Abstract
The Antillean manatee (Trichechus manatus manatus) occupies the tropical coastal waters of the Greater Antilles and Caribbean, extending from Mexico along Central and South America to Brazil. Historically, manatees were abundant in Mexico, but hunting during the pre-Columbian period, the Spanish colonization and throughout the history of Mexico, has resulted in the significantly reduced population occupying Mexico today. The genetic structure, using microsatellites, shows the presence of two populations in Mexico: the Gulf of Mexico (GMx) and Chetumal Bay (ChB) on the Caribbean coast, with a zone of admixture in between. Both populations show low genetic diversity (GMx: NA = 2.69; HE = 0.41 and ChB: NA = 3.0; HE = 0.46). The lower genetic diversity found in the GMx, the largest manatee population in Mexico, is probably due to a combination of a founder effect, as this is the northern range of the sub-species of T. m. manatus, and a bottleneck event. The greater genetic diversity observed along the Caribbean coast, which also has the smallest estimated number of individuals, is possibly due to manatees that come from the GMx and Belize. There is evidence to support limited or unidirectional gene flow between these two important areas. The analyses presented here also suggest minimal evidence of a handful of individual migrants possibly between Florida and Mexico. To address management issues we suggest considering two distinct genetic populations in Mexico, one along the Caribbean coast and one in the riverine systems connected to the GMx.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
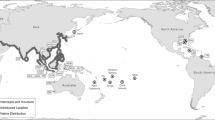
The National Manatee Conservation Program, implemented in 2001, divided Mexico’s manatee (Trichechus manatus manatus) population into three management units (MU) (SEMARNAT 2001) (Fig. 1). These MUs were based on the geographic distribution of the manatees, the different regional threats, local research capacities, regional socio-cultural factors, and similarities in habitat characteristics including water quality and availability that has led to a protection plan for the region (SEMARNAT 2001). An understanding of the manatee population structure and diversity in Mexico, based on population genetics, will provide a valuable tool to improve the scientific information lending support for management actions, such as the new National Manatee Conservation Action Plan for Mexico (PACE manati). That plan is expected to be adopted this year (2011). In Mexico, manatees have been protected since 1922 (Diario Oficial 01/20/1922), listed as a threatened species in 1991 under Federal Law (SEDUE 1991) and since 1994 has been classified as a species at risk of extinction by the Mexican Government (SEMARNAT 2002). At the international level, Antillean manatees were classified as vulnerable by the International Union for Conservation of Nature (IUCN) Red List from 1982 to 2007 and endangered since 2008 (Deutsch et al. 2008). West Indian manatees have also been listed as an endangered species by the Convention on International Trade in Endangered Species (CITES 2009) since 1975.
The Antillean manatee occupies the tropical coastal waters of the Greater Antilles and Caribbean, extending from Mexico along Central and South America to Brazil (Husar 1978; Lefebvre et al. 1989, 2001). The distribution of manatees in Mexico includes the coastal and wetland systems of the Gulf of Mexico (GMx) and the Caribbean coast (Fig. 1). The majority of manatees inhabit the wetland systems of Veracruz, Tabasco, Chiapas and Campeche states along the GMx, where a conservative estimate of population size is between 500 and 1,500 manatees (Olivera-Gómez 2006). It is estimated that approximately 200 to 250 manatees reside in Quintana Roo region (Morales-Vela and Padilla-Saldívar 2011) and much fewer reside within the Yucatan state (Morales-Vela et al. 2003). In Quintana Roo, most manatees occur in Chetumal Bay (ChB) with an estimate of about 100 to 150 manatees (Morales-Vela and Padilla-Saldivar 2009a). In Ascencion Bay (AB) the estimate is smaller, with approximately 25 manatees counted during a single aerial survey in July of 2009 (Landero-Figueroa 2010). ChB and AB are located approximately 335 km apart (shore distance). North of the Yucatan Peninsula no estimates have been provided and manatee abundance is presumed to be very low (Morales-Vela et al. 2003).
Previous genetic studies on the West Indian manatee suggest that their mitochondrial DNA (mtDNA) variability ranges between one (Florida) to as many as eight haplotypes (Colombia) per country (Garcia-Rodriguez et al. 1998; Vianna et al. 2006). Genetic studies of manatees in Mexico, from Tabasco to Quintana Roo indicate differences among the populations. The biggest difference is between the GMx, where only one haplotype (J) is found, and the Caribbean coast where three haplotypes (J, A, A4) have been identified (Medrano-González et al. 1997; Castañeda-Sortibrán unpublished data). However, the small sample size from the GMx (n = 4) at the time of this mtDNA study was not sufficient to provide significant results.
Historically, manatees were abundant in Mexico from Tamaulipas to the Yucatan Peninsula, but hunting for food during the pre-Columbian period by Mayans (McKillop 1985), the Spanish colonization and throughout the history of Mexico (Durand 1983), has resulted in the significantly reduced population occupying Mexico today. Even as recently as 1983, it was reported that manatees were occasionally harpooned in the region north of Quintana Roo as a source of food (Gallo-Reynoso 1983). Direct hunting is rare now in the GMx, but manatees caught in nets are still sometimes killed for food, with children reporting that they had eaten fresh manatee meat recently (Olivera-Gómez 2006). Fishermen have also declared that the manatee population decline in the Northern and Western coasts of the Yucatan Peninsula was caused by a number of factors, including hunting for local consumption, entanglement as a result of higher net-fishing activities in rivers, and habitat destruction and coastal construction due to human population growth and hurricane impacts (Morales-Vela et al. 2003). The last documented manatee hunting activities on the Northeast coast of the Yucatan Peninsula occurred in the 1960–1970s, but since then opportunistic poaching has continued into the 1990s (Morales-Vela et al. 2003). In the GMx other factors such as natural or artificial closing of drainages from freshwater ecosystems have restricted manatee access to vegetation, increased water temperature, and have resulted in increases in pollution, sedimentation and eutrophication due to agricultural and poultry runoff (Olivera-Gómez 2006).
The high level of anthropogenic and habitat destruction pressure on the manatee population may have caused a population bottleneck, which is known to reduce genetic diversity. Typically, habitat loss and degradation increase the rate of fragmentation resulting in the isolation of small populations and leads to an increased probability for inbreeding and unstable demographics (Frankham et al. 2002).
Herein, we present a fine-scale population structure of manatees in Mexico using analysis of microsatellite DNA markers. The genetic diversity and population structure were compared to a geographically close conspecific, the Florida manatee (Trichechus manatus latirostris), which has a larger estimated population size. The results of this microsatellite DNA marker study were compared with those obtained previously from an unpublished Mexican mtDNA study.
Methods
Microsatellite DNA amplification and fragment analysis
We analyzed 94 samples from different regions of Mexico including the Peninsula of Yucatan MU (ChB: 51, AB: 15), the central GMx MU (Tabasco: 15, Chiapas: 5 and Campeche: 1) and the north of the GMx MU (Veracruz: 7) (Fig. 1). Blood or skin tissue from the tail was collected from wild manatees captured for health assessment and radio tagging studies. Skin tissue was collected from carcasses recovered by manatee research projects throughout Mexico. Blood from captive manatees was also utilized for this study because their original rescue location was known. To resolve population structure, we included the genotypes of 95 individuals from Florida based on the estimated population size in each of the four recognized MU’s in that state (Pause 2007).
Blood and tissue samples were preserved with lysis or tissue buffer respectively (lysis buffer: 100 mM Tris–HCl, 100 mM EDTA, 10 mM NaCl, 1.0% SDS (White and Densmore 1992); SED tissue buffer: saturated NaCl; 250 mM EDTA pH 7.5; 20% DMSO (Amos and Hoelzel 1991; Proebstel et al. 1993)). DNA extractions, amplifications and fragment analysis were performed at the University of Florida, ICBR Genetic Analysis Laboratory in Gainesville, Florida, USA and at the US Geological Survey, Southeast Ecological Science Center Conservation Genetics Laboratory in Gainesville, FL, USA.
DNA extractions were carried out using either a standard phenol–chloroform protocol (Hillis et al. 1990) or the DNeasy tissue extraction kit (Qiagen, Valencia, CA, USA). Polymerase chain reaction (PCR) amplifications were performed for each sample for each of 13 microsatellite loci previously designed for manatees: TmaA02, TmaE02, TmaE08, TmaE11, TmaE26, TmaF14, TmaM79 (Garcia-Rodriguez et al. 2000), TmaSC13, TmaE7, TmaH13, TmaE14, TmaK01, TmaJ02 (Pause et al. 2007). Amplifications were performed in Biometra UNOII, UNO-Thermoblock, T-Gradient thermocyclers (Biometra®, Göttengen, Germany) or a PTC-200 (MJ Research, Waltham, MA) thermocycler using the following conditions: 95°C for 5 min, 35 cycles of 95°C for 30 s, with the specific annealing temperature as listed in the original publication for each primer, with the exception of TmaM79 = 54°C; TmaA02 = 56°C; TmaE02, TmaE11, TmaE26 and TmaF14 = 58°C; TmaE08 = 60°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. Amplifications were performed in a total volume of 15 μL, with 10 ng target DNA, 1× Sigma PCR Buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl, 0.001% gelatin), MgCl2 as indicated in the original publications, 0.2 mM each dNTP, 0.04 units of Sigma JumpStart Taq polymerase (Sigma–Aldrich, St. Louis, MO, USA), 0.25 μM each primer, and bovine serum albumin (BSA), where indicated (Garcia-Rodriguez et al. 2000; Pause et al. 2007). For fragment analysis, the forward primers were labeled with the fluorescent dyes HEX or 6-FAM for processing and visualization on an ABI 3730xl Automated DNA Analyzer. Fragment data from the PCR products were collected from the ABI 3730xl and analyzed using GeneMarker 1.5 (SoftGenetics 2008) to determine allele sizes. Allele sizes were standardized using previously analyzed Florida samples as the baseline.
Data analysis
Convert (Glaubitz 2004) was used to convert the data into different input file formats. We used Structure, version 2.3.1, (Pritchard et al. 2000) to identify possible subpopulation designations, without an a priori assignment of the overall population structure in Mexico. The data from Mexico and Florida were analyzed together. The admixture model was used and the number of populations (K) was set from 1 to 10 with a burn-in period of 100,000 iterations, followed by 1,000,000 Monte Carlo Markov Chain iterations. Five independent analyses were simulated for each value of K. The value of K with the lowest posterior probability was identified as the optimum number of subpopulations, as recommended by the Structure manual. Geneclass2 (Piry 2004) and Whichrun version 4.1 (Banks and Eichert 2000) were used to test individuals attributed to a different population than the geographic location assigned by Structure. GenAlEx 6.2 (Peakall and Smouse 2006) was used to calculate genetic distance between individuals which were used on a Principal Coordinate Analysis (PCA).
Estimates of the effective population size were made by NeEstimator using the Linkage Disequilibrium algorithm (Peel et al. 2004). GenAlEx 6.2, GenePop 3.4 (Raymond and Rousset 1995; Rousset 2008) and Arlequin 3.1 (Excoffier et al. 2005) were used to compare and determine the genetic diversity and genetic differentiation including allelic richness NA and NE, heterozygosity observed (Ho) and expected (He), estimate of population subdivision FST and RST and inbreeding coefficients (FIS). Arlequin was used to check for deviation from Hardy–Weinberg equilibrium. GenePop webversion was used to examine for Linkage Disequilibrium. The presence of null alleles was analyzed with Micro-checker (Oosterhout et al. 2004). The presence of a potential bottleneck was estimated using Bottleneck examining the heterozygosity excess (Cornuet and Luikart 1996).
Results
Results from the three different software packages (GenAlEx, Genepop, and Arlequin) were compared and similar results were observed for genetic diversity and population structure as had been previously published. Therefore, only the results from GenAlEx are presented in Tables 1 and 3.
Population structure
Results for the Structure analysis identified that k = 3 was the appropriate number of clusters (Fig. 2) based on the manual’s recommendation of the lowest posterior probability LnP(D). Most individuals were assigned to a cluster with Q > 80%. In all simulations, Florida manatees were assigned to a separate population cluster (92.8% assignment) and the GMx and ChB populations were assigned to a cluster that corresponded to their geography (89.0 and 84.6% respectively). Samples collected from AB were not as clearly delineated, and were clustered with either the GMx (56.1%) or ChB (41.6%).
Proportions of ancestry for individuals were assessed without a priori information using Bayesian clustering via Structure. This graphic represents the best fit of the data, where three population clusters are clearly distinguished (K = 3). Manatees from Florida consistently grouped separately from the manatees from Mexico. Manatees from the Chetumal Bay (ChB) cluster together and those from the Gulf of Mexico (GMx) river systems clustered together. Samples from Ascencion Bay (AB) suggest it may be a mixing zone for the Mexican manatee populations
The Structure analysis suggests that there were some individuals that may have had mixed ancestry based on this analysis. Interestingly, some of the AB individuals with a high percentage of ancestry corresponding to the GMx cluster do not share the GMx’s unique haplotype. Instead, these individuals’ haplotypes corresponded to the AB/ChB regions. Four manatees sampled from ChB show high percent ancestry with the Florida cluster (between 59% and 76%). One of them was a female manatee carcass sampled in ChB which shared a 59% ancestry with the Florida population. This manatee died of natural causes (birthing complications) and her calf was not recovered. Another was a female juvenile who shared 75.6% ancestry with Florida but her haplotype did not correspond with the Florida haplotype (Castañeda-Sortibrán pers. comm). One male manatee, who was rescued as a calf in March 2002 and is now in captivity in Veracruz, also shares a high percent ancestry with Florida samples (51%). One female manatee from Veracruz shares ancestry with the GMx samples (44%) and ChB region samples (47%). Two manatees from Florida share a high percent ancestry (45 and 60%) with the GMx individuals. Results from Geneclass2 and Whichrun analysis corroborate these assignments to populations other than their geographic location (Table 1). The Veracruz samples have lower ancestry values and cannot be totally attributed to Florida or the GMx as they appear to be mixtures of both populations on Geneclass2 and Whichrun analysis (Table 1).
The pairwise FST value between ChB and AB is low but significant (FST = 0.047), and RST is not significant (RST = 0.016) (Table 2). All pairwise FST and RST values are presented in Table 2. Significant levels of subdivision were observed between the GMx and all other regions with FST and RST, and among all regions with FST. All values were significant when calculated between the three regions: the Caribbean coast (including ChB and AB), the GMx and Florida (data not show) but most of RST values were not significant when calculated by separating the Caribbean coast into two areas: ChB and AB and compared with the two other geographic regions: the GMx and Florida (Table 2). A higher level of differentiation (via FST) was observed between ChB and the GMx than between ChB and Florida. This does not correspond to the Structure analysis, which consistently grouped Florida into its own cluster, separate from ChB. A handful of individuals had microsatellite genotypes that clustered with Florida, but overall, the group is distinct from Florida. This can also be observed in the PCA from genetic distance that shows separate clusters for Florida and Mexico (Fig. 3). The null hypothesis tested for heterozygosity excess using the Wilcoxon test, with Bottleneck software, provided (P = 0.0004 and P = 0.07) probabilities under Two-Phase mutation (TPM) and Step-wise mutation (SMM) respectively, with a normal L-shaped distribution for ChB population. For the GMx and AB populations the Wilcoxon test indicates a probability of 0.40 (TPM) and 0.17 (SMM) with a shifted mode distribution for the GMx and 0.046 (TPM) and 0.10 (SMM) with a shifted mode distribution for AB. For Florida, the Wilcoxon test shows a probability of 0.02 (TPM) and 0.55 (SMM) with a normal L-shaped distribution.
Genetic diversity
No evidence of null alleles was identified in either the Mexico or Florida populations. After 78 comparisons and a sequential Bonferroni correction, no linkage disequilibrium was observed (overall α = 0.05). All loci were in Hardy–Weinberg equilibrium (HWE) after a sequential Bonferroni correction for all Mexican populations but TmaE02 was not within HWE for Florida. The error rate was determined by re-genotyping 12% of the samples. No errors were detected. Estimates of the mean number of alleles, effective number of alleles, and heterozygosity expected and observed, FIS, estimated population size and effective size for each determinate population (ChB, AB, the GMx and Florida) are presented in Table 3. Private alleles were found in Florida (n = 16), ChB (n = 2) and AB (n = 4).
Discussion
The GMx, with extended riverine and lagoon systems in Tabasco, Veracruz, Chiapas and Campeche, is a very favourable habitat for manatees. It has abundant vegetation, fresh water and areas where manatees can evade hunters. The Caribbean coast also represents very suitable habitat, especially ChB and AB, which are protected areas. ChB is the largest marine protected area at the state level in Mexico and is shared with Belize. AB is part of the Biosphere Reserve of Sian Ka’án, one of the largest Federal marine reserves in Mexico.
Manatees are known to travel long distances along coastlines with suitable habitat (e.g. a Florida manatee was observed as far north as Rhode Island), as well as infrequent travel across open ocean (Alvarez-Alemán et al. 2010; Deutsch et al. 2003; Fertl et al. 2005; Reep and Bonde 2006). Using existing tools, genetic analyses of Florida manatees have suggested little population structure throughout Florida (McClenaghan and O’Shea 1988; Garcia-Rodriguez et al. 1998; Pause pers. com.). Although manatees are capable of travelling along the shoreline between the GMx and the Caribbean coast, seasonal migration in response to cold weather is not necessary for survival in Mexico as it is in Florida.
Radio tagging studies in ChB illustrated movement by five males and one female along the shoreline between discrete manatee habitats in ChB and Belize (Morales-Vela et al. 2007). One of these males was also observed participating in a mating group in Belize (Auil-Gomez pers. comm). The movement pattern from ChB to Belize was very similar as manatees made directed, continuous moves along the shoreline between discrete manatee habitats (Morales-Vela et al. 2007). None of the 19 manatees tagged with Argos-linked GPS radio tags, either from ChB or AB showed movements north of the initial tagging site (Morales-Vela and Padilla-Saldívar 2009b). The natural morphological features of ChB, oriented in the south of Mexico, easily connect ChB with both the Caribbean Sea and coastal Belize. An extensive coral reef barrier forms a natural marine corridor that may promote easier movement of the manatees between ChB to the south along the coast of Belize, than to the north of the Yucatan Peninsula. The Caribbean group is represented by ChB and AB, with a significant but low FST between these two locations. AB appears as a mixture of the southern portion of the Caribbean coast (ChB) and the GMx manatees, which is why the FST between ChB and AB was significant but low (Fig. 2). It seems that migrants from the GMx breed in AB and do not continue south towards ChB. Mothers and calves are often observed in AB, which hosts suitable and abundant habitat for manatees. The mixture found in AB with the presence of individuals with both ancestral roots was not observed in the GMx region. The higher genetic diversity found in the Caribbean coast when compared to the GMx are consistent with results from the mtDNA data. Analyses of mtDNA analysis identified three haplotypes along the Caribbean coast, one common to Florida, another in common with Belize and the last shared a haplotype unique to the GMx (Castañeda-Sortibrán unpublished data). The Caribbean Cluster is geographically closer to Florida (approximately 800 km) than to the GMx (approximately 1,150 km).
A high level of genetic diversity in a population is thought to increase the probability of surviving a catastrophic event (Frankham et al. 2002). In this study, the mean number of observed alleles was very low and corresponds to less than what is predicted for wildlife populations that have been hunted or have been significantly fragmented (DiBattista 2008). However, these data describing low variability are consistent with other West Indian manatee populations throughout their range (Hunter et al. 2010, Pause pers. comm.). The lower genetic diversity, based on the number of alleles and heterozygosity found in the GMx, is probably due to a combination of a founder effect and a bottleneck event. This is likely due to manatees occupying the northern range of the subspecies and the extensive historical exploitation experienced during and since the pre-Columbian period up to the 1960–1970s. That depletion of manatee stocks resulted in the reduced population numbers observed today (Durand 1983; McKillop 1985; Medrano-González et al. 1997). The population with the greatest genetic diversity observed along the Caribbean coast also has the smallest estimated number of individuals (Table 3). This is possibly due to manatees that come from the GMx and Belize. Some breeding contribution of manatees from Belize is suggested by recent radio tagging data (Morales-Vela and Padilla-Saldívar 2009b) and by the presence of shared mtDNA haplotypes (Vianna et al. 2006). The proximity of the large Belize manatee population, only 200–300 km south of ChB, may explain the dispersal route of the Caribbean coast manatees. Having a large population close by with an easy access route, makes it easier for the manatees to travel south than to try to make the longer journey to the GMx. There is evidence from radio tagging studies where some male manatees move from ChB to south Belize and return (Morales-Vela et al. 2007). However mtDNA shows different matrilineal trends in haplotype dispersal patterns in Belize and Mexico (Vianna et al. 2006) indicating that ChB and the Belize populations are not one well mixed population but have persisted independently over time. The genetic diversity along the Caribbean coast may have been recently influenced by potential migration from the growing manatee population in Florida as suggested by the few individuals that share remote ancestry with the Florida population.
The Belize population shows a stronger separation from Florida (FST = 0.141) (Hunter et al. 2010) than the Mexican populations, with FST = 0.096 between the Mexican Caribbean coast and Florida and FST = 0.106 between the GMx population and Florida. Belize and the Caribbean coast show a different pattern of haplotype distribution with two haplotypes in common and one private haplotype. The Florida haplotype, which is absent in Belize, yet is present in Mexico, confirms the closer relationship between Florida and Mexico than between Florida and Belize as confirmed by microsatellite data.
Current collaborative studies utilizing samples from Mexico, Belize, Puerto Rico, Florida and other Caribbean countries will be completed in 2011–2012 to further assess the phylogeography and dispersal patterns of West Indian manatees. The effective population size is lower than the estimated population size for manatees in Mexico, which may be a reflection of the reduction in population size due to prior hunting pressure. The low Ne of the manatee population in the GMx may be due to a founder effect resulting in low genetic diversity, recent bottleneck events, and migratory limitations due to climate change and glacial periods. This could also be an artefact of the small sample size for the GMx. More samples are urgently needed from this area.
Manatees in the GMx population tend to stay within the inland river systems, not in the coastal waters, and the group is likely fragmented by urban and agricultural development. An area of 140 km in the northeastern region of the Yucatan Peninsula is unsuitable habitat for manatees due to urban development, and the lack of vegetation and fresh water sources, resulting in a likely barrier to gene flow between the GMx and Caribbean coast (Medrano-González et al. 1997; Morales-Vela et al. 2003). Close to this zone, there are also significant coastal fishing activities and poaching (Morales-Vela et al. 2003). Examination of oceanographic structures shows a differentiation between the GMx and the Yucatan Peninsula (Laura Carrillo, pers. comm.; Merino 1997). This barrier may explain the low gene flow between the GMx and the Caribbean coast. Nevertheless, a group of six large manatees were observed in 2006 along the northeast coast of the Yucatan Peninsula (Reyes-Mendoza and Morales-Vela 2007). It is not known if these manatees were transient or resident.
The analyses presented here suggest minimal evidence of a handful of individual migrants possibly between Florida and Mexico. One Florida manatee and six potential second generation migrants from Florida were detected in Mexico, which raises the question of whether migration or breeding occurs between the two subspecies located in Florida and Mexico. Two manatees from Florida share a high percent ancestry (45 and 60%) with the GMx individuals suggesting individuals may be second generation migrants. One of these manatees was a female with a known record of Florida maternal lineage from photo-identification. She was born in the Naples, FL area in 1995 and captured in Port of the Islands, FL in February 2001. The other was the carcass of a male calf found in August 1997 in the Banana River on the east coast of Florida (Beck, USGS, pers. comm.). One possibility is that migrants could be from Cuban waters; however, no genetic data are currently available from manatees in Cuba. Major currents go from Mexico to Florida and travel close to Cuba. Events such as tropical storms and hurricanes can change the currents and can separate manatees from the coast, taking them offshore and subject to drift (Langtimm and Beck 2003). In that way, currents may also allow manatees to travel to Mexico from Florida. Manatees are able to survive short open water travels, and adults have no major natural predators so they will be able to survive incidental open sea travel. For example a known long-term resident female from the Florida manatee photo-identification catalog was sighted in Cuba in 2007 (Alvarez-Alemán et al. 2010). This manatee successfully travelled across open sea illustrating potential for population expansion. Reports of Florida manatees in the northern GMx indicate that they also can travel as far west as the southern Texas coast following the shoreline (Laguna Madre and Rio Grande) (Fertl et al. 2005). Historical distribution of manatees in Tamaulipas, Mexico which is north of the GMx shows a possible link that may exist between the GMx and Florida for manatees to travel (Lazcano-Barrero and Packard 1989). This leads to the question of successful reproduction between the two subspecies (T. m. manatus and T. m. latirostris) and the possible positive (increased variability, heterosis) or negative consequences (outbreeding depression) of breeding.
Conclusion/conservation application
The genetic structure of manatees in Mexico indicates two clusters with gene flow from the GMx to the Caribbean coast with no migration from the Caribbean back towards the GMx. This movement pattern could not be detected using mitochondrial DNA as there is only one haplotype (J) present in the GMx which is also present in the Caribbean. The three haplotypes found in the Caribbean are also found in the GMx, Florida and Belize. The individuals from AB appear as a mixture of the populations from ChB and the GMx. Additional evidence should be collected to determine the connectivity of other Central American populations of T. m. manatus, as well as the connectivity between the two subspecies.
To address management issues, we suggest considering two distinct genetic populations in Mexico, one along the Caribbean coast and one in the riverine systems connected to the GMx. There is evidence to support unidirectional or limited gene flow between these two important areas. Special attention for conservation in the AB population would result in a healthier genetic stock, as this region is a source for potential mixture between the two primary genetic clusters.
Based on this information it is important to maintain the natural migration routes of manatees from the GMx to the Caribbean area. This can be accomplished by conserving suitable manatee habitat along the north and east coasts of the Yucatan Peninsula, enforcing protection, reducing poaching and helping local initiatives that increase public education. Ongoing tourism and urban development projects along those coasts are a major issue of concern.
All of the captive manatees in Mexico were from the GMx region where they were originally rescued. However, most of them are in facilities located along the Caribbean coast. In the event of their release, returning them to the GMx would help maintain distinct populations. Also, if these facilities continue breeding manatees in captivity, a program that minimizes inbreeding and prevents inbred individuals or manatees with parents from different genetic populations from being released into the wild population must be considered. Genetic tools to assess lineages are currently being evaluated (Nourisson 2011).
References
Alvarez-Alemán A, Beck CA, Powell JA (2010) First report of a Florida manatee (Trichechus manatus latirostris) in Cuba. Aquat Mamm 36(2):148–153
Amos B, Hoelzel AR (1991) Long-term preservation of whale skin for DNA analysis. Rep Int Whal Comm Special Issue 13:99–103
Banks MA, Eichert W (2000) WHICHRUN (Version 3.2): a computer program for population assignment of individuals based on multilocus genotype data. J Hered 91:87–89
CITES (2009) Appendices I, II and III. In: The CITES Appendices. Convention on International Trade in Endangered Species of Wild Fauna and Flora. http://www.cites.org/eng/app/appendices.shtml. Accessed 29 Oct 2009
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for inferring recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Deutsch CJ, Reid JP, Bonde RK, Easton DE, Kochman HI, O’Shea TJ (2003) Seasonal movements, migratory behavior and site fidelity of West Indian manatees along the Atlantic Coast of the United States. Wildlife Monographs 151:1–77
Deutsch CJ, Self-Sullivan C, Mignucci-Giannoni A (2008) Trichechus manatus. In: The IUCN Red List of Threatened Species Version 2009.1. International Union for Conservation of Nature and Natural Resources. http://www.iucnredlist.org/details/22103/0. Accessed 29 Oct 2009
DiBattista JD (2008) Patterns of genetic variation in anthropogenically impacted populations. Conserv Genet 9(1):141–156
Durand J (1983) Ocaso de sirenas, esplendor de manatíes, 2nd edn. Fondo de cultura económica, México
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fertl DA, Schiro J, Regan GT, Beck CA, Adimey N, Price-May L, Amos A, Worthy GAJ, Crossland R (2005) Manatee occurrence in the northern Gulf of Mexico, West of Florida. Gulf Caribb Res 17:69–94
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Gallo-Reynoso JP (1983) Notas sobre la distribución del manati (Trichechus manatus) en las costas de Quintana Roo. An Inst Biol Univ Nac Auton Mex 53(1982), Ser Zool (1):443–448
Garcia-Rodriguez AI, Bowen BW, Domning D, Mignucci-Giannoni AA, Marmontel M, Montoya-Ospina RA, Morales-Vela B, Rudin M, Bonde RK, McGuire PM (1998) Phylogeography of the West Indian manatee (Trichechus manatus): how many populations and how many taxa? Mol Ecol 7:1137–1149
Garcia-Rodriguez AI, Moraga-Amador D, Farmerie W, McGuire P, King TL (2000) Isolation and characterization of microsatellite DNA markers in the Florida manatee (Trichechus manatus latirostris) and their application in selected Sirenian species. Mol Ecol 9:2161–2163
Glaubitz JC (2004) CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Hillis DM, Larson A, Davis SK, Zimmer EA (1990) Nucleic acids III: sequencing. In: Hillis DM, Moritz C (eds) Mol Syst. Sinauer Associates, Sunderland, pp 318–370
Hunter ME, Auil-Gomez NE, Tucker KP, Bonde RK, Powell J, McGuire PM (2010) Low genetic variation and evidence of limited dispersal in the regionally important Belize manatee. Anim Conserv 13:592–602
Husar LS (1978) Trichechus manatus. Mamm Sp 93:1–5
Landero-Figueroa MM (2010) Distribución potencial del manatí (Trichechus manatus manatus) en Bahía de la Ascensión, Quintana Roo. Master dissertation. CINVESTAV-Merida, Mexico
Langtimm CA, Beck CA (2003) Lower survival probabilities for adult Florida manatees in years with intense coastal storms. Ecol Appl 13:257–268
Lazcano-Barrero MA, Packard JM (1989) The ocurrence of manatees (Trichechus manatus) in Tamaulipas, Mexico. Mar Mamm Sci 5(2):202–205
Lefebvre LW, Marmontel M, Reid JP, Rathbun GB, Domning DP (2001) Status and biogeography of the West Indian manatee. In: Woods CA, Sergile FE (eds) Biogeography of the West Indies: patterns and perspectives, 2nd edn. CRC Press, Boca Raton, pp 425–474
Lefebvre LW, O'Shea TJ, Rathbun GB, Best RC (1989) Distribution, status, and biogeography of the West Indian manatee. Biogeography of the West Indies:567–610
McClenaghan LR, O’Shea TJ (1988) Genetic variability in the Florida manatee (Trichechus manatus). J Mamm 69:481–488
McKillop HI (1985) Prehistoric exploitation of the manatee in the Maya and circum-Caribbean areas. World Archaeol 16:337–353
Medrano-González L, Morales-Vela B, García-Rodríguez A, Robles-Saavedra R, Baker S (1997) Análisis preliminar de la variación del DNA mitocondrial y del complejo mayor de histocompatibilidad en la laguna de Catazajá, Chiapas, y en la bahía de Chetumal, Quintana Roo. In: Morales-Vela B, Medrano-González L (eds) Variación genética del manatí (Trichechus manatus), en el sureste de México y monitoreo con radiotransmisores en Quintana Roo. Informe final. El Colegio de la Frontera Sur, Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (H164), Chetumal, pp 12–38. http://www.conabio.gob.mx/institucion/proyectos/resultados/InfH164.pdf.pdf. Accessed 29 Oct 2009
Merino M (1997) Upwelling on the Yucatan Shelf: hydrographic evidence. J Mar Syst 13:101–121
Morales-Vela B, Padilla-Saldivar JA (2009a) Aspectos biológicos de los manatíes en el sur de Quintana Roo. In: Espinoza-Ávalos J, Islebe GA, Hernández-Arana HA (eds) El sistema ecológico de la bahía de Chetumal/Corozal: costa occidental del Mar Caribe. El Colegio de la Frontera Sur. Chetumal, Quintana Roo, pp 115–123. http://w2.ecosur-qroo.mx/cna/julio/libbahia.pdf. Accessed 20 Feb 2010
Morales-Vela B, Padilla-Saldívar JA (2009b) Demografía, ecología y salud de la población de manatíes (Trichechus manatus manatus) en Quintana Roo, y su variación y representación genética en México. Informe Técnico Final Proyecto SEMARNAT/CONACYT 2002-C01-1128. El Colegio de la Frontera Sur, Chetumal, 259 p
Morales-Vela B, Padilla-Saldívar JA (2011) El manatí. La sirena del Caribe. In: Pozo C, Armijo-Canto N, Calmé S (eds) Riqueza biológica de Quintana Roo. Un análisis para su conservación. Tomo 1. ECOSUR, CONABIO, Gob. Edo. Q. R., PPD, México, pp 248–255
Morales-Vela B, Padilla-Saldívar JA, Mignucci-Giannoni AA (2003) Status of the manatee (Trichechus manatus) along the northern and western coasts of the Yucatán Peninsula, México. Caribb J Sci 39(1):42–49
Morales-Vela B, Padilla-Saldívar JA, Reid J, Butler S (2007) First records of long-distance manatee movements between Mexico and Belize. Sirenews 47:12–14
Nourisson C (2011) Estructura genética de los manatíes en México. Tesis de doctorado, El Colegio de la Frontera Sur, Chetumal, Quintana Roo, México. 121pp
Olivera-Gómez LD (2006) Estado actual del manatí (Trichechus manatus) en humedales del sur del Golfo de México. In: Primer Simposio para la Biología y Conservación del manatí antillano en Mesoamérica. Loma Linda University. http://resweb.llu.edu/rford/research/Manatees/ProgramayResumenes.pdf. Accessed 30 Oct 2009
Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Pause KC (2007) Conservation genetics of the Florida manatee, Trichechus manatus latirostris. PhD dissertation. University of Florida, USA
Pause KC, Nourisson C, Clark A, Kellogg ME, Bonde RK, McGuire PM (2007) Polymorphic microsatellite DNA markers for the Florida manatee (Trichechus manatus latirostris). Mol Ecol Notes 7:1073–1076
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Peel D, Ovenden JR, Peel SL (2004) NeEstimator: software for estimating effective population size. In: Fisheries. Queensland Government, Department of Primary Industries and Fisheries. http://www.dpi.qld.gov.au/cps/rde/dpi/hs.xsl/28_6908_ENA_HTML.htm. Accessed 3 Nov 2009
Piry S (2004) GeneClass2: A software for genetic assignment and first-generation migrant detection. J Her 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proebstel DS, Evans RP, Shiozawa DK, Williams RN (1993) Preservation of nonfrozen tissue samples from a salmonine fish Brachymystax lenok (Pallas) for DNA analysis. J Ichthyol 9:9–17
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reep RL, Bonde R (2006) The Florida manatee. The University Press of Florida, Gainesville, FL
Reyes-Mendoza O, Morales-Vela B (2007) New observations of manatees off the northern coast of Quintana Roo, Mexico. Sirenews 47:14
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
SEDUE (1991) Acuerdo por el que se establecen los criterios ecológicos CT-CERN-001-91 que determinan las especies raras, amenazadas, en peligro de extinción o sujetas a protección especial y sus endemismos, de la flora y la fauna terrestres y acuáticas en la República Mexicana. In: Diario Oficial de la Federación, 17 de mayo de 1991. Gobierno Constitucional de los Estados Unidos Mexicanos. México, pp 7–36
SEMARNAT (2001) Proyecto de conservación, recuperación y manejo del manatí Trichechus manatus en México. Ser PREP num 11. Secretaría de Medio Ambiente y Recursos Naturales, México
SEMARNAT (2002) Norma Oficial Mexicana NOM-059-SEMARNAT-2001 Protección Ambiental—Especies nativas de México de Flora y Fauna Silvestres—Categorías de Riesgo y Especificaciones para su Inclusión, Exclusión o Cambio—Lista de Especies en Riesgo. In: Diario Oficial de la Federación, 6 de marzo de 2002, Segunda Sección. Gobierno Constitucional de los Estados Unidos Mexicanos. México, pp 1–80
SoftGenetics (2008) GeneMarker, the biologist friendly software. In SoftGenetics, software powertools for genetic analysis. http://www.softgenetics.com/GeneMarker.html. Accessed 30 Oct 2009
Vianna JA, Bonde RK, Caballero S, Giraldo JP, Lima RP, Clark A, Marmontel M, Morales-Vela B, de Souza MJ, Parr L, Rodríguez-López M, Mignucci-Giannoni AA, Powell JA, Santos FR (2006) Phylogeography, phylogeny and hybridization in trichechid sirenians: implications for manatee conservation. Mol Ecol 15:433–447
White PS, Densmore LD (1992) Mitochondrial DNA isolation. In: Hoezel AR (ed) Molecular genetic analysis of populations: a practical approach. IRL Press, Oxford University Press, New York, pp 29–58
Acknowledgments
This study is part of the doctoral dissertation research of the first author (CN), who was funded by a grant from the Secretaria de Relaciones Exteriores de México and by the Ministère des Affaires Etrangères Française—EGIDE. Financial support for training in Florida was provided by the Marine Mammal Commission. The project was funded by SEMARNAT/CONACYT (Project 2002-C01-1128) and Dolphin Discovery for manatee captures and genetic analysis, as well as the US Geological Survey-Sirenia Project for genetic analysis. Mexican Government Research Permits and sample collection authorizations were obtained from DGVS # 03144; 04513; 03670 and 03675. Samples were collected under scientific collection permits NUM/SGPA/DGVS/03144, NUM/SGPA/DGVS/04513, NUM/SGPA/DGVS/03670/06, SGPA/DGVS/04060/06; SGPA/DGVS/01103/07, SEDUM/SSMA/DGPPE/0191/2004 and RBSK OFICIO CUN 037/04. Samples were transported to the research laboratories in the US under CITES export permits MX22523, MX24463, MX28578, MX26485, MX38416, MX34645, MX31591 and CITES import permits 06US808447/9 and 07US808447/9. Samples were processed in Florida under authority of USFWS wildlife research permit MA791721 issued to the USGS Sirenia Project. All sample collection was done under approval of IACUC standards. Samples from the Gulf of Mexico area came from various projects and facilities: Dolphin Discovery, Veracruz Aquarium, Xcaret, ViaDelphi and Universidad Juárez Autónoma de Tabasco. We thank Dr. Margaret Hunter of the USGS Southeast Ecological Science Center for advice and help in the laboratory with various software packages. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nourisson, C., Morales-Vela, B., Padilla-Saldívar, J. et al. Evidence of two genetic clusters of manatees with low genetic diversity in Mexico and implications for their conservation. Genetica 139, 833–842 (2011). https://doi.org/10.1007/s10709-011-9583-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-011-9583-z