threonine kinase domain (Song et al. 1995). Xa21 was the first disease resistance gene cloned from rice (Mew 1987), which was isolated from the wild rice species Oryza longistaminata. Because of its broad spectrum resistance against Xoo (Wang et al. 1996), the Xa21 gene has been drawing the attention of rice breeders since it was found, and Xa21-mediated immunity has been well-studied (Park et al. 2010).
There is evidence to support that Xa21 is a member of a multi-gene family located on rice chromosome 11 (Ronald et al. 1992; Song et al. 1995). This family has seven members which were cloned and grouped into two classes based on sequence similarity (Song et al. 1997). Like Xa21, Xa21-D encodes a receptor-like protein carrying LRR motifs and displays partial resistance (Wang et al. 1998), demonstrating that the LRR domain is responsible for race-specific pathogen recognition. Within the Xa21 family, there is also evidence of recombination both in intergenic and intragenic regions which are highly conserved. Therefore, gene duplication and diversification are very common during the evolution of the Xa21 gene family, and these processes are thought to be associated with the creation of novel resistance phenotypes.
There are >1,000 RLKs and >300 LRR–RLKs members identified in rice (Shiu et al. 2004; Tang et al. 2010). Most of them are involved in a variety of cellular signaling processes responding to diverse extracellular signals. Other than disease resistance, RLKs play important roles in many biological processes, including development and growth, in both animals and plants (Fantl et al. 1993; Becraft et al. 1996; Stein et al. 1996; Torii et al. 1996; Li and Chory 1997). As an active motif, LRR domain commonly exists in plants and is usually combined with other motifs. The LRR region tends to be highly variable and subject to positive selection in resistance genes, consistent with host-pathogen co-evolution (Parniske et al. 1997). In the Xa21 gene family, LRRs of different members may have evolved to recognize different pathovars and may confer altered resistance phenotypes (Song et al. 1997). Moreover, comparison of Xa21 and Xa21-D reveals that there are many non-synonymous substitutions accumulated in the LRR regions resulting from adaptive evolution (Wang et al. 1998).
Although there have been many studies on the Xa21 gene, most of them are concerned with its activation and expression (Park et al. 2008, 2010; Xu et al. 2006; Song et al. 2006). Song et al. have proposed a model for the evolution of Xa21 family members in rice (Song et al. 1997), but the evolutionary history and pattern of Xa21 genes are largely unclear. Recently, several gramineous and a few rice genomes were sequenced. These genomes provided a great opportunity for us to investigate the inter-species and intra-species evolutionary patterns of Xa21 homologs. In this study, we identified variable copy numbers of Xa21 homologs (3–17) in four gramineous genome sequences and analyzed the syntenic blocks of Xa21 homologs, which may help us to understand the origin and evolutionary history of these genes in gramineous species. We also investigated the genetic structure, variations of copy number of these genes within rice species, and the nucleotide diversity between Xa21-like orthologs. Our work may contribute to better understanding of the selection upon Xa21-like genes within rice species and between gramineous species.
Materials and methods
Whole-genome sequenced species
The fully sequenced gramineous genomes included one sorghum (Sorghum bicolor), one maize (Zea Mays), one Brachypodium (Brachypodium distachyon) and eight rice (Oryza sativa) genomes. Among the eight sequenced rice genomes, five were publicly available and the other three were unpublished. Nipponbare (O. sativa L. ssp. japonica, Release 6.1) whole-genome map-based sequences were downloaded from the International Rice Genome Sequencing Project (IRGSP) (International Rice Genome Sequencing Project 2005; http://rgp.dna.affrc.go.jp/E/IRGSP/download.html). Two whole genomes assembled by shotgun sequencing, 93-11 and PA64s (O. sativa L. ssp. indica), were obtained from the Beijing Genomics Institute (BGI) database (http://rise2.genomics.org.cn/page/rice/index.jsp). GLA4 (indica) and NK58 (japonica) whole-genome sequences obtained by sequencing-by-synthesis technology were downloaded from the National Center for Genome Resources (NCGR) (Huang et al. 2010; http://www.ncgr.ac.cn/scientific_data.asp). The other three resequenced indica genomes, MH63, SH527 and IR24, obtained using high-throughput sequencing technology from Illumina, were kindly provided by Ping Li (Sichuan Agricultural University, China).
Assembly and gene models of the other three gramineous genomes, Sorghum (S. bicolor, v1.0; Paterson et al. 2009), maize (Z. Mays cv B73; Schnable et al. 2009), and Brachypodium (B. distachyon, v2.0), were obtained from the Joint Genome Institute (JGI) (http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html), MaizeSequence.org (http://www.maizesequence.org/index.html) and http://www.Brachypodium.org/, respectively. Arabidopsis thaliana sequences were downloaded from the Arabidopsis information resource (TAIR) (The Arabidopsis Genome Initiative 2000; http://www.arabidopsis.org/).
Identification of Xa21 homologous genes
The amino acid and nucleotide sequences of XA21 described by Song et al. (1995, 1997), as well as the other six family members (Xa21-A1, -A2, -C, -D, -E, -F; Song et al. 1997) were downloaded from GenBank. We used the amino acid sequence of XA21 as a query to find homologs or orthologs in rice (Nipponbare, 9311 and PA64s), maize, sorghum, Brachypodium and A. thaliana genomes using TBLASTN search. The threshold expectation value was set to 1E-150, and the other numerical options were left at default values. The candidate sequences were further surveyed to determine whether they encoded kinase and LRR motifs using the Pfam database v24.0 (E value cut-off of 10−4).
Xa21 homologs in the resequenced rice genomes were retrieved by mapping of reads to the Nipponbare, 9311 and PA64s sequences used as references (SHORE, http://sourceforge.net/projects/shore/files). Aligned reads were picked up with a cut-off of minimum 90 % identity over a read. Only uniquely aligned reads (reads mapped to unique locations in these reference sequences) were retained and low-quality base sites (base-quality Q score in Phred scale <20) were removed.
Sequence alignment and data analysis
The amino acid sequences were first aligned by using the MUSCLE program with default options (Edgar 2004), and then MEGA v5.0 (Tamura et al. 2011) was used to manually correct the alignments. The resulting amino acid sequence alignments were then used to guide the alignments of the nucleotide coding sequences (CDSs). Nucleotide diversity (π) with Jukes and Cantor correction (Lynch and Crease 1990) was calculated using DnaSP v5.0 (Librado and Rozas 2009). To detect positive selection, we estimated the ratios of non-synonymous (Ka) to synonymous (Ks) nucleotide substitutions rates, also known as Ka/Ks, on the full-length coding sequences (CDS) and core regions of the LRR domain. The LRR core regions are ××L×L×× motif (L = Leu or other aliphatic amino acid; × = any amino acid) including the solvent-exposed residues (Wang et al. 2011). Phylogenetic analysis based on the bootstrap neighbor-joining (NJ) method with a Kimura 2-parameter model was performed using MEGA v5.0. The stability of internal nodes was assessed by bootstrap analysis with 1,000 replicates.
The Xa21 homologs and their flanking ~100 genes were used to investigate the gene collinearity in orthologous regions between rice and other gramineous species. A gene pair was generated by TBLASTN search of the most similar CDS sequence in other species against that in rice. The relationships between gene pairs were visualized by GenomePixelizer software (http://www.gnu.org/).
Results
Identification of Xa21 homologs in Gramineae and Arabidopsis
Using the amino acid sequence of XA21 (GenBank ID: AAC80225.1) and its reported homologs (Song et al. 1995, 1997) as queries, 52, 34, 15, 14 and 5 candidates of Xa21 homologs were detected in rice (Nipponbare), sorghum, Brachypodium, maize and A. thaliana, respectively. Subsequently, these candidate amino acid and CDS sequences were aligned using the MUSCLE program (see Sect. “Materials and Methods”). Based on the nucleotide sequence alignments, a phylogenetic tree was constructed by the bootstrap NJ method with a Kimura 2-parameter model. In this phylogenetic tree, the five candidate Xa21 homologs in A. thaliana were clustered within the same clade with a 100 % bootstrap value, and had the highest similarity in the A. thaliana genome with Xa21 homologs in the gramineous species. Interestingly, of these five Arabidopsis genes, one (At5g20480) was identified as the EFR gene, which has been confirmed to recognize the surrogate peptide elf18 of bacterial elongation factor (EF)-Tu (Kunze et al. 2004). When we used the five Arabidopsis Xa21 homologs as an outgroup to define Xa21 homologs in gramineous species, within the Xa21 clade, 17, 7, 7 and 3 Xa21 homologs were identified in rice, Brachypodium, sorghum and maize, respectively (Table 1; Fig. 1). All of these genes were also further verified to encode LRR and Pkinase proteins using the Pfam database.
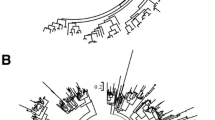
Phylogenetic tree of Xa21 homologs from four gramineous species and A. thaliana genomes. The gramineous clade in the phylogenetic tree was divided into four major subclades based on three criteria, which were high bootstrap values (100 %), high nucleotide similarity within subclade (≥60 %) and species number in each subclade (≥3 out of 4 species). The two outgroups were genes from A. thaliana. Outgroup 1 contained EFR genes which had maximum similarities toXa21 in A. thaliana. Outgroup 2 consisted of the FLS2 gene encoding a receptor which was reported to recognize Ax21-derived peptides. The underlined genes were those which had no synteny blocks between the flanking regions of these genes and the corresponding regions of the Xa21 homologs in rice genome
The LRR–RLK, FLS2 in Arabidopsis is the pattern-recognition receptor for bacterial flagellin (Gomez-Gomez and Boller 2000). Interestingly, the FLS2 protein has been confirmed to also mediate the perception of Xanthomonas Ax21 secreted peptides (Danna et al. 2011). Using the five Arabidopsis Xa21 homologs (EFR clade) as outgroup 1 and Arabidopsis FLS2 sequence as outgroup 2, a phylogenetic tree was reconstructed (Fig. 1) together with 34 gramineous Xa21 homologs and seven publicly available Xa21 sequences (Song et al. 1995, 1997). Within the gramineous Xa21 clade, four distinct subclades (subclades 1–4) were detected with 100 % bootstrap value, ≥60 % nucleotide similarity within subclade and at least three gramineous species in each subclade (Fig. 1; Table 1). It was clear that the Xa21 homologs were distributed unevenly in each subclade or gramineous species, and the rice genome in subclade 1 had the largest copy number (eight Xa21 homologs in two clusters; Table 1; Fig. 1), suggesting that frequent gene duplication or loss may contribute to the copy number variations in gramineous species.
Syntenic block analysis of Xa21 homologs among gramineous species
The progenitors of rice, Brachypodium, sorghum, and maize split ~60 mya. While the sorghum and maize share a common ancestor at ~25 mya, the progenitors of rice and Brachypodium split ~45 mya (Gaut 2002). As shown in Fig. 1, after the split of dicots and monocots, four distinct Xa21-like subclades survived in gramineous species, suggesting that Xa21-like genes may have expanded in gramineous species or their corresponding genes have been lost in Arabidopsis. To further investigate the orthologous relationships and the copy number variations of Xa21 homologs in gramineous species, the analysis of collinear regions at Xa21 homologous loci was performed together with their flanking genes (see Materials and methods for details). In the rice genome, 17 Xa21 homologs located at six independent loci were distributed in all four phylogenetic subclades, whereas fewer Xa21-like genes and loci were found in the other three grass genomes (Fig. 1). Therefore, the six rice Xa21 homologous loci (Table 1) were used as references to investigate their syntenic relationship among gramineous species.
Five apparent syntenic regions, including 16 rice Xa21 homologs, were detected among these species (Fig. 2 and S2). For one rice Xa21 homolog, Os10g19160, we did not find its corresponding syntenic regions in the other three grass species. In subclade 1, two independent syntenic regions (subclade 1a and 1b in Fig. 2 and S2, respectively) were detected. In the syntenic region of subclade 1a, the orthologous regions were found in all of the four grass species, whereas Xa21 homologs were detected only in rice and Brachypodium (Fig. 2), indicating that the Xa21 homologs in this subclade have a common ancestor(s) at least in the BEP clade (Fig. S1). However, it was difficult to determine whether the syntenically homologous Xa21 was lost in the Andropogoneae clade (Fig. S1) or these Xa21 homologs were translocated to this region in the ancestor of the BEP clade after these two grass clades split. Interestingly, in this subclade, there were seven and three Xa21 homologs in the rice and Brachypodium genomes, respectively (Fig. 2), suggesting frequent number variation after their split. On the other hand, Bd4g16130 and Bd4g08300 had ~94 % nucleotide identity in the Brachypodium genome, which was significantly higher than the identity (73 %) between Bd4g16130 and Bd4g16070. However, Bd4g16130 and Bd4g16070 were located in the same cluster of the syntenic region in subclade 1a (Fig. 2), while Bd4g08300 was found far away from those two homologous members, implying that this copy may have recently translocated from the syntenic region to another region. In subclade 1b, one syntenic Xa21 homolog was found in each grass genome except maize (Fig. S2), suggesting that this Xa21 homolog was lost in maize after the sorghum/maize split. In the other three syntenic regions (Fig. 2 and S2), similarities in the variable copy number via duplication or loss, and frequent gene translocation of Xa21 homologs, were also found in each subclade between species (Fig. S2).
Gene collinearity in orthologous regions between rice and other gramineous species. Genes are indicated as black dots. Orthologous genes are connected by blue lines. The genes marked with red circles represent Xa21 homologs. a The syntenic relationship of the subclade 1a locus in Fig. 1 between rice and Brachypodium, b between rice and sorghum, and c between rice and maize. (Color figure online)
Variations of Xa21 homologs among eight whole-genome sequenced rice lines
Using the whole-genome annotated data, 17, 15 and 21 Xa21 homologs were identified in the Nipponbare, PA64s and 93-11 rice genomes, respectively (Table 2). Using the 53 rice Xa21 genes in these reference sequences, 17, 17, 21, 21, 21 and 21 homologs were assembled and identified from the resequenced reads of GLA4, NK58, MH63, SH527 and IR24, respectively (Table 2), indicating that the numbers of Xa21 homologs were slightly different among these rice genomes.
Using these 150 identified Xa21 homologs and the seven publicly available Xa21 family genes (Song et al. 1997), an intra-species phylogenetic tree was constructed. Using a 100 % bootstrap value, ≥3 rice lines and only one copy in each rice line as grouping criteria, 27 distinct subclades were grouped in this tree (Table 2). Variations in presence/absence (P/A) of Xa21 homologs between rice lines were clearly observed in each subclade (Table 2). A phylogenetic tree was constructed to examine the variations in P/A of Xa21 homologs in each subclade of each rice line (Fig. 3). Three distinct haplotypes were observed based on the patterns of P/A polymorphism in Xa21 homologs. The MH63, IR2, SH527 and 9311, indica rice lines were all grouped in the same clade, indicating that these genomes shared the same P/A polymorphism haplotype in these Xa21 homologs. Nipponbare and NK58 are both japonica rice varieties and clustered together, sharing the same P/A haplotype with the GLA4 indica rice line. Interestingly, the P/A polymorphism haplotype in the PA64s genome was intervenient between the two haplotypes above, which is consistent with the report that PA64s resulted from multiple crosses and that its genome is 55, 25 and 20 % similar to that of indica, japonica and javanica, respectively (http://rise2.genomics.org.cn/page/rice/index.jsp).
Grouping of eight rice lines based on the P/A status of Xa21 homologs in 27 subclades. This tree was constructed by the discrete morphology (parsimony) method using the programs PARS of the PHYLIP package v3.6. The diversity of each subclade can be seen in Table 2
However, when using P/A polymorphisms in the 27 subclades to construct a phylogenetic tree, six distinct groups were found (Fig. S4). Group I included eight subclades, and one member in each of the eight rice genomes was detected in each subclade, suggesting that these genes are orthologs and have a conservative evolutionary pattern in the copy number variations. However, in the other five groups, including 19 subclades, Xa21 homologs were absent in one to five out of the eight rice lines in each subclade (Fig. S4 and Table 2), suggesting the rapidly generated P/A variations in Xa21 homologous alleles in rice lines.
Nucleotide polymorphisms and selective pressure of Xa21 homologs among rice lines
Since Xa21 homologous genes in each subclade may be potential orthologs in different rice lines, the nucleotide diversity within each subclade was used to evaluate the evolutionary rate of these genes. In the 27 Xa21 homologous subclades, the average nucleotide diversity (π) was 0.020 between rice lines, ranging from 0.0 (subclade 4) to 0.122 (subclade 15; Table 2), which was approximately two-fold higher than that (1.15 %) in the comparison of genomes between Nipponbare and 9311 (Tang et al. 2006). Surprisingly, the average π of subclades in groups 1, 2 and 3 were 0.0314, 0.0405 and 0.0473, respectively, which were approximately 6 to 27-fold higher than those in the other three groups (Table 2).
In order to determine the factors leading to the high level of nucleotide diversity, the genetic structure of these rice lines was investigated. The above results showed that three P/A haplotypes of the Xa21 homologs were observed in the eight whole-genome sequenced rice lines. Therefore, the nucleotide diversity was calculated within and between these three P/A haplotypes (Table S1). In the three high level nucleotide diversity groups (groups 1–3), the average π were 0.0012–0.0112 within haplotypes, which were similar to or slightly lower than the diversity observed between rice genomes (Tang et al. 2006), but were significantly lower than their corresponding diversities among all rice lines or divergences (Dxys) between P/A haplotype groups within the same subclades (t-test, P < 0.01; Table S1). On the other hand, the average Dxys between P/A haplotype groups were significantly higher than their corresponding diversities among all rice lines within the same subclades (t-test, P < 0.01; Table S1), suggesting that the genetic structure of rice lines may contribute substantially to the high level of nucleotide diversity and play an important role in the rapidly generated variations between these Xa21-like orthologs.
To detect selective pressure, the Ka/Ks substitution rate was calculated within the 27 rice subclades among the eight rice lines (Table 2). When calculating the Ka/Ks using the full CDS region, only three out of 27 subclades had a Ka/Ks ratio over one. However, in the ××L×L×× motif of the LRR region (core region of LRR), which is assumed to be a determinant of recognition specificity for Avr factors (Ellis et al. 1999), eight out of 27 subclades had Ka greater than Ks (Table 2). The functional Xa21 and Xa21-D genes, cloned from wild rice (O. longistaminata) and shown to confer resistance to Xoo (Song et al. 1995), were clustered in subclade 10 (Table 2). Interestingly, significantly higher Ka/Ks ratios were detected in the core region of the LRR in this subclade, suggesting that these genes selectively accumulated non-synonymous amino acid substitutions for resistance to pathogens.
To further analyze the selective pressure on the Xa21 subclade in rice, another 54 publicly available Xa21 allelic sequences were downloaded from GenBank (AY885769–AY885800 and DQ374726–DQ374747), including 25 O. sativa (14 indica and 11 japonica), 19 Oryza rufipogon, 3 Oryza glumipatula, 3 Oryza meridionalis, 2 Oryza nivara, 1 Oryza longistaminata and 1 Oryza barthii. When comparing the available Xa21 allelic sequences from 60 lines of cultivated and wild rice, a high proportion (23.6 %) of pairwise comparisons was also detected under positive selection in the LRR regions. Moreover, three alleles could be considered as potential candidates for resistance genes to Xoo strains according to the phylogenetic tree constructed by the 60 alleles (Fig. S5).
Discussion
Ancient origin and rapid accumulation of variations in Xa21 homologs in gramineous species
In plants, multi-layer defenses are employed to defend themselves against pathogenic organisms. The primary layer of immunity, the pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), relies on recognizing the conserved microbial molecules that act as signatures of a whole class of microbes (Ausubel 2005). Both FLS2 and EFR encode a transmembrane RLK consisting of LRRs in the putative extracellular domain, which bind conserved bacterial peptide PAMPs flg22 (derived from flagellin) and elf18/elf26 (derived from elongation factor Tu), respectively (Zipfel et al. 2006; Gomez-Gomez and Boller 2000; Albert et al. 2010). FLS2 orthologs can be found in a wide range of monocotyledonous and dicotyledonous plant species, whereas EFR appears to be limited only to Arabidopsis. In addition, the protein encoded by the FLS2 homologous gene (OsFLS2, Os04g52780 in Nipponbare) in rice has also been confirmed to be capable of recognizing flg22 (Takai et al. 2008). In our study, only one syntenic region with one FLS2 homologous gene in each grass genome was clearly detected (Fig. S3), indicating that FLS2 homologous genes may be evolutionarily conserved and may play a basic defense role in plants (Takai et al. 2008).
On the other hand, the proteins inducing the second layer of immunity, e.g., plant intracellular nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins, initiate strong defense responses upon recognition of specific pathogen effector molecules (DeYoung and Innes 2006). Our previous studies have shown that NBS-LRR genes are extremely diverse and evolve rapidly within species or between closely related species (Zhang et al. 2009; Chen et al. 2010; Li et al. 2010; Yue et al. 2012). By comparing the four whole-genome sequenced gramineous species, considerable variation between species has been observed with only 3.93 % of NBS-LRR conforming to a conserved family, compared with 96.1 % conservation in selected housekeeping genes, indicating that the striking lack of conservation most likely reflects rapid gene diversification as a result of pathogen-mediated selection pressures (Li et al. 2010).
Similarly, XA21 proteins induce the second layer of immunity and initiate defense responses upon recognition of the specific secreted protein Ax21 (Lee et al. 2009). Interestingly, the Arabidopsis EFR resistance gene showed the highest similarity with Xa21 and its homologs in gramineous species in this study (Fig. 1), suggesting that Xa21 homologs have an ancient origin. In addition, in contrast with the evolutionary pattern of FLS2 alleles, frequent gene duplication and/or loss and gene translocation were found in Xa21 homologs among the four closely related gramineous species. Compared to Brachypodium genome, Gene duplication and translocation events in rice were detected in 4 syntenic regions (Fig. 2 and S2). Therefore, the rapid gene diversification of Xa21 homologs may be an important strategy for species to adapt to the quickly changing spectrum of species-specific pathogens.
Strongly positive selection and potentially resistant candidates of rice Xa21 alleles
Many NBS-LRR genes (e.g., RPP13, RPP8, Pib, L, RGC2) have exceptionally high levels of polymorphism (Kuang et al. 2004; Dodds et al. 2006; Ding et al. 2007; Jiang et al. 2007). The extreme diversity of these loci was often the result of positive selection, under which the ratio of its non-synonymous (Ka) to synonymous (Ks) substitutions rate was greater than one. Our data also showed that eight out of 27 Xa21 homologous subclades had Ka greater than Ks in the ××L×L×× motif among the eight whole-genome sequenced rice lines (Table 2), suggesting the occurrence of positive selection on these genes. Interestingly, in subclade 10, which included the functional resistant genes, Xa21 and Xa21-D, only non-synonymous substitutions were found in the ××L×L×× motifs, indicating strongly positive selection on this locus.
In addition, by comparing the sequences of 60 rice lines, Tang et al. (2006) revealed that the silent nucleotide diversity of Xa21 genes among rice cultivars was ~0.002, approaching our results in subclade 10 (Table S1). They also found evidences of negative selection on this locus which showed negative values of Tajima’s D. However, the LRR regions, which participate in recognizing pathogens, are usually under positive selection. Therefore, we calculated the Ka/Ks ratios within the LRR regions between every two of the 60 rice lines. As expected, pairwise comparisons showed that approximately 23.6 % had Ka/Ks ratios greater than one in the LRR regions. On the other hand, in the ××L×L×× motif of the LRRs, seven non-synonymous and no synonymous substitutions were detected among these sequences, further confirming that strong positive selective forces acted on this region, which may be a determinant for recognition of specific pathogenic effectors.
When a phylogenetic tree was constructed using the ××L×L×× motif sequences by the bootstrap NJ method, 60 Xa21 sequences were grouped in a distinct mixed clade (Fig. S5), including cultivated and wild rice. No substitution was detected in the sequences of this clade. However, for the other five Xa21 sequences scattered outside of this clade in the tree (Fig. S5), only non-synonymous substitutions were detected compared with the 60 clustered sequences or with each other. Interestingly, the functional resistance genes Xa21 and Xa21-D were among those five Xa21 sequences, suggesting that the non-synonymous substitutions may be an important signature for resistance to Xoo strains. Therefore, the other three Xa21 sequences with the selective accumulation of the non-synonymous substitutions (Fig. S5), DQ374727 from O. meridionalis, AY885771 from O. longistaminata and DQ374729 from O. glumipatula, may be candidates for resistance genes to Xoo strains. However, this hypothesis needs further experimental testing.
References
Albert M, Jehle AK, Mueller K, Eisele C, Lipschis M, Felix G (2010) Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J Biol Chem 285(25):19035–19042
Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6(10):973–979
Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273(5280):1406–1409
Chen Q, Han Z, Jiang H, Tian D, Yang S (2010) Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J Mol Evol 70:137–148
Danna CH, Millet YA, Koller T, Han SW, Bent AF, Ronald PC, Ausubel FM (2011) The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc Natl Acad Sci USA 108(22):9286–9291
DeYoung BJ, Innes RW (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7(12):1243–1249
Ding J, Cheng HL, Jin XQ, Araki H, Yang YH, Tian DC (2007) Contrasting patterns of evolution between allelic groups at a single locus in Arabidopsis. Genetica 129(3):235–242
Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103(23):8888–8893
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11(3):495–506
Fantl WJ, Johnson DE, Williams LT (1993) Signaling by receptor tyrosine kinases. Annu Rev Biochem 62:453–481
Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154(1):15–28
Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5(6):1003–1011
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42(11):961–967
Initiative TheArabidopsisGenome (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436(7052):793–800
Jiang HY, Wang CC, Ping L, Yang SH, Tian DC (2007) Pattern of LRR nucleotide variation in plant resistance genes. Plant Sci 173(2):253–261
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16(11):2870–2894
Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16(12):3496–3507
Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326(5954):850–853
Leung H (2008) Stressed genomics—bringing relief to rice fields. Curr Opin Plant Biol 11(2):201–208
Li JM, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90(5):929–938
Li J, Ding J, Zhang W, Zhang YL, Tang P, Chen JQ, Tian DC, Yang SH (2010) Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Genet Genomics 283(5):427–438
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Lynch M, Crease TJ (1990) The analysis of population survey data on DNA sequence variation. Mol Biol Evol 7(4):377–394
Mew TW (1987) Current status and future-prospects of research on bacterial-blight of rice. Annu Rev Phytopathol 25:359–382
Park CJ, Peng Y, Chen XW, Dardick C, Ruan DL, Bart R, Canlas PE, Ronald PC (2008) Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol 6(9):1910–1926
Park CJ, Han SW, Chen XW, Ronald PC (2010) Elucidation of XA21-mediated innate immunity. Cell Microbiol 12(8):1017–1025
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91(6):821–832
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob ur R, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457(7229):551–556
Ronald PC, Albano B, Tabien R, Abenes L, Wu KS, Mccouch S, Tanksley SD (1992) Genetic and physical analysis of the rice bacterial-blight disease resistance locus, Xa21. Mol Gen Genet 236(1):113–120
Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du FY, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen WZ, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He RF, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin JK, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren LY, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han YJ, Lee H, Li PH, Lisch DR, Liu SZ, Liu ZJ, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang LX, Yu Y, Zhang LF, Zhou SG, Zhu Q, Bennetzen JL, Dawe RK, Jiang JM, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Shiu SH, Karlowski WM, Pan RS, Tzeng YH, Mayer KFX, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16(5):1220–1234
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270(5243):1804–1806
Song WY, Pi LY, Wang GL, Gardner J, Holsten T, Ronald PC (1997) Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9(8):1279–1287
Song WY, Wang YS, Pi LY, Chen XH, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li LC, Benny U, Oard J, Ronald PC (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18(12):3635–3646
Stein JC, Dixit R, Nasrallah ME, Nasrallah JB (1996) SRK, the stigma-specific S locus receptor kinase of Brassica, is targeted to the plasma membrane in transgenic tobacco. Plant Cell 8(3):429–445
Takai R, Isogai A, Takayama S, Che FS (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol Plant Microbe Interact 21(12):1635–1642
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang T, Lu J, Huang J, He J, McCouch SR, Shen Y, Kai Z, Purugganan MD, Shi S, Wu CI (2006) Genomic variation in rice: genesis of highly polymorphic linkage blocks during domestication. PLoS Genet 2(11):e199
Tang P, Zhang Y, Sun XQ, Tian DC, Yang SH, Ding J (2010) Disease resistance signature of the leucine-rich repeat receptor-like kinase genes in four plant species. Plant Sci 179(4):399–406
Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8(4):735–746
Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC (1996) The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv oryzae isolates in transgenic plants. Mol Plant Microbe In 9(9):850–855
Wang GL, Ruan DL, Song WY, Sideris S, Chen LL, Pi LY, Zhang SP, Zhang Z, Fauquet C, Gaut BS, Whalen MC, Ronald PC (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10(5):765–779
Wang J, Tan S, Zhang L, Li P, Tian D (2011) Co-variation among major classes of LRR-encoding genes in two pairs of plant species. J Mol Evol 72(5–6):498–509
Xu WH, Wang YS, Liu GZ, Chen XH, Tinjuangjun P, Pi LY, Song WY (2006) The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor-like kinase. Plant J 45(5):740–751
Yue JX, Meyers BC, Chen JQ, Tian D, Yang S (2012) Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 193(4):1049–1063
Zhang YL, Wang J, Zhang XH, Chen JQ, Yang SH, Tian DC (2009) Genetic signature of rice domestication shown by a variety of genes. J Mol Evol 68(4):393–402
Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125(4):749–760
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30970198), the Fundamental Research Funds for the Central Universities (1116020808), the Key project of Chines Ministry of Education (109071) and Qing Lan Project to S.Y.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shengjun Tan, Dan Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, S., Wang, D., Ding, J. et al. Adaptive evolution of Xa21 homologs in Gramineae. Genetica 139, 1465–1475 (2011). https://doi.org/10.1007/s10709-012-9645-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-012-9645-x