Abstract
Chromosome painting (CP) with a probe of B chromosome obtained by microdissection and fluorescence in situ hybridization (FISH) with probes of As51 satellite DNA, C o t−1 DNA, and 18S and 5S rDNA confirmed sharing of some repetitive DNA but not rDNA between A and B chromosomes in the fish Astyanax scabripinnis. Meiotic analysis revealed a pachytene B chromosome bivalent nearly half the size of its mitotic configuration, suggesting a self-pairing of B chromosome arms. Such an isochromosome nature of somatic B chromosome was further evidenced by CP and FISH. All the findings obtained suggest (i) intraspecific origin of B chromosome, and (ii) evolutionary enrichment of repetitive DNA classes, especially those contained in the C o t−1 and the As51 probes, in B chromosome. However, the precise origin of B chromosome in the present species remains to be elucidated by further molecular cytogenetic analysis because of painting of some A chromosome regions with the B chromosome-derived probe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Supernumerary or B chromosomes are additional genetic elements found in the chromosome complement of about 15% of eukaryotic species. A number of hypotheses have been raised to explain the origin, frequency and evolution of B chromosomes, such as derivation from autosomes followed by gene silencing, heterochromatinization, and accumulation of repetitive DNA and transposons (Camacho et al. 2000). Moreover, B chromosomes may differ morphologically from A chromosomes, display a non-Mendelian inheritance pattern and exhibit accumulation mechanisms (Jones and Rees 1982; Beukeboom 1994; Camacho 2005).
Astyanax scabripinnis belongs to the order Characiformes and is listed among the Incertae sedis in the family Characidae (Lima et al. 2003). Some populations of this species have B chromosomes (Moreira-Filho et al. 2004). The shape and number of these chromosomes may vary in different populations of A. scabripinnis. However, the occurrence of a B macrochromosome similar in size to the first chromosome pair is most common (Néo et al. 2000). Meiotic studies suggest that B chromosomes in A. scabripinnis exhibit non-Mendelian behaviour and form univalents during meiosis through pairing between chromosome arms (Mestriner et al. 2000).
Repetitive DNA elements have been mapped for the B macrochromosome in A. scabripinnis. Mestriner et al. (2000) identified a satellite DNA enriched in A+T (59%), with monomeric units of 51 bp, in the genome of this species, named As51. Fluorescence in situ hybridization (FISH) demonstrated that this DNA is mainly located in the distal heterochromatin of several acrocentric A chromosomes, in the short arms of chromosome 10, where the main nucleolus organizer region (NOR) is located in this species, and interstitially in the B chromosome (Mestriner et al. 2000). The symmetric location of As51 in both arms of the metacentric B chromosome, along with their autopairing during meiosis, led these authors to suggest that this B is an isochromosome originated through misdivision of an acrocentric chromosome. However, they could not ascertain the precise A chromosome from which the B arose, and they suggested B chromosome microdissection as a possible valuable tool for this task. Here we perform the microdissection of the metacentric B chromosome found in the same population of A. scabripinnis analysed by Mestriner et al. (2000), and perform chromosome painting in conjunction with FISH trying to get new insights into its origin by comparing meiotic behaviour and repetitive DNA composition with those in A chromosomes.
Materials and methods
Ten specimens of A. scabripinnis (4 males and 6 females) were collected at Córrego das Pedras population, belonging to the Paraíba do Sul River Basin, located in the municipality of Campos do Jordão (state of São Paulo, Brazil) (−22°43′332″ and −45°33′074″). For cytogenetic analysis, at least 30 mitotic metaphase cells were analysed per individual. The specimens were identified and deposited in the Museum at Universidade Estadual de Londrina with voucher number (MZUEL 5655).
Mitotic chromosomes were obtained from cells of the anterior kidney using the air-drying procedure (Bertollo et al. 1978). Meiotic preparations were made according to Kligerman and Bloom (1977), with some modifications (Bertollo et al. 1978). The chromosome preparations were submitted to conventional Giemsa staining and C banding (Sumner 1972) for the determination of diploid number, chromosome morphology, distribution of heterochromatin and presence of B chromosomes. Silver impregnation was performed according to Howell and Black (1980) for chromosome location of active nucleolus organizer regions in meiotic pachytene cells previously subjected to FISH.
Chromosomes were classified into metacentric (m), submetacentric (sm), subtelocentric (st) and acrocentric (a) based on arms ratio (Levan et al. 1964), and were arranged in order of decreasing size within each class. For B chromosome analysis, probes of the B chromosome (B-probe) and repetitive DNAs (C o t−1 DNA and As51) were obtained as described below.
Microdissection of B chromosomes
B chromosomes were obtained by microdissection of C-banded mitotic metaphase cells, using an inverted IX51 microscope (Olympus) equipped with a Transferman mechanical micromanipulator (Eppendorf) (Vicari et al. 2010). The recovered B chromosomes were then transferred to a microtube and submitted to ‘degenerated oligonucleotide primer—polymerase chain reaction’ (DOP-PCR). The DOP-PCRs followed the procedure described by Telenius et al. (1992) with slight modifications (Vicari et al. 2010). The products were submitted to electrophoresis in 1% agarose gel, observing an expected DNA smear between 100 and 400 bp.
A second amplification reaction was performed using 100 ng of the first PCR product in a final volume of 50 μl. The reaction was made up of buffer (200 mM of Tris, pH 8.4 and 500 mM of KCl), 1× Taq DNA polymerase (Invitrogen), 2 mM of MgCl2, 400 μM of dNTP, 2 μM of DOP primer 5′-CCGACTCGAGNNNNNNATGTGG-3′ and 2 U of Taq DNA polymerase. Amplification was performed in a thermocyler (Biocycler) under the following program: 3 min at 94°C; 35 cycles of 90 s at 90°C, 90 s at 52°C and 90 s at 72°C; followed by post-cycling extension for 5 min at 72°C.
Acquisition of total repetitive DNA probe based on Cot–1 kinetic reassociation
Total genomic DNA from a specimen of A. scabripinnis without B chromosomes was used in the C o t−1 DNA procedure (Zwick et al. 1997) adapted for the acquisition of repetitive DNA in fish (Ferreira and Martins 2007; Vicari et al. 2010). Genomic DNA at a concentration of 500 ng/μl in a NaCl (0.3 mol/l) solution was autoclaved (121°C, 1,034 × 105 Pa) for 5 min to obtain fragments between 100 and 2,000 bp. Aliquots of 50 μl of fragmented DNA were then submitted to denaturation at 95°C for 10 min, placed on ice for 10 s and incubated at 65°C for 3 min for reannealing of the strips. Next, 1 U nuclease S1 (Promega), 1 × buffer [20 mM of Tris–HCl (pH 7.5 at 25°C), 0.1 mM of ZnCl2, 50 mM of NaCl and 50% (v/v) glycerol] of the enzyme were added to 1 μg of DNA and left for 8 min at 37°C. The repetitive fraction of this DNA was recovered by immediately freezing it in liquid nitrogen and performing phenol–chloroform extraction. Following precipitation, the total repetitive DNA (C o t−1-probe) was re-suspended in 20 μl of ultrapure H2O for subsequent in situ localization.
Preparation of DNA-probes
For chromosome painting with the B-probe, the DNA microdissected from the B chromosome was labelled through a DOP-PCR procedure using a modified biotin-14-dATP nucleotide (Invitrogen). The PCR mixture consisted of 1× Taq DNA polymerase buffer (Invitrogen), 2 mM of MgCl2, 40 μM of dTTP, dGTP and dCTP, 20 μM of dATTP, 20 μM of biotin-14-dATP, 2 μM of DOP primer and 2 U of Taq DNA polymerase.
Four repetitive DNA probes (As51 satellite DNA, total C o t−1, 18S rDNA and 5S rDNA) were mapped by FISH on A. scabripinnis A and B chromosomes. The As51, C o t−1 and 18S rDNA probes were labelled through nick translation using digoxigenin-16-dUTP (Dig Nick Mix—Roche) and the 5S rDNA probe was labelled with biotin through nick translation using biotin-16-dUTP (Nick Translation Biotin—Roche).
Fluorescence in situ hybridization (FISH)
Chromosome painting and FISH were performed under conditions of high stringency (2.5 ng/μl of probe, 50% formamide, 2× SSC, 10% dextran sulphate) followed the general procedure described by Pinkel et al. (1986). The detection of the signal was performed with the Avidin-FITC (Sigma) and anti-digoxigenin-rhodamine (Roche) antibodies. The chromosomes were counterstained with DAPI (0.2 μg/ml) in Vectashield mounting medium (Vector) and analysed using an epifluorescence microscope (Olympus BX41) coupled to a DP71 imaging system (Olympus).
Results
The cytogenetic analysis of the specimens of A. scabripinnis from the population of Campos de Jordão (state of São Paulo, Brazil) exhibited 2n = 50 chromosomes (6 m + 22sm + 10st + 12a) according to Salvador and Moreira-Filho (1992) for the standard complement and inter-individual variation of 0 to 1 B chromosome showing similar size and shape to the first chromosome pair, in resemblance to previous findings (Mestriner et al. 2000).
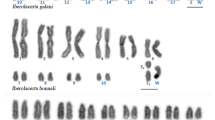
Double CP and FISH analysis performed with the B and As51 probes, respectively, on mitotic metaphase cells from B-lacking individuals showed pericentromeric and distal location of the B probe in many A chromosome pairs (Fig. 1a). The As51 satellite DNA probe was localised in 14 chromosome sites (Fig. 1c). These two probes co-located in some chromosomes, and this was confirmed by FISH in pachytene cells (e.g. pairs 1, 10, 20, 24 and B chromosome, Fig. 3). Double CP and FISH analysis with the B and As51 probes on metaphase cells from B-carrying individuals revealed that the B chromosome was completely labelled, in addition to the above mentioned pericentromeric and distal sites in some A chromosome pairs (Fig. 1b). The As51 satellite DNA was located in a large portion of the B chromosome, with the exception of the terminal and centromeric regions (Fig. 1b, d). The images overlapping showed the B probe and As51 sites in synteny (Fig. 1f).
Mitotic metaphases of A. scabripinnis submitted to double CP-FISH without and with one B chromosome (a, c, e and b, d, f, respectively). In a, b chromosome location of the B-probe and c, d location of the As51-probe. Note that the B chromosome is completely labeled with the B probe and, almost completely, with the exception of the centromeric and distal regions e merging of a and c images and f merging b and d images. Bar: 10 μm
Double CP and FISH analysis using the B and C o t−1 probes, respectively, showed that the repetitive DNA sequences contained in these probes were located in the pericentromeric and distal regions of most A chrom–osomes, and also in the B chromosome (Fig. 2c). The B-probe co-localised with the C o t−1 probe in some A chromosomes (e.g. no. 24), but not in others. Remarkably, the B-probe painted the whole B chromosome (Fig. 2a) whereas the C o t−1 probe hybridized at five sites symmetrically placed in both B chromosome arms (Fig. 2b).
Double CP and FISH analysis with the As51 (Fig. 3a, c) and B- (Fig. 3b, c) probes in pachytene cells revealed that these probes were located on one chromosome being about half the size of the largest bivalent (the first metacentric pair), thereby confirming the self-pairing of the supernumerary element (see Mestriner et al. 2000), as well as some smaller sites in bivalents no. 1, 10, 20 and 24 (Fig. 3a, b, c). The Ag-NOR staining, performed to the same preparations where double CP-FISH had previously been made, allowed identifying the bivalent showing an active NOR, i.e. no. 10 (Fig. 3d).
Double FISH analysis with 18S and 5S rDNA probes failed to reveal their presence on the B chromosome (Fig. 4). In the A chromosomes, the 18S rDNA was found in distal regions of pairs 4, 10 and in just one homologue of 16 pair in all cells analyzed (Fig. 4a), whereas the 5S rDNA was located at six proximal/interstitial sites (Fig. 4b), on chromosomes other than those carrying the 18S rDNA (Fig. 4c).
Discussion
The A. scabripinnis species complex exhibits variation in the diploid number ranging from 46 to 50 chromosomes (Moreira-Filho and Bertollo 1991). In a comparative cytogenetic analysis, Vicari et al. (2008a) have recently inferred that 2n = 50 chromosomes is plesiomorphic in the Astyanax scabripinnis complex and Robertsonian translocation rearrangements gave rise to populations with a smaller diploid number. However, B chromosomes are found in different populations of this species complex (Moreira-Filho et al. 2004) and it remains unclear whether this additional element originated once or at different times in the evolutionary history of these Neotropical fish.
Origin and meiotic behaviour of the B chromosome in Astyanax scabripinnis
The conventional analysis of gamete cells from 2n = 50 B-lacking individuals demonstrated normal meiotic behaviour, complete pairing of the 25 bivalents in pachytene cells, and formation of one to two chiasmata per bivalent in diplotene cells. In spermatogonial metaphases from B-carrying individuals, the B macrochromosome was heteropycnotic with a size similar to that of the largest bivalent (first metacentric pair). However, only one large bivalent corresponding to the first pair was observed in cells with 26 elements in pachytene cells. This suggests that the B macrochromosome is arm-to-arm autopaired at pachytene, thus showing half the size of the largest bivalent. This was confirmed by painting with the B-probe at pachytene, and supports the hypothesis that the B chromosome in A. scabripinnis is an isochromosome, proposed by Mestriner et al. (2000).
The origin of supernumerary chromosomes is most often unknown and conflicting theories have been raised at this respect. Most authors accept the hypothesis that these chromosome segments emerge from elements of the standard complement (Camacho and Cabrero 1987; Palomeque et al. 1993; Jesus et al. 2003; Artoni et al. 2006). Other theories state that the B chromosome is not derived from such elements and a heterologous origin is proposed to explain the emergence of the giant B chromosome in Alburnus alburnus, in which this chromosome is made up of dispersed retrotransposon repetitive DNA (Ziegler et al. 2003).
In A. scabripinnis, the origin of the B macrochromosome was initially attributed to events of chromosome non-disjunction based on comparative inferences of the size and morphology of the B chromosome in relation to the first chromosome pair of the standard complement (Salvador and Moreira-Filho, 1992). Currently, the most accepted theory proposes that the B chromosome in A. scabripinnis originated from the formation of an isochromosome from the acrocentric pair no. 24 (Mestriner et al. 2000; Néo et al. 2000; Moreira-Filho et al. 2004). This hypothesis is based on the presence of an interstitial heterochromatic band in the long arm of this pair, which, after the formation of the isochromosome, is believed to have given rise to interstitial As51 sites at symmetric distances in relation to the centromere in both arms of the B chromosome (Mestriner et al. 2000).
In the present study, the localisation of the B-probe revealed the highly repetitive composition of this additional genome element, which maintains similarity with small centromeric or distal regions in several chromosomes of the standard complement. Double CP-FISH with the B-probe and As51 satellite probe (As51-probe) revealed that the B chromosome has huge amounts of As51, with the exception of distal and centromeric regions. Double CP-FISH revealed co-localization of the B and As51 probes in the terminal region of the chromosomes 1, 10 (NOR bearing), 20 and 24, especially when observed at pachytene. One of these chromosomes was the acrocentric pair no. 24, which exhibited presence of the As51 DNA in an interstitial region of the long arm and hybridized with the B-probe in the pericentromeric region.
The isolation of the C o t−1 probe from a specimen with no B chromosome and its subsequent in situ localisation on A. scabripinnis chromosomes revealed pericentromeric and distal signals in several chromosomes, including the acrocentric pair no. 24 and the B chromosome. The physical mapping of these repetitive probes is consistent with B chromosome origin from chromosome no. 24 through isochromosome formation. But the present results also demonstrate that the B macrochromosome in A. scabripinnis share some DNA sequences (e.g. As51 satellite DNA and repetitive centromeric and distal DNA) with some other members of the A chromosome complement. Therefore, the precise A chromosome from which the B chromosome arose cannot be firmly inferred with the molecular cytogenetic information provided here, and it will surely be necessary to get information at DNA sequence level.
Differentiation of the B chromosome in Astyanax scabripinnis
The location of C-positive heterochromatin found in our specimens is consistent with previous descriptions (revised in Moreira-Filho et al. 2004), with pericentromeric C-bands in all chromosomes, and polymorphic terminal bands in most acrocentric chromosomes. Moreover, C banding reinforces the former conclusion that the B chromosome is completely heterochromatic and is made up mostly of As51, with the exception of the terminal and centromeric regions (data not shown). Amplification and dispersal mechanisms of repetitive sequences (e.g. As51) should have taken place in the B chromosome after its origin, since it is much richer than A chromosomes in this kind of repetitive elements.
Besides evidencing the heterogeneity of the heterochromatin contained in the B chromosome, the localisation of B chromosome and repetitive DNA probes in meiotic cells at pachytene provided interesting information on the molecular diversification of the B chromosome. First, the B chromosome arms paired each other giving rise to a structure similar to a bivalent. However, in contrast to Mestriner et al. (2000), we found no evidence for the occurrence of chiasmata in the autopaired B’s. Gray (2000) described an ectopic recombination mechanism among copies of repetitive elements dispersed in non-homologous chromosomes and therefore not dependent upon crossing-over, and Vicari et al. (2008b) used this mechanism to explain the co-localisation of repetitive As51 DNA and 18S rDNA in 14 large heterochromatic domains in Astyanax janeiroensis. Thus, this mechanism might promote heterochromatinization through the dispersion of repetitive DNA sequences, especially As51, among the self-paired arms of B chromosomes without the need for crossing-over, provided that this kind of ectopic exchanges would occur between B chromosome arms. Thus, new mutations in DNA sequences in one B chromosome arm could spread to the other arms, thereby increasing B chromosome heterochromatinization and molecular diversification in relation to the standard complement.
Distribution and chromosome localisation of rDNA in Astyanax scabripinnis
The co-localization of 18S rDNA and As51 satellite sites is a well-documented feature in the genus Astyanax. Mestriner et al. (2000) found that the NOR site on the short arm of submetacentric chromosome 10 is co-located with As51 satellite DNA in a population of A. scabripinnis from Campos de Jordão (state of São Paulo, Brazil). In the present study, in the same population, despite a huge amount of As51 satellite DNA, no rDNA site was observed by FISH on the B chromosome, and the co-localisation of 18S rDNA and As51 satellite sites was also restricted to the short arm region of chromosome 10. These results might suggest that this region of chromosome 10 in A. scabripinnis does not perform exchanges of DNA sequences with other As51 chromosome domains on non-homologous A chromosomes and the B chromosome. Alternatively, it is conceivable that the huge amounts of As51 in the B chromosome obscure small amounts of 18S rDNA that are thus imperceptible with the FISH technique. However, the presence of rDNA sites in B chromosomes has rarely been reported in fish. The only exceptions are the presence of 18S rDNA in the B microchromosome in Moenkhausia sanctaefilomenae (Dantas et al. 2007) and the B chromosome in the African ciclid Haplochromis obliquidens (Poletto et al. 2010).
In the present study, five 18S rDNA sites were located in situ in the short arms of submetacentric pair 10 and the terminal region of submetacentric pair 4, and an additional site located in a subtelecentric chromosome. The number of sites and location of 18S rDNA are generally multiple and variable in Astyanax (Néo et al. 2000; Ferro et al. 2001; Mantovani et al. 2005; Vicari et al. 2008a, b) and, at times, co-located with As51 satellite DNA (Mestriner et al. 2000; Vicari et al. 2008b). Twenty-two 18S rDNA sites were reported in A. janeiroensis, and, remarkably, 14 of them were inactive and co-located with As51 satellite DNA (Vicari et al. 2008b), suggesting a possible role of As51 in the epigenetic regulation of rDNA expression in this species. DNA transposition mechanisms have frequently been proposed to explain the high variation found in the number and location of 18S rDNA clusters in Astyanax (Fernandes and Martins-Santos 2006; Vicari et al. 2008b), although specific studies are needed to check this hypothesis.
In contrast to the 18S rDNA, chromosome localization of the 5S rDNA tends to be highly conserved in Astyanax (Martins and Galetti 2000; Mantovani et al. 2005; Vicari et al. 2008a). In this genus, 5S rDNA is generally located in the proximal region of the long arm of one acrocentric and one metacentric pair (Almeida-Toledo et al. 2002; Mantovani et al. 2005; Vicari et al. 2008a) or, in some cases, only in the acrocentric pair (Vicari et al. 2008b). However, the occurrence of more than two pairs has been described in some populations of A. scabripinnis (Ferro et al. 2001). The present study has demonstrated the occurrence of three chromosome pairs with 5S rDNA labelling. The acrocentric and metacentric chromosomes with proximal labelling are homologues to those described elsewhere (Almeida-Toledo et al. 2002; Mantovani et al. 2005; Vicari et al. 2008a) and the acrocentric pair with terminal labelling on the long arm could be considered a an apomorphy for this population.
In conclusion, our present analysis of somatic and germinal cells confirms that the B macrochromosome in A. scabripinnis arose through misdivision and isochromosome formation from an A chromosome. Our results are consistent with B origin from the acrocentric pair 24, but some uncertainty emerges from some similarities with other A chromosomes.
References
Almeida-Toledo LF, Ozouf-Costaz C, Foresti F, Bonillo C, Porto-Foresti F, Daniel-Silva MFZ (2002) Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenet Genet Res 97:229–233. doi:10.1159/000066609
Artoni RF, Vicari MR, Endler AL, Cavallaro ZI, Jesus CM, Almeida MC, Moreira-Filho O, Bertollo LAC (2006) Banding pattern of A and B chromosomes of Prochilodus lineatus (Characiformes, Prochilodontidae), with comments on B chromosomes evolution. Genetica 127:277–284. doi:10.1007/s10709-005-4846-1
Bertollo LAC, Takahashi CS, Moreira-Filho O (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Braz J Genet 1:103–120
Beukeboom LW (1994) Bewildering Bs: An Impression of the 1st B-Chromosome Conference. Heredity 73:328–336. doi:10.1038/hdy.1994.140
Camacho JPM (2005) B chromosomes. In: Gregory TR (ed) The evolution of the genome. Academic Press, New York, pp 223–286
Camacho JPM, Cabrero J (1987) New hypotheses about the origin of supernumerary chromosome segments in grasshoppers. Heredity 58:341–343. doi:10.1038/hdy.1987.60
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. R Soc 355:163–178. doi:10.1098/rstb.2000.0556
Dantas ESO, Vicari MR, Souza IL, Moreira-Filho O, Bertollo LAC, Artoni RF (2007) Cytotaxonomy and karyotype evolution in Moenkhausia Eigenmann, 1903 (Teleostei, Characidae). Nucleus 50:509–522
Fernandes CA, Martins-Santos IC (2006) Mapping of the 18S and 5S ribosomal RNA genes in Astyanax altiparanae Garutti & Britski, 2000 (Teleostei, Characidae) from the upper Paraná river basin. Brazil. Genet Mol Biol 29:464–468
Ferreira I, Martins C (2007) Physical chromosome mapping of repetitive DNA sequences in Nile tilapia Oreochromis niloticus: Evidences for a differential distribution of repetitive elements in the sex chromosomes. Micron 39:411–418. doi:10.1016/j.micron.2007.02.010
Ferro DAM, Néo DM, Moreira-Filho O, Bertollo LAC (2001) Nucleolar organizing regions, 18S and 5S in Astyanax scabripinnis (Pisces, Characidae): population distribution and functional diversity. Genetica 110:55–62. doi:10.1023/A:1017963217795
Gray YH (2000) It takes two transposons to tango: transposable element-mediated chromosomal rearrangements. Trends Genet 16:461–468. doi:10.1016/S0168-9525(00)02104-1
Howell WM, Black DA (1980) Controlled silver-staining of nucleolus organizer regions whit a protective colloidal developer: a l-step method. Experientia 36:1014–1015
Jesus CM, Galetti PM Jr, Valentini SR, Moreira-Filho O (2003) Molecular characterization and chromosomal location of two families of satellite DNA in Prochilodus lineatus (Pisces, Prochilodontidae), a species with B chromosomes. Genetica 118:25–32. doi:10.1023/A:1022986816648
Jones RN, Rees HB (1982) Chromosomes. Academic Press, New York
Kligerman AD, Bloom SE (1977) Rapid chromosome preparation from solid tissues of fishes. J Fish Res Board Can 43:266–269
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
Lima FCT, Malabarba LR, Buckup PA, Silva JFP, Vari RP, Harold A, Benine R, Oyakawa OT, Pavanelli CS, Menezes NA et al (2003) Genera Incertae Sedis in Characidae. In: Reis RE, Kullander S, Ferraris CJ Jr (eds) Check list of the freshwater fishes of South and Central America, 1st edn. Edipucrs, Porto Alegre, pp 106–169
Mantovani M, Abel LDS, Moreira-Filho O (2005) Conserved 5S and variable 45S rDNA chromosomal localization revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123:211–216. doi:10.1007/s10709-004-2281-3
Martins C, Galetti PM Jr (2000) Conservative distribution of 5S rDNA loci in Schizodon (Pisces, Anostomidae) chromosomes. Chromosome Res 8:353–355. doi:10.1023/A:1009243815280
Mestriner CA, Galetti PM Jr, Valentini SR, Ruiz IRG, Abel LDS, Moreira-Filho O, Camacho JPM (2000) Structural and functional evidence that a B chromosome in the characidae fish Astyanax scabripinnis an isochromosome. Heredity 85:1–9. doi:10.1046/j.1365-2540.2000.00702.x
Moreira-Filho O, Bertollo LAC (1991) Astyanax scabripinnis (Pisces, Characidae): A species complex. Braz J Genet 14:331–357
Moreira-Filho O, Galetti PM Jr, Bertollo LAC (2004) B chromosomes in the fish Astyanax scabripinnis (Characidae, Tetragonopterinae): An overview in natural populations. Cytogenet Gen Res 106:230–234. doi:10.1159/000079292
Néo DM, Moreira-Filho O, Camacho JPM (2000) Altitudinal variation for B chromosome frequency in the fish Astyanax scabripinnis. Heredity 85:136–141. doi:10.1046/j.1365-2540.2000.00744.x
Palomeque T, Chica E, Díaz de La Guardia R (1993) Supernumerary chromosome segments in different genera of Formicidae. Genetica 90:17–29. doi:10.1007/BF01435174
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938
Poletto AB, Ferreira IA, Martins C (2010) The B chromosomes of the African cichlid fish Haplochromis obliquidens harbour 18S rRNA gene copies. BMC Genetics 11:2010. doi:10.1186/1471-2156-11-1
Salvador LB, Moreira-Filho OB (1992) chromosomes in Astyanax scabripinnis (Pisces, Characidae). Heredity 69:50–56. doi:10.1038/hdy
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 74:304–306
Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725. doi:10.1016/0888-7543(92)90147-K
Vicari MR, Noleto RB, Artoni RF, Moreira-Filho O, Bertollo LAC (2008a) Comparative cytogenetics among species of the Astyanax scabripinnis complex. Evolutionary and biogeographical inferences. Genet Mol Biol 31:173–179. doi:10.1590/S1415-47572008000200002
Vicari MR, Artoni RF, Moreira-Filho O, Bertollo LAC (2008b) Colocalization of repetitive DNAs and silencing of major rRNA genes. A case report of the fish Astyanax janeiroensis. Cytogenet Gen Res 122:67–72. doi:10.1159/000151318
Vicari MR, Nogaroto V, Noleto RB, Cestari MM, Cioffi MB, Almeida MC, Moreira-Filho O, Bertollo LAC, Artoni RF (2010) Satellite DNA and chromosomes in Neotropical fishes: methods, applications and perspectives. J Fish Biol 76:1094–1116. doi:10.1111/j.1095-8649.2010.02564.x
Ziegler CG, Lamatsch DK, Steinlein C, Engel W, Schartl M, Schmid M (2003) The giant B chromosome of the cyprinid fish Alburnus alburnus harbours a retrotransposon-derived repetitive DNA sequence. Chromosome Res 11:23–25. doi:10.1023/A:1022053931308
Zwick MS, Hanson RE, McKnight TD, Islam-Faridi HM, Stelly DM, Wing RA, Price JH (1997) A rapid procedure for the isolation of C0t–1 DNA from plants. Genome 40:138–142. doi:10.1139/g97-020
Acknowledgments
The authors are grateful to the Brazilian environmental protection agency IBAMA (Instituto Brasileiro do Meio Ambiente e Recursos Naturais Renováveis) for authorization to catch the specimens in nature (IBAMA/MMA/SISBIO license number 15117). The study was funded by the Brazilian fostering agencies CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico; process number 473448/2007-6), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; process number 087/2007), FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and the Araucária Foundation (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná; process number 17217/2009).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vicari, M.R., de Mello Pistune, H.F., Castro, J.P. et al. New insights on the origin of B chromosomes in Astyanax scabripinnis obtained by chromosome painting and FISH. Genetica 139, 1073–1081 (2011). https://doi.org/10.1007/s10709-011-9611-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-011-9611-z