Abstract
Cassava is a tropical crop and grown for its tuberous starchy roots. In Africa it is mainly cultivated by small-scale farmers who observe, select and name their cassava varieties based on morphology, food, social and economic interest. Here we have used an interdisciplinary approach involving farmer interviews, genetic markers and morphological descriptors to study the composition of cassava varieties on small-scale farms in 11 villages located in three districts in Uganda, the genetic structure within and between these varieties and their morphology. The composition of local, newly introduced and improved varieties differed widely between villages and districts. The Ugandan farmers in our study seemed to adopt improved varieties to a greater extent when there was a nearby market, prevalence of disease epidemics and good extension service. We found considerable genetic variation both within and between cassava varieties though the variation was larger between varieties. However, most local and improved varieties showed predominating genotypes at many loci. Accessions of commonly grown varieties meeting farmers’ preferences could therefore be selected and implemented in future breeding programmes involving development, dissemination and adoption. The like-named varieties in different villages were genetically similar, demonstrating farmers’ ability to differentiate and maintain the same variety over large areas. However, some varieties with different names in different villages showed both genetic and morphological similarity, suggesting that farmers may rename plants when they are introduced into their fields. The large differences found in variety and genetic composition between villages and districts in Uganda may be a result of the diverse needs and growing conditions characteristic for traditional farming system. This suggests that efforts to conserve and increase the genetic diversity in farmers’ fields will require policies tailored to each area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cassava (Manihot esculenta Crantz) is one of Africa’s major staple crops, feeding about 200 million people (Nweke 2004). It is mainly grown for its starchy tuberous roots though the leaves can also be used as vegetables. The roots also contain variable levels of cyanogenic glucosides. Varieties with high levels of cyanogenic glucosides (>1000 mg hydrogen cyanide (HCN) equivalent kg−1 dry weight) are called bitter and need to be processed for safe consumption, whereas varieties with low levels are called sweet. Cassava is tolerant to drought and low pH. It gives reasonable yields in soils with low nutrient content and does not require high management costs compared to other major food crops. Cassava is therefore an attractive crop for poor resource farmers. Because of cassava’s inherent tolerance to stressful environments, where other food crops would fail, cassava is often considered a food-security crop against famine and makes it one of the most useful and important crops in unstable environments in Sub Saharan Africa.
Cassava is mainly grown by small-scale farmers who chose and name their cassava according to complex motivations based on morphology, culinary attributes, social and economic interests. A group of plants identified by farmers under a single name is here referred to as a variety. In Africa the small-scale farmers derive new varieties from farmers of the same or different villages and sometimes from breeding programmes. Farmers mainly propagate cassava vegetatively through stem cuttings obtained from mature plants in their fields. However, small-scale farmers are also known to incorporate cuttings obtained from volunteer seedlings produced by sexual reproduction (Elias et al. 2001b; Fregene et al. 2003). The observed performance of a cassava variety in the local environment and farming system with respect to the farmers preferences determines whether it will continue to be cultivated. In the process of observing new planting material the name of a variety may change as the needs and choices may be specific to different farmers. This may result in the same genotype having different names or different genotypes having the same name in different farmers’ fields. In addition, mixing of varieties may arise from farmers’ misclassification. Therefore, a variety defined by a farmer may be complex and consist of a diverse genetic component.
In traditional agricultural systems it is common to find different cassava varieties in the farmers’ fields, which may be sweet, bitter or both. The proportion of different varieties in the field may, however, be highly dynamic with high turnover. The farmers’ decision over which varieties to grow is highly influenced by conditions such as biotic stress. For instance, in the Amazonian region of Peru there is a turnover of cassava varieties every 15 years due to pest and disease accumulation in the propagules (Salick 2001). Turnovers of varieties also occurred in Uganda as a result of severe epidemics of cassava mosaic disease (CMD) in the 1920s and late 1980s (Otim-Nape et al. 1997, 2001). The earlier breeding programmes of selections from Amani, Tanzania, released in Uganda as the Bukalasa series in the early 1960s (Jennings 1994) and the selections from the International Institute of Tropical Agriculture (IITA) starting from the 1980s resulting in the release of the Namulonge–Serere (NASE; Otim-Nape et al. 1997), have contributed to the high turnovers of cassava varieties in Uganda.
Scientific breeding of cassava began only recently compared with other major crops and is conducted by international and national research centres. Small-scale farmers have, however, been the engine behind cassava and variety development for thousands of years and continue today to actively select the planting material sourced from other cassava farmers, breeding programs and occasionally from sexually reproduced seedlings. The cultivated cassava on small-scale farmers’ fields could therefore be looked upon as both the production and breeding populations of cassava. The local breeding occurring over generations may have resulted in varieties that are adapted to the farmers’ needs for consumption, marketing requirements and cultivation conditions. A problem with scientific breeding in general is that the focus is mainly on broad adaptability so that a variety will produce a high average yield over a range of environments and years. Unfortunately, candidate genetic material that produces very good yields in one area, but poor yields in another, tends to be quickly eliminated from the breeder’s gene pool. Yet, this may be exactly what the small-scale farmers in some areas need. Traditional farming systems, which are characterized by unstable environments and resource poor farmers with diverse needs, would greatly benefit from maintaining a high genetic diversity within their crops for food security. Professional breeders, often working in relative isolation from farmers, have sometimes been unaware of the multitude of preferences, beyond yield, and resistance to diseases and pests, of their target farmers. This may result in a low adoption rate of improved varieties by the farmers in some areas. As an attempt to solve this problem participatory plant breeding (PPB) projects involving small-scale farmers are developing. In these projects breeders work closely with farmers at different stages of the research process to develop cassava varieties tailored to these farmers’ needs and to the requirements of the growing conditions of their cassava. For any successful breeding programme involving development, dissemination and adoption of new varieties it is of great importance to have knowledge about the genetic composition of the varieties grown by the target farmers and also to know which genotypes continue to be of interest to these farmers since their production system may change over time.
The evolution of crops that takes place in farmers’ fields in traditional agricultural systems may be underestimated especially in vegetatively propagated crops, and plant varieties are assumed to be conserved without evolution over long periods of time. Our recent results from a broad-scale study on the genetic diversity of cassava in Uganda showed, however, a high genetic diversity both in areas with high and low CMD incidence (Balyejusa Kizito et al. 2005). The genetic differentiation within and among varieties was, however, not investigated in that study. We found it, therefore, interesting to investigate the differentiation of Ugandan farmers’ varieties and test whether these varieties are of uniform genetic composition. Here we report the composition of small-scale farmers’ varieties in different villages in Uganda, how the genetic diversity of cassava is structured within and between these varieties and how morphologically distinct they are. In addition, we discuss how the Ugandan small-scale farmers maintain and differentiate their varieties. We have chosen an interdisciplinary approach in which farmer interviews on their naming of varieties was combined with genetic marker and morphological descriptions of cassava varieties. The interdisciplinary approach has been used in a few other cassava studies on small-scale farmers’ varieties in Malawi (Mkumbira et al. 2003) and Guyana (Elias et al. 2000, 2001a). Our findings will provide a unique basis for the understanding of genetic composition of Ugandan farmers’ varieties that can be used for developing strategies for cassava breeding programs with regard to improvement, multiplication and dissemination of cassava varieties.
Materials and methods
Study areas
Three districts where cassava is an important crop in the farming system were chosen for the study: Kumi, Luwero and Hoima (Fig. 1). Kumi district is located in the eastern part of Uganda and previous studies in the area have shown that the area experienced high CMD epidemics (Otim-Nape et al. 1997). Consequently there was an increase of activities by different Non Governmental Organisations (NGOs) and the National Cassava Breeding Programme to supply cassava varieties. The villages visited were Atiira, Apama-Oteteen, Kachaboi and Omolokonyo. In Luwero district, central part of Uganda, the villages selected were near the Namulonge Agricultural and Animal Production Research Institute (NAARI). Some of the farmers have participated in on-farm trials of varieties developed by the National Cassava Breeding Programme especially after the recent CMD outbreak. The villages visited were Kabembe, Kibanga, Nattyole and Vvumba. Hoima district, in the western part of Uganda, has been affected by CMD to a lesser extent than Luwero and Kumi districts and has not had many activities from the NASE breeding program (Otim-Nape et al. 1997). The villages studied were Kyeramya, Kyarubanga-A and Kyarubanga-B.
Key informant interviews
Prior to collection of cassava material in the eleven villages, initial information on cassava cultivation in each district was gathered from key officers at governmental and development organizations in individual interviews at the district offices. In each village interviews were held in groups ranging from 3 farmers in Attira, Kumi to 17 farmers in Kyeramya, Hoima to gather information about the preferences, utilization and naming of their most commonly grown varieties. A list of guidelines was developed for the group meetings for purposes of consistency and uniformity. The interviews were carried out in a semi-structured informal manner and were based on methods according to Sperling and Ashby (1997) and Salick et al. (1997). In addition, we made direct observation on the growing and use of cassava in the villages. After these interviews one farmer per village, known to be growing cassava for a long time, was purposely selected for depth interviews. These farmers were asked to give a general description and use of their varieties and how the varieties differed from each other. The source of planting material was classified into four categories: (1) local—varieties which have been grown continuously for over one farmer generation in the area (Berthaud et al. 2001), (2) newly introduced—varieties acquired from outside the village (relatively recently within the last farmer generation), (3) improved—varieties from the Bukalasa or NASE cassava breeding programmes and (4) unknown—plants for which the farmers did not have a name.
Plant material
Each farmer in the group interviews identified their most commonly grown varieties and was asked to give a cutting (of about 1 m) of the respective variety, here called an accession. All accessions collected from each village and district are listed in Table 1. Three to five accessions were given per farmer. Each accession was divided into four cuttings. Three cuttings of each accession were planted in an experimental field at Namulonge Agricultural and Animal Research Institute (NAARI) for morphological description while the fourth cutting of the same accession was planted in a screenhouse at NAARI for molecular marker analysis.
Molecular marker analysis
All accessions collected from the farmers (Table 1) and an additional accession of each of the improved varieties NASE1, NASE2, NASE3, NASE5, NASE8, NASE10 and NASE12 from NAARI were subjected to molecular marker analysis. DNA was isolated from young leaf tissue following the method by Doyle and Doyle (1987). Eleven microsatellite markers (SSRY4, SSRY9, SSRY19, SSRY51, SSRY64, SSRY82, SSRY103, SSRY148, SSRY151, SSRY164 and SSRY181) with high polymorphism information content (PIC) were selected from 67 markers from an earlier study (Fregene et al. 2003). PCR (polymerase chain reaction) was carried out as described by Mba et al. (2001) using 10 ng of DNA per reaction. The PCR product was denatured and electrophoresed on 6% polyacrylamide gels using Bio-Rad Vertical Sequencing System (Bio-Rad Inc., USA) and visualized by silver staining according to the Promega manufacturer’s guide. Allele sizes were determined based upon an internal gel molecular marker size standard. Scoring was done manually and with the aid of computer software ‘Quantity One’ (Bio-Rad Inc.). In addition, a few plants with known genotypes from the Centro Internacional de Agricultura Tropical (CIAT), Cali, Colombia, were used as controls on each gel.
Morphological description
Three cuttings of each accession given by the farmers were randomly planted together in one block consisting of three rows in an experimental field at NAARI. Because of an unusually early beginning of the dry season many accessions did not sprout or died back. The accessions that survived were studied for 15 morphological traits of which 14 were characterized into different classes as described in Table 6. In order to estimate the trait variation within a variety only the 21 varieties with three or more accessions reaching maturity (9 months after planting) are presented in the table, given a total of 180 accessions. At sprouting the colour of first fully expanded leaf (CEL), the colour of young shoot (CYSH), the colour of leaf vein (CLV) and the pubescence of young leaves (PUYL) were checked. At 6 months after planting leaves on the mid-section part of each plant was scored for the number of leaf lobes per leaf (NLL), the position in which the leaf lobes are held (POLL), the shape of leaf lobes (SHLL), the length of the petiole (LEPE) and the colour of petioles (CPE). In addition, the number of petioles (NPE) on the entire plant was recorded. Finally, at 9 months after planting the stem morphology was checked for the growth habit of stem (GHST), the pubescence of young stem (PUYST), the colour of mature stem (CMST), the prominence of leaf scars (PRLSC) and the height of first apical branch (HFAB). The studied traits are among other traits recommended by the International Plant Genetic Resources Institute (IPGRI) for cassava germplasm characterization and adapted by the National Cassava Programme premised at NAARI.
Statistical analysis
Genetic differentiation within and between varieties was quantified by F-statistic estimators F IS, F ST and F IT (Wright 1965) as described by Weir and Cockerham (1984) using FSTAT 2.9 (Goudet 1995). Pair-wise distances between the varieties in the different villages based on Nei’s genetic distance (Nei 1972) were calculated using the software package PHYLIP version 3.65 (http://evolution.gs.washington.edu/phylip.html). These pair-wise estimates were used to construct a dendrogram based on the Neighbour-joining method (Saitou and Nei 1987) and a dendrogram based on the unweighted paired group method with the arithmetic mean (UPGMA) implemented in the PHYLIP package version 3.65. The original data set of allele frequencies were resampled with 1000 bootstraps and a consensus tree was constructed. In order to look for a correlation between genetic and geographic distances between villages a test of isolation by distance (Slatkin 1993) was performed using a Mantel test (Sokal and Rohlf 1995) in the GENSURVEY program (Vekemans and Lefèbvre 1997).
Results
Composition and naming of farmers’ varieties
All together 288 accessions were given by the farmers from 11 villages in Luwero, Kumi and Hoima districts. Based on the farmers’ interviews these accessions represented as much as 49 different varieties named by the farmers and 44 unnamed accessions (Table 1). In general farmers grew a mixture of their local varieties (varieties that have been grown continuously for over one farmer generation in the area, Berthaud et al. 2001), varieties acquired from outside the village (relatively recently within the last farmer generation), improved varieties which they identified as coming from the Bukalasa or NASE cassava breeding programmes, and plants with unknown origin and without a name. The percentage of these categories based on the names given by farmers is plotted in Fig. 2. In Hoima district about two thirds were said to be local varieties, while only 4% were said to be from the Bukalasa breeding programme. The Hoima farmers did not identify any accession to be from the NASE programme. In Kumi on the other hand, as much as 40% were identified to be from the NASE programme but no accessions were said to be from the Bukalasa programme. Only 23% of the accessions were identified by the Kumi farmers as local varieties. A similar pattern was found in Luwero where more than half of the accessions were said to be from the NASE programme and only 17% were identified as local varieties. The highest frequency of newly introduced varieties (23%) and the lowest frequency of accessions with unknown origin (7%) were found in Hoima. For example, in Kyeramya village in Hoima none out of 78 accessions were unnamed by the farmers. All unknown and almost all named accessions were identified as sweet by the farmers.
The naming of the varieties, both local and improved, was specific to the districts except the improved Bukalasa and SS4 variety names that were common to both Kumi and Luwero districts (Table 1). Some of the improved varieties such as NASE2 and NASE3 have been renamed in some villages. Many varieties were found in two or more villages, often in the same district, while some were only found in one village. None of the varieties was found in all villages in a district. This resulted in a high number of varieties named by the farmers and a small number of accessions collected for many varieties.
In general there was no indication that farmers purposely used the same variety name for different phenotypes except in Kyeramya village in Hoima where the farmers differentiated two phenotypes of the variety Nyakabiriti. These were labelled as Nyakabiriti-1 and Nyakabiriti-2, respectively, during collection (Table 1). However, no distinct morphological differences were found between cuttings of these two varieties when grown in an experimental field (Table 7).
Number of alleles and percentage of polymorphic loci
The number of alleles found at each locus ranged from 3 at SSRY148 and SSRY181 to 7 alleles at SSRY9, SSRY82 and SSRY151 with a mean of 4.9 alleles per locus (Table 2). Percentage of polymorphic loci and mean number of alleles per locus are presented in Table 3 for varieties with 4 or more accessions given by the farmers. The small number of accessions given by the farmers of some varieties may represent a limited part of the genetic variation existing in these varieties. The genetic analysis of these varieties is therefore intended to be heuristic rather than definitive. The lowest frequency of polymorphic loci (45.5%) and the lowest mean number of alleles per locus (2.0) were found in the local variety Bamunaanika, while the three newly introduced varieties Emusugut, Nyakakwa and Nyalanda, and the local variety Kidimo had 100% polymorphic loci and on average 3.4 to 3.7 alleles per locus (Table 3). Both the local and improved varieties showed an average of 2.6 alleles per locus while the three introduced varieties showed an average of 3.5.
Genetic differentiation within and between varieties
Genetic differentiation within and between varieties was quantified by F-statistics as described by Weir and Cockerham (1984) for varieties that had 4 or more accessions (Table 3). Of the total diversity (F IT = 0.236) most diversity was found among varieties (F ST = 0.250) compared to the within-variety diversity (F IS = −0.021) showing that the varieties were genetically differentiated from each other. The local varieties were more greatly differentiated (F ST = 0.275) than either the newly introduced (F ST = 0.172) or improved (F ST = 0.164) varieties. However, all the variety categories showed large confidence intervals for the F ST values indicating that some varieties are genetically more similar than others. The within-variety diversity differed among the local, newly introduced and improved varieties (F IS = −0.138, 0.130 and 0.051, respectively). There was an excess of heterozygotes within the local varieties as indicated by the negative F IS value while the positive F IS values showed a deficiency of heterozygotes within the newly introduced and improved varieties. However, the confidence intervals of the within-variety diversity ranged from negative to positive F IS values for all three variety-categories indicating a difference in the genetic structure of varieties within each category.
All local, newly introduced and improved varieties consisted of several multilocus genotypes (Table 4) and in general no clones (accessions with identical multilocus genotype) were found. For most varieties all accessions within a variety differed in at least one marker locus. Accessions within a variety with an identical multilocus genotype were only found in 2 of the 21 varieties studied, namely Nigeria and Nyakabiriti-2. Interestingly, none of the farmers’ NASE accessions showed identical multilocus genotype with any of the single accessions of the NASE varieties collected from NAARI. The newly introduced varieties Emusugut and Nyakakwa showed a high number of different genotypes. No predominating genotype was found at any loci in Emusugut and only 2 out of 11 loci showed predominating genotype in Nyakakwa. However, most of the other varieties had predominating genotypes at many loci. The highest number of predominating genotypes was found in the local varieties Nyakabiriti-1, Nyakunyaku (10 out of the 11 loci) and Nyakabirit-2 (9 out of the 11 loci). About two thirds of the varieties had five or more loci with predominating genotypes.
The pattern of variation found differed between varieties and marker loci (Table 4). For example, the local varieties Mulyandongo and Nyakabiriti-2 showed little variation at the marker loci SSRY82 and SSRY164, while great variation was found in the local variety Kidimo within the same loci. The marker loci SSRY9 and SSRY181 showed the same predominating genotype within ten and nine different varieties, respectively, while SSRY4 showed no predominating genotypes in all but two varieties.
The 44 unknown accessions given by the farmers showed all different multilocus genotypes. Twenty-nine of these showed unique multilocus genotypes not similar to any of the varieties named by the farmers, while 6 accessions showed similar multilocus genotypes to the introduced varieties (3 to Emusugut, 2 to Nyakakwa and 1 to Nyalanda), 5 accessions showed similarity to some of the varieties in the NASE breeding program (3 to TME14 and NASE 10, 1 to Nigeria and 1 to No. 00057) and 4 accessions had similar multilocus genotypes to some of the local varieties (2 to Bamunaanika, 1 to Kidimao and 1 to Mulyandongo).
Genetic distances between farmers’ varieties
Because missing genotype data is not allowed in analysis with the phylogeny software package PHYLIP version 3.65 (http://evolution.gs.washington.edu/phylip.html), handling both allele frequencies and bootstrap resampling, only 165 out of 244 named accessions by the farmers and one additional NAARI accession were analyzed. These varieties were divided into 32 groups depending on variety and village. Nei’s genetic distances (Nei 1972) between varieties in the different villages are presented in Table 5. Most varieties given the same name in different villages showed among the closest genetic similarity, for example the local varieties Kidimo and Nyakunyaku in two villages in Hoima district and the improved variety Nigeria in three villages in Kumi district. The like-named local accessions differentiated into two phenotypes by the farmers, here called Nyakabiriti-1 and Nyakabiriti-2 in Kyeramya village, Hoima, showed relatively small genetic distance. On the other hand, some accessions given different names by the farmers such as the local varieties Ebwanateraka and Emulai in different villages in Kumi district were also genetically similar. Also Nyakabiriti-1 and Nyakabiriti-2 showed similarity with the relatively newly introduced Nyalanda variety within the same village. As expected, relatively close genetic similarity was also found between some of the improved varieties in the NASE breeding programme such as SS4 in Apama-Oteteen village in Kumi district, released as NASE4 and the Nigerian variety, released as NASE3 in several villages in Kumi. Interestingly, SS4 in Apama-Oteteen village and NASE3 from NAARI showed no genetic distance. NASE3 from NAARI showed also genetic similarity between NASE3 in Apama-Oteteen village. In addition, Vumba in Nattyole village in Luwero, released as NASE12, showed relatively small genetic distances to Nigeria and 00057 from among the on-farm trials in the NASE breeding programmes.
Relatively close genetic similarity was found between the improved Bukalasa and the local variety Bamunaanika in the village Nattyole in Luwero district. Other local varieties in different villages showed larger genetic distances to Bukalasa. This may suggest that Bamunaanika has a Bukalasa origin and is younger than the other local varieties or that the farmers in Nattyole misclassified the Bukalasa and Bamunaanika accessions. Due to missing genotype data only Bukalasa and Bamunaanika from one village, Nattyole, could be analyzed and it is therefore difficult to further discuss the genetic similarity between the two varieties.
When comparing the varieties in the NASE breeding program with non-improved varieties, NASE3 and Nigeria showed the smallest genetic distance with the newly introduced variety Emusugut in the village Apama-Oteteen in the Kumi district. Most other non-improved varieties showed considerable distances between the varieties in the NASE breeding program. The largest genetic distances were found between the improved variety in the Bukalasa breeding program, developed in Amani, Tanzania and in Uganda, released in the 1960s, and the improved varieties in the NASE breeding program (NASE, Nigeria, SS4, TC1), starting from the 1980s at IITA. This suggests that the Bukalasa and the NASE varieties have different origin. However, additional Bukalasa accessions from other villages need to be analyzed to prove this. Even though most non-improved varieties showed large genetic distances to the NASE-varieties (average D = 0.67) and many of the NASE varieties showed relatively small genetic distances (average D = 0.40) no clustering of major groups of varieties were supported by bootstrap resampling with the Neighbor-joining (Saitou and Nei 1987) or the UPGMA methods based on Nei’s genetic distance (1972) implemented in PHYLIP version 3.65 (data not shown). The average genetic distance between varieties within villages ranged from 0.40 in Kyeramya, Hoima district to 0.96 in Kachaboi, Kumi district. The relatively large genetic distances suggest that many varieties grown by farmers within the same village were genetically distinct. When comparing the genetic variability among villages the accessions in nearby villages seemed in general to be more genetically similar than accessions in villages far apart since there was a positive significant correlation between genetic and geographic distances among villages (r = 0.195, P = 0.002) found by the Mantel test (Sokal and Rohlf 1995) in the GENSURVEY program (Vekemans and Lefèbvre 1997).
Morphological description
Table 7 presents the morphological class with the highest frequency for the respective trait in each variety. In addition, the average of all cuttings and accessions of each variety for the trait HFAB is given. Plants given the same name by the farmers were grouped together irrespective of the village they came from. In order to estimate the trait variation within a variety only the varieties with three or more accessions surviving in the field are presented here. Cuttings of the same accession showed the same morphology class in respective traits. Seven of the traits (CEL, CYSH, CLV, LEPE, CPE, CMST and HFAB) showed more variation within and among varieties than the other traits (PUYL, NLL, POLL, SHLL, NPE, GHST, PUYST and PRLSC). Four of the traits (CEL, CPE, CMST and HFAB) showing more variation within and among varieties and two of the traits (SHLL and GHST) showing less variation were said to be important by the farmers in differentiating their varieties (Table 6).
The genetically similar local varieties Ebwanateraka and Emulai showed similar morphology. Also the like-named and genetic similar accessions differentiated into two phenotypes by the farmers, here called Nyakabiriti-1 and Nyakabiriti-2, had similar morphology. However, the morphology class, which showed the highest frequency in several traits important by the farmers in differentiating their varieties differed between the genetically similar local variety Bamunaanika and the improved Bukalasa variety. Most of the improved varieties in the NASE breeding program, showing genetic similarity, differed also in at least two traits. The relatively newly introduced variety Nyalanda showed a distinct morphology in CEL and CPE compared to all other varieties.
The morphological variation found within and between varieties could be an effect of both genotype differences and differences in the growing conditions in the experimental field, and the phenotypic expression may differ in the farmers’ fields. The individual effect of these parameters could, however, not be estimated without a randomized complete block design with several blocks and replicates of each accession in each block (Chahal and Gosal 2002). Since this design was not used here we believe it would be inappropriate to further analyse the morphological variation with multivariate statistical methods.
Discussion
Evolution of crops that takes place in farmers’ fields in traditional agricultural systems is often underestimated especially in vegetatively propagated crops, and plant varieties are assumed to be conserved without evolution over long periods of time. Knowledge related to farmer management in influencing the genetic diversity of their crops is limited even though it is of major importance for developing strategies for breeding programs with regard to improvement, multiplication and dissemination of varieties. In this study we have focused on the composition and genetic structure of cassava varieties in farmers’ fields in Uganda and tested the hypothesis that very little variation is found within these varieties.
Genetic diversity
We found considerable genetic variation both within and between cassava varieties though the variation was larger between varieties. Very few clones were found among the accessions collected from different farmers and villages. This may be surprising considering that cassava is mainly vegetatively propagated. The large genetic variation within varieties may be a result of farmers’ selection of volunteer seedlings produced by spontaneous sexual recombination. Most unnamed accessions showed multilocus genotypes which were very different from the multilocus genotypes found in any of the varieties identified by the farmers. This may further illustrate farmers’ selection of spontaneous seedlings with unknown origin. The extent to which the African farmers incorporate seedlings into the planting population may therefore be underestimated. However, a small number of accessions and only one accession of each variety were collected per farmer in this study. This study has therefore not investigated whether there are clones within farmers’ fields. Large genetic variation within varieties has also been found in the small-scale farming communities in Guyana (Elias et al. 2001a) and Brazil (Second et al. 1997; Sambatti et al. 2001).
We found that the pattern of variation differed between varieties so that some varieties showed extensive variation within almost all marker loci such as the newly introduced varieties Nyakakwa and Emusugut, while many local varieties showed predominating genotypes in most loci (Table 4). The finding that some loci showed extensive variation within varieties may indicate that these loci are not linked to any trait of interest for the farmers whereas loci that showed less variation and predominating loci within varieties may be linked to traits of interest. Also varieties grown by small-scale farmers in a village in Guyana showed differing levels of variation within them, some were monomorphic while others had high intra-varietal variability (Elias et al. 2001b).
Differences in levels of variation in the Ugandan farmers’ varieties may be a result of a combination of their history, the source of each variety and farmers’ management. At some point varieties are brought into farmers’ villages from neighbouring communities and sometimes from breeding programmes. Before being given to farmers improved varieties usually have undergone cycles of selection based on the respective breeding objectives. Ultimately improved varieties go through selection cycles for morphological uniformity, which would most likely result in more genetically homogenous varieties. Among the few Bukalasa accessions identified by the farmers no identical multilocus genotypes were found. In Uganda the bred Bukalasa varieties were released in the early 1960s (Otim-Nape et al. 1994) and have since been subject to various evolutionary forces such as gene exchange between farmers, gene flow and human and natural selection within farmers’ fields. One might therefore not expect to find high genetic homogeneity within the Bukalasa varieties.
Even though less variation was found in the improved NASE varieties than in the introduced varieties surprisingly no clones were found in the NASE varieties given by the farmers. Furthermore, none of the farmers’ NASE accessions showed identical multilocus genotype with any of the single accessions of the NASE varieties collected from NAARI. For further studies it would be interesting to more thoroughly compare the genetic constitution of the NASE varieties within the breeding programme with the genetic constitution of farmers’ NASE varieties in both within and outside target areas to shed light on the short-term effect of farmers’ management on the genetic changes within varieties.
In contrast to varieties from breeding programmes, varieties introduced from farmers outside the village may come from various sources and a variety consisting of plants given the same name may therefore consist of more variability as was seen for Emusugut and Nyakakwa varieties (Table 4). Local varieties not related to the improved varieties probably start as varieties introduced from different areas outside the village. All varieties, local, improved or newly introduced undergo selection on farmers’ fields. Phenotypes that do not meet the farmers’ preferences get eventually eliminated and the genetic variation within a variety may reduce with time. The varieties that are grown in the Ugandan farmers’ fields may therefore be at different stages of selection with the old local varieties being at more advanced levels of selection than the newly introduced varieties.
Our findings on the intra-varietal diversity differ from cassava studies in Malawi where a single multilocus genotype predominated within each of the ten most grown local varieties and showed a wide distribution in the cassava growing areas in the country, although varieties went by different names in different areas (Mkumbira 2002; Mkumbira et al. 2003). The clonality of cassava varieties in Malawi may be explained by governmental intervention efforts in the late 1980s in response to mealy bug infestation that focused on the identification, recommendation of specific genotypes of some local varieties and the rapid distribution of cleaned and improved cuttings thereof (Mkumbira 2002; Haggblade and Zulu 2003). The use of tissue culture in the multiplication scheme may also have helped to perpetuate the selected clones from each variety together with an efficient distribution system. Varieties identified by the Malawian farmers that were not among the ten most grown varieties showed extensive genetic variation within varieties (Mkumbira et al. 2003).
Farmers’ maintenance and differentiation of varieties
In spite of intra-varietal variation, the large genetic differentiation (Table 3) and genetic distances (Table 5) found in general among Ugandan farmers’ varieties show farmers ability to maintain and differentiate their varieties. A closer genetic similarity among accessions in nearby villages suggests occurrence of exchange of planting material between farmers in nearby villages. The exchange of planting material may lead to misclassification of accessions. However, varieties given the same name by farmers in different villages were genetically similar (Table 5), showing that the farmers are able to maintain the same variety over larger areas. Based on our interviews with the Ugandan farmers, certain morphological traits such as HFAB and CPE and culinary attributes such as taste of a plant seemed to be of most importance to differentiate and name varieties. Thus, plants that showed similar morphology would most likely be given the same name in the same village. Plants with unfamiliar morphology, which were found to have multilocus genotypes different from any of the accessions identified by the farmers, were unnamed but continued to be grown if they met other farmer’s demands such as taste. In fact, all unknown varieties were sweet. However, when cuttings obtained from farmers were grown in an experimental field, morphological variation in several traits was found within varieties. If the morphological heterogeneity within varieties is maintained within farmers’ fields the identification of plants by the farmers may be difficult and may lead to misclassification in naming and increased heterogeneity within varieties. Some varieties with different names were genetically similar. Based on the morphological description of cuttings obtained from farmers some of these varieties showed the same morphology while others differed in several morphological traits considered to be important by the farmers in differentiating varieties. Varieties which were genetically and morphologically similar but had different names in different villages may suggest that farmers rename plant material when it is introduced to the farmers’ fields.
Composition of varieties
We found a difference in the composition of local, newly introduced and improved varieties in the Ugandan villages and districts and many varieties had names specific to the district and some even to the village. The difference in variety composition may reflect farmers’ decisions made under different circumstances such as the access to urban markets, influence from government extension programmes, biotic stress such as pest and diseases in the farmers’ fields and innovativeness of farmers in acquiring new planting material.
The farmers in Hoima district did not grow any of the most recently released NASE varieties and a very low frequency of the older bred Bukalasa varieties. In comparison with the farmers in the two other districts Luwero and Kumi, the Hoima farmers grew a larger frequency of newly introduced varieties, suggesting that the Hoima farmers have relied more on other farming communities for sources of planting material than the breeding programmes. Farmers in Kumi and Luwero on the other hand have had more access to planting material from the cassava breeding programme and therefore seem to rely more on planting material from there than from other farming communities. Access to urban markets by both Luwero and Kumi farmers has also largely influenced their production choices in that they may have chosen to use more material from the breeding programme since improved varieties are usually higher yielding than local varieties. Kumi is a major dried cassava supplier to market (Otime-Nape et al. 1997) and Luwero supplies fresh cassava. Additionally, the recent CMD epidemic in Kumi and Luwero led to interventions by government and NGOs to restore cassava production and counter the heavy variety losses that were caused by the disease with improved varieties. The recently released NASE varieties seem to have had high adoption rates in these areas, showing farmers motivation to adopt new varieties from the breeding programme. The dissemination policy and time are important factors in the adoption rate therefore further studies in time would give a more conclusive reflection of the adoption of improved varieties in areas near and distant from urban markets.
In conclusion, our results showed large genetic differentiation among varieties. However, genetic and morphological variation was also found within local, newly introduced and improved varieties and almost no clones were found within varieties. This shows the complexity of defining a farmer’s variety and the importance of combining genetic analysis with farmers’ naming in determining the composition of farmers’ varieties and understanding the role of farmers management on the cassava genetic diversity. However, most local and improved varieties had predominating genotypes at many loci. Accessions of commonly grown varieties meeting farmers’ preferences could therefore be selected with the same interdisciplinary approach used in this study and implemented in future breeding programmes in the country. We also found that the composition of local, newly introduced and improved varieties differed between villages and districts. The Ugandan farmers in our study seemed to adopt improved varieties to a greater extent when there was a nearby market, high CMD incidence and good extension service. Accordingly, breeders can strongly affect the composition of farmers’ varieties by controlling the number and dissemination of improved varieties, which may lead to a loss of local varieties and a reduction of genetic diversity. However, as traditional farming systems, which are characterized by unstable environments and diverse needs of farmers, would greatly benefit from maintaining a high genetic diversity within their crops for food security it is of great importance to prevent loss of genetic diversity in the farmers’ fields. Therefore the large differences found in variety and genetic composition between villages and districts in Uganda suggest that efforts to conserve and increase genetic diversity in farmers’ fields will require policies tailored to each area. In addition, as the genetic constitution within a variety and the composition of varieties on small-scale farms may change over time the national programs for gene conservation and breeding need to consider the dynamics of genetic change.
References
Balyejusa and Kizito E, Bua A, Fregene M, Egwang T, Gullberg U, Westerbergh A (2005) The effect of cassava mosaic disease on the genetic diversity of cassava in Uganda. Euphytica 146:45–54
Berthaud J, Clement JC, Emperaire L, Louette D, Pinton F, Sanou J, Second G (2001) The role of local-level gene flow in enhancing and maintaining genetic diversity. In: Cooper HD, Spillane C, Hodgkin T (eds) Broadening the genetic base of crop production. CABI, Wallingford, Oxon, UK, pp 81–103
Chahal GS, Gosal SS (2002) Principles and procedures of plant breeding: biotechnological and conventional approaches. Alpha Science International Ltd. Pangbourne, UK
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Elias M, Panaud O, Robert T (2000) Assessment of genetic variability in a traditional cassava (Manihot esculenta Crantz) farming system, using AFLP markers. Heredity 85:219–230
Elias M, Penet L, Vindry P, MacKey D, Panaud O, Robert T (2001a) Unmanaged sexual reproduction and the dynamics of genetic diversity of a vegetatively propagated crop plant, cassava (Manihot esculenta Crantz), in a traditional farming system. Mol Ecol 10:1895–1907
Elias M, McKey D, Panaud O, Anstett MC, Robert T (2001b) Traditional management of cassava morphological and genetic diversity by the Makushi Amerindians (Guyana, South America): perspectives for on-farm conservation of crop genetic resources. Euphytica 120:143–157
Fregene MA, Suarez M, Mkumbira J, Kulembeka H, Ndedya E, Kullaya A, Mitchel S, Gullberg U, Rosling H, ADixon GO, Dean R, Kresovich S (2003) Simple sequence repeat marker diversity in cassava landraces: genetic diversity and differentiation in an asexually propagated crop. Theor Appl Genet 107:1083–1093
Goudet J (1995) FSTAT (vers. 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Haggblade S, Zulu B (2003) The recent cassava surge in Zambia and Malawi. Paper 9. InWent/IFPRI/NEPAD/CTA conference: Successes in African Agriculture, pp 7–12
Jennings DL (1994) Breeding for resistance to African cassava mosaic geminivirus in East Africa. Trop Sci 34:110–122
Mba REC, Stephenson P, Edwards K, Melzer S, Mkumbira J, Gullberg U, Apel K, Gale M, Tohme J, Fregene MA (2001) Simple Sequence Repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theor Appl Genet 102:21–31
Mkumbira J (2002) Cassava development for small-scale farmers. PhD thesis. Agraria 365. Swedish University of Agricultural Science
Mkumbira J, Chiwona-Karltun L, Lagercrantz U, Mahungu NM, Saka J, Mhone A, Bokanga M, Brimer L, Gullberg U, Rosling H (2003) Classification of cassava into ‘bitter’ and ‘cool’ in Malawi: from farmers’ perception to characterisation by molecular markers. Euphytica 132:7–22
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nweke F (2004) New challenges in the cassava transformation in Nigeria and Ghana. Discussion paper No. 118. Environment and Production Technology Division, International Food Policy Research Institute
Otim-Nape GW, Bua A Baguma Y (1994) Accelerating the transfer of improved production technologies: Controlling African cassava mosaic virus disease epidemics in Uganda. Afr Crop Sci J 2:479–485
Otim-Nape GW, Bua A, Thresh JM, Baguma Y, Ogwal S, Semakula GN, Acola G, Byabakama B, Martin A (1997) Cassava mosaic virus disease in Uganda: the current pandemic and approaches to control. NARO/DFID. Chatham, UK: Natural Resources Institute, pp 33–38
Otim-Nape GW, Alicai T, Thresh JM (2001) Changes in the incidence and severity of cassava mosaic virus disease, varietal diversity and cassava production in Uganda. Ann Appl Biol 138:313–327
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salick J, Cellinese N, Knapp S (1997) Indigenous diversity of cassava: generation, maintenance, use and loss among the Amuesha, Peruvian upper Amazon. Econ Bot 51:6–19
Salick J (2001) Diversity, evolution and domestication within Manihot. Fifth International Scientific Meeting of the Cassava Biotechnology Network November 4–9, 2001, St. Louis, Missouri USA, PS1–03
Sambatti JBM, Martins PS, Ando A (2001) Folk taxonomy and evolutionary dynamics of cassava: a case study in Ubatuba—Brazil. Econ Bot 55:93–105
Second G, Allem AC, Mendes RA, Carvalho LCJB, Emperaire L, Ingram C (1997) Molecular marker (AFLP)-based Manihot and cassava numerical taxonomy and genetic structure analysis in progress: implications for their dynamic conservation and genetic mapping. Afr J Root Tuber Crops 2:140–144
Slatkin M. (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research. 3rd edn. Freeman and Co., New York. pp 887 ISBN: 0-7167-2411-1
Sperling L, Ashby J (1997) Participatory plant breeding: emerging models and future development. In: Tripp R (eds) New seeds and old laws. Regulatory reform and the diversification of national seed systems. IT publications, London, pp 198–213
Vekemans X, Lefèbvre C (1997) On the evolution of heavy-metal tolerant populations in Armeria maritima: evidence from allozyme variation and reproductive barriers. J Evol Biol 10:175–191
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1965) The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution 19:395–420
Acknowledgement
We thank U. Gullberg for guidance in the planning of the study, with farmer interviews and discussions, Y. Baguma (NAARI) for comments on an earlier version of the manuscript, J. Onsingaada, J. Orone and H. Nanyonjo for assistance in cassava cultivation. This work was supported by a grant from the Swedish International Development Cooperation Agency/Development for Research Cooperation (Sida/SAREC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Balyejusa Kizito, E., Chiwona-Karltun, L., Egwang, T. et al. Genetic diversity and variety composition of cassava on small-scale farms in Uganda: an interdisciplinary study using genetic markers and farmer interviews. Genetica 130, 301–318 (2007). https://doi.org/10.1007/s10709-006-9107-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-006-9107-4


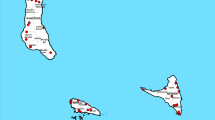

 ) in Hoima, Kumi and Luwero districts
) in Hoima, Kumi and Luwero districts