Abstract
Pearlscale angelfish Centropyge vrolikii is a kind of protogynous hermaphrodite fish with a natural sexual reversion. Under appropriate social conditions, a female fish can transform into a male fish spontaneously. It is an important prerequisite for artificial breeding to understand the process of its gonadal development and sexual reversion. Gonadal development is regulated by many sex-related genes. In this study, we used unreferenced RNA-Seq technology to sequence the ovary at the perinucleolus stage (OII), ovary at the yolk vesicle stage (OIV),IV and testis (T), respectively; screened the gonadal differential expression genes (DEGs); and analyzed the expression of these genes in different developmental stages of ovary and different sex gonads. The results showed that a total of 142,589 all-unigene samples were assembled, and gene annotation was performed by COG, GO, KEGG, KOG, Pfam, Swissprot, eggNOG, and NR functional database. Comparative analysis revealed that there were 1919 genes that were up-regulated and 1289 genes were down-regulated in comparison to OIV vs OII, while there were 3653 genes that were up-regulated and 2874 genes were down-regulated in comparison of OIV vs T, there were 3345 genes that were up-regulated and 2995 genes were down-regulated in comparison of the OII vs the T. At the same time, the results verified by RT-qPCR were consistent with the variation trend of transcriptome data. Among the results, amh, sox9b, dmrt1, dmrt2, cyp11a, cyp17a, and cyp19a were significantly expressed in the testes, while sox3, sox4, sox11, sox17, and hsd3b7 were significantly expressed in the ovaries. And, the expression of the amh, sox9b, dmrt2, and dmrt1 were low in the OII and OIV, while significantly increased during the ovotestis in the hermaphroditic period (OT), and finally reached the highest level in pure testis after sex reversal. The expression of sox3, sox4, hsd3b7, sox11, and sox17 was significantly reduced during the hermaphroditic period (OT). These results suggested that these genes may play an important role in the process of sex reversal. This study is helpful to further understand the molecular regulation mechanism of gonadal development and sexual reversion in Pearlscale angelfish and also provide important clues for future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Centropyge vrolikii, a kind of protogynous hermaphrodite fish, is commonly known as the black tail angelfish (Fernandez-Silva et al. 2018), which is mainly distributed in the Indian Ocean and the Pacific Ocean and lives near coral reefs, and it is one of the important marine ornamental fish species with huge market potential (Dibattista et al. 2016). The variety of Centropyge, various colors, and body shapes of Centropyge were deeply favored by consumers. At present, the fish of Centropyge is obtained mainly by natural fishing; however, the natural production of Pearlscale angelfish has been decreasing year by year because of the increase of market demand and the destruction of ecological environment, which requires to be payed more attention to environmental protection and artificial breeding.
Gonadal development and sex determination have always been the focus of developmental biology, which is greatly significant for the study of the development and evolution of species. Fish sex determination and differentiation are influenced by multiple genetic, physiological, and environmental factors (Devlin and Nagahama 2002). Sexual reversion is common in marine fishes of tropical and subtropical species, such as tongue sole fish (Cynoglossus semilaevis) (Shao et al., 2014), the cichlid fishes (Neochromis omnicaeruleus) (Lande et al., 2001), orange-spotted grouper (Epinephelus coioides) (Chen et al., 2019b), Archocentrus myrnae (Michael 2007), medaka (Oryzias latipes) (Hayasaka et al., 2019), the coral reef fishes (Nakashima et al., 2000; Sakai et al., 2003; Gust 2004; Barros et al., 2017; Maxfield and Cole 2019), and so on. Gonadal development systems in different individuals are regulated by different sex-determining gene, and the expression levels of these genes vary greatly in different tissues. Many genes for sex determining have been identified in fish. Foxl2 and sox9a are the ovarian-specific genes confirmed by morphological and mRNA localization analysis which play a decisive role in ovarian development; dmrt1 and amh are the main genes for testes tissue formation and maintaining; and foxl3 seems playing an important role in oogenesis, spermatogenesis, and gonads structure in Kryptolebias marmoratus (Qu et al., 2020). Amh gene has been demonstrated to be expressed in sexual dimorphism during sexual differentiation (Yoshinaga et al., 2004), which may initiate the process of testicular differentiation and participate in the early development and maintenance of testes (Wang et al., 2020). At high temperature, the high expression of amh and gsd gene led to the decrease expression of cyp19a gene, which resulted in masculinization of female flounder (Paralichthys olivaceus) (Wang et al., 2017; Yang et al., 2020). Dmrt1 is the main male sex determination genes. Dmrt1 and sox9b mainly expressed in the testes in Nile tilapia (Oreochromis niloticus), and the expression of sox9b in testes is reduced by knocking out the dmrt1 genes, which showed that the male sexual differentiation factor dmrt1 can have positive regulation of sox9b gene transcription and affect the gender differentiation in O. niloticus (Wei et al., 2019). Foxl2, tends to play a deterministic role in sex differentiation as opposed to dmrt1, is a key gene in ovarian and testicular differentiation (Trukhina et al., 2013). By analyzing the expression and localization of foxl2 and dmrt1 in gonadal development of Oxyeleotris marmorata using in situ hybridization, it was revealed that foxl2 may be involved in the establishment of oocyte polarity and ovarian differentiation, while dmrt1 is strongly expressed in spermatogonial cells and spermatogonial cells with testis-bias expression; this suggests that they may play an important role in the sexual differentiation and germ cell development of O. marmorata (Liu et al., 2020). There are also genes involved in reproduction that have been studied, such as Cbx2 in medaka (Chao et al., 2020), zona pellucida 1 (zp1) in Sparus aurata (Forner-Piquer et al., 2019), sox3 (Gao et al., 2015) and sox7 (Yu et al., 2018) in P. olivaceus, and so on. So far, many studies on gonadal development have found that most of these genes may involve in gonadal development in different species (Yue et al., 2015). Based on this information, we screened the differential expression genes (DEGs) in gonads at different stages in Pearlscale angelfish using RNA-Seq technology for further analysis of the function of these genes.
As a new transcriptome research technology, RNA-Seq technology has gained more and more attention due to its accuracy, efficiency, and speed. RNA-Seq technology not only annotates and maps the fish transcriptome, but also helps us understand the biological information of fish, such as immunology, toxicology, physiology, evolutionary biology, and developmental biology (Qian et al., 2014). Transcriptome sequencing was performed on ovaries and testes of Amur sturgeon (Acipenser schrenckii) to obtain 26 gene families related to reproduction and sex (Jin et al., 2015). GnRH was highly expressed in gonads and brains during reproductive seasons through the transcriptomic analysis in male and female Gymnocypris przewalskii (Tian et al., 2019). By transcriptomic sequencing, the relevant genes which may be involved in reproduction and gonadal development were identified, and their gender dimorphism was also analyzed in glands of Scatophagus argus. The results showed that dmrt1, amh, gsdf, Wt1a, sox9b, and Nanos2 were testis-bias genes, while foxl2, gdf9, bmp15, sox3, Zar1, and Flgla were mainly expressed in ovaries (He et al., 2019). The genes of testes and ovaries in A. schrenckii were screened through full-length transcriptome sequencing and comparative transcriptomic analysis; it was found that the number of ovary-specific expression unigenes was greater than those of testis (Zhang et al., 2020b). Among them, testis-biased genes amh and gsdf, as well as ovary-biased genes foxl2 and cyp19a, were screened out, suggesting that these genes may play an important regulatory role in spermatogenesis and oogenesis (Zhang et al., 2020b).
Pearlscale angelfish is one kind of fish with natural sexual reversion. Under appropriate social conditions, a female fish can transform into a male fish spontaneously. The studies of gonadal development and sex reversion are the important contents in the artificial breeding of Pearlscale angelfish. In this study, the ovaries at different stages and testes of Pearlscale angelfish were sequenced by transcriptome high-throughput sequencing technology, and the key genes affecting the development of gonad were screened out and verified by RT-qPCR, which suggesting that they may play a regulatory role in the development of gonad of Pearlscale angelfish. These results not only provide a new perspective on gonadal development of small sea angelfish, but also have important significance for artificial breeding of Pearlscale angelfish.
Materials and methods
Ethics statement
The study was approved by the Key Laboratory of Healthy Mariculture for East China Sea Ministry of Agriculture and Fisheries College Jimei University. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Tissue separation and preservation
Individuals of Pearlscale angelfish were collected from the Aquatic Product Experiment Center of Fisheries College Jimei University (Xiamen China). A total of 48 individual fishes were taken and carefully dissected for the ovary at the perinucleolus stage (OII), ovotestis in the hermaphroditic period (OT), the ovary at the yolk vesicle stage (OIV), and testis (T) which were naturally transformed from female’s ovaries. Part of gonads (0.5 cm3) was quickly fixed in 4% paraformaldehyde (PFA) at 4 ℃ for 24 h for further use; other gonads were collected in RNAlater (Ambion) and stored at 4 ℃ for 24 h and then transferred to – 80 ℃ for storage until RNA extraction.
Preparation and observation of tissue sections
The fixed gonadas (PFA) were dehydrated with gradient ethanol, transparented with xylene, embedded with paraffin, and sliced consecutively with the thickness of the Sects. 6 μm. Then, the sections were stained with hematoxylin–eosin (H.E) and sealed with neutral gum. The images were observed and photographed using a Nikon DS-FI2 microscope.
RNA extraction and transcriptome sequencing
RNA was extracted from OII, OT, OIV, and T (three samples per stage) using the Total RNA Kit II (OMEGA, USA). RNA concentration and purity were determined using the NanoDrop1000 spectrophotometer (Thermo Scientific, MA, USA). Samples with A260/280 values of 1.8 ~ 2.0 could be utilized for downstream experiments. The integrity of the RNA was tested by 1.0% agarose gel electrophoresis. Then, one part of RNA (OII, OT, OIV, and T) performed reverse transcription immediately, and others of RNA (OII, OIV, and T) were sent to Biomarker Technologies Co., Ltd. (Beijing, China) for quality testing and transcriptome sequencing.
Transcription sequence acquisition and gene annotation
The cDNA library was sequenced based on sequencing by synthesis (SBS) technology by Illumina Hiseq high-throughput Sequencing platform and then assembled sequentially after obtaining high-quality sequencing data. Trinity software first broke sequencing reads into shorter fragments (k-mer), then extended these small fragments into longer fragments (contig), and made use of the overlap between these fragments to obtain contig collections (component). Finally, De Bruijn graph method and sequencing read information were used to identify transcription sequences in each contig collection, separately. BLAST (Altschul et al., 1997) software was used to compare the unigene sequence with the NR, Swiss-Prot, GO, COG, KOG, eggNOG4.5, and KEGG Orthology database. KOBAS2.0 was used to obtain the KEGG Orthology result of unigene in KEGG. After predicting the amino acid sequence of unigene, HMMER software was used to compare it with the Pfam (Finn et al., 2010) database to obtain the annotation information of unigene.
Screening and enrichment of differential expression genes
To compare differences in gene expression in Pearlscale angelfish ovaries and testis of different stages, a method described by Benjamini–Hochberg was adopted to evaluate the tag frequency in the different RNA-Seq libraries. The threshold P value in multiple tests was determined by measuring the FDR (false discovery rate). FDRs, with less than 0.01 and an absolute value of the log2 ratio, are more than 1, which were used as the threshold to evaluate significant differences in gene expression. DEGs were then subjected to enrichment analysis by GO (gene ontology) functional group, KEGG pathways, COG (cluster of orthologous groups of proteins) database, and EggNOG database; moreover, the volcano plot was used to show the number of DEGs in different groups, so as to quickly check the expression level difference of genes between two groups of samples and its statistical significance.
RT-qPCR verification of transcriptome sequencing results
In order to verify the accuracy of differential gene expression by transcriptome sequencing, 13 genes are screened for RT-qPCR, primers as shown in Table 4. The trace amount of genomic DNA was removed from RNA samples by DNAse I treatment. The first strand of cDNA was then synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA) from 1.0 μg total RNA samples. The product of reverse transcription was diluted with sterile water by 10 times and then stored at − 20 ℃ as a RT-qPCR template. The amplification of cDNA was based on the QuantiFast SYBR Green PCR Kit (Qiagen, Nangjing, China) on 384-well PCR plates. The 10 μL reaction system was adopted: SYBR Green Real-Time PCR Master Mix was 5 μL, the positive and negative primers were 0.25 μL (10 μmoL/L), the cDNA template was 4.5 μL, and the reaction was conducted at 95 ℃ and denatured for 5 min. Then, the temperature was as follows: 95℃ for 10 s, 59 ℃ for 10 s, 72 ℃ for 10 s and circulation for 45 times. After the amplification curve was completed, the quantitative PCR was cooled down, and the fluorescence value was read. Then, the solution curve was analyzed to ensure a single product of each pair of quantitative primers. In this experiment, there were 4 biological replicates for each tissue and 3 technical replicates for each sample. Rps29 was used as a reference gene. The 2−ΔΔCt method was used to calculate the relative fold change of the gene expression level.
Result
Biopsy results of ovary at different stages
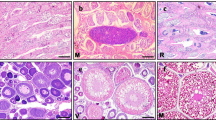
The structure of gonads was studied by histological method for determining the stage of gonad development. The results are shown in Fig. 1. The ovary at the perinucleolus stage (OII) is dominated by the oocytes at stage II with small cell volume and no yolks (Fig. 1A); the ovary at the yolk vesicle stage (OIV) is dominated by oocytes at stage IV with large volume and rich yolks (Fig. 1B); when male germ cells begin to appear in the ovary, this marks the beginning of the gonad reversal; it gradually becomes hermaphrodite and form ovotestis (OT) with coexistence of male and female germ cells (Fig. 1C); the oocytes in the gonad completely are atrophied and disappeared (Fig. 1D), and then, the male germ cells of each stage are further differentiated; at this time, the gonad has transformed into pure testis, which marks the successful transformation of the female into a functional male; the mature testes (T) contain a large number of mature sperms (Fig. 1E).
Histological structure of gonad at different stages in Pearlscale angelfish. A Histological structure of ovary at the perinucleolus stage (OII). B Histological structure of ovary at the yolk vesicle stage (OIV). C and D Histological structure of ovotestis in the hermaphroditic period (OT). E Histological structure of mature testes (T)
Assembly of transcriptome sequencing data
Through 1.0% agarose gel electrophoresis, it was found that the RNA quality of each sample was good (see Fig. S1 in Supplementary) and could be used for downstream experiments. Illumina HiSeq platform was used for transcriptome sequencing of OII, OIV, and T from Pearlscale angelfish. The samples were divided into three groups: OII group with three parallel samples (OII -1, OII -2, OII -3), OIV group with three parallel samples (OIV -1, OIV -2, OIV -3), and T group with three parallel samples (T-1, T-2, T-3). After sequencing quality control, a total of 65.39 GB clean data were obtained, and the Q30 base percentage of each sample was not less than 91.98%. After the removal of low quality and subsequences, the clean reads are further analyzed, and the read number of OII -1, OII -2, OII -3, OIV -1, OIV -2, OIV -3, T-1, T-2, and T-3 is shown in Table 1. The reads are mapped to the reference genome sequence obtained for OII, OIV, and T and are shown in Table 1. GC content of all samples is shown in Table 1. Overall, 87.19%, 83.16%, and 80.87% of the reference genome sequence were mapped to OII, OIV, and T, respectively. The clean data of each sample is sequenced with the assembled transcript or unigene library, and the alignment results are compared as shown in Table 1. The reads that are written to the transcript or unigene are described as mapped reads shown in Table 2, and those that are mapped will be used for subsequent analysis. The number of all-unigene is 142,589; the total length is 222,841,991; the average length is 1562.83; and N50 is 2442. The distribution of all-unigene sequence length in the transcriptome of OII, OIV, and T is shown in Fig. S2 in Supplementary.
The results of all-unigene function annotation
A total of 142,589 all-unigene samples were assembled from the transcriptome of OII, OIV, and T in Pearlscale angelfish, and compared to the functional database of COG, GO, KEGG, KOG, Pfam, Swissprot, eggNOG, and NR, respectively (Table 3). The results of annotation of unigenes by the four databases (COG, KEGG, SwissProt, and NR) are depicted by an E-value threshold of 1e-5 (1 × 10−5) in the Venn diagram (Fig. 2). The results of all-unigene annotated by NR can be further transferred to the GO database which includes three levels: biological process, cellular component, and molecular function. The biological process contains cellular process, single-organism process, and metabolic process; the cellular component contains cell, cell part, membrane, membrane part, and organelle; the molecular function contains binding, catalytic activity, transporter activity, and signal transporter activity (Fig. 3). The COG database was used for functional annotation of unigene, and 25 COG functional classification information of gene homologous were obtained (see Fig. S3 in Supplementary). The BLAST top-hit species distributions for matches between the assembled unigenes of Pearlscale angelfish and gene entries in the NCBINR database showed that 26.28% of unigenes had significant similarity to Larimichthys crocea (8,019), followed by Lates calarifer (14.21%), Seriola dumeril (8.04%), and Seriola lalandi (6.30%) (Fig. 4).
Analysis of differential expression genes
By comparing the transcriptome of OII, OIV, and T in Pearlscale angelfish, genes with a difference multiple of more than two and a Q-value < 0.001 were identified as significantly differential expression genes. Comparing OII and OIV, 1919 genes were up-regulated, while 1289 genes were down-regulated; comparing OIV and T, 3653 genes were up-regulated, while 2874 genes were down-regulated; comparing OII and T, 3345 genes were up-regulated, while 2995 genes were down-regulated (Fig. 5). GO functional annotation classification of differential expression genes was conducted (see Fig. S4 in Supplementary). In organisms, different gene products coordinate with each other to perform biological functions. Pathway annotation analysis of differential expression genes is helpful to further clarify gene functions. The annotation results of differential expression genes were classified according to the pathway types in KEGG (Fig. 6). The differential expression genes were classified according to KEGG pathway annotations, and the first 20 KEGG pathways with the lowest Q-value were turned into enrichment bubble maps (see Fig. S5 in Supplementary). The KEGG pathway included oxidative phosphorylation, Fanconi anemia pathway, homologous recombination, spliceosome, nucleotide excision repair, RNA polymerase, apoptosis, p53 signaling pathway, basal transcription factors, DNA replication, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, and cytosolic DNA-sensing pathway. And, compared with OIV and OII, the top three KEGG pathways were oxidative phosphorylation, Fanconi anemia pathway, and homologous recombination; compared with OIV and T, the top three KEGG pathways were cell cycle, cytokine-cytokine receptor interactio,n and regulation of actin cytoskeleton; and compared with OII and T, the top three KEGG pathways were cell cycle, DNA replication, and homologous recombination.
Volcano map of differential expression genes and numbers of DEGs in each comparison. A Volcano map of differential expression genes in OIV vs OII. B Volcano map of differential expression genes in OIV vs T. C Volcano map of differential expression genes in OII vs T. D Numbers of DEGs in each comparison. Note: Each point in the differential expression volcano map represents a gene, and the abscissa represents the logarithm value of the differential multiple of the expression amount differential expression of a gene in the two samples, the larger the absolute value, and the greater the differential multiple of the expression amount between two samples. The ordinate represents the negative pair value of the error detection rate, the larger the value, the more significant the differential expression, and the more reliable the screened differential expression genes. Green and red dots represent genes with significant expression differences, green represents down-regulated gene expression, red represents up-regulated gene expression, and black dots represent genes with no significant expression differences
Enrichment and distribution of differential expression gene KEGG pathway of Pearlscale angelfish. A Enrichment and distribution of differential expression gene KEGG pathway in OIV vs OII. B Enrichment and distribution of differential expression gene KEGG pathway in OIV vs T. C Enrichment and distribution of differential expression gene KEGG pathway in OII vs T
Sexual dimorphic biological pathway
To identify biological pathway that shows sexual dimorphism in Pearlscale angelfish, the number of sex-biased genes was counted in different category of pathways. It shows that the number of up-regulated genes in most of metabolic pathways is much more in female than that in male (Fig. 5D). And the significantly divergent pathways of male and female gene numbers mainly include lipid metabolism, signal transduction, translation, and cell growth death. Among them, ovarian steroidogenesis, estrogen signaling pathway, progesterone-mediated oocyte maturation, prolactin signaling pathway, GnRH signaling pathway, oocyte meiosis, TGF-beta signaling pathway, steroid hormone biosynthesis, and Wnt signaling pathway play an significant part in gonadal development. By analyzing this, we found the signaling pathway, ovarian steroidogenesis (shown as Fig. S6 and Table S1 in Supplementary), and estrogen signaling pathway (shown as Fig. S7 in Supplementary). They are likely to influence their sexual dimorphism.
RT-qPCR verification
Twelve differential expression genes were selected from the transcriptome of Pearlscale angelfish for RT-qPCR verification (Primers were shown in Table 4). A single band was obtained for ovaries and testes using rps29 as a reference gene (Fig. 7). Among them, in OIV vs OII, the mRNA expression levels of amh, sox9b, dmrt2, and cyp17a were increased, while that of dmrt1, sox3, sox4, sox11, sox17, cyp11a, cyp19a, and hsd3b7 were reduced; the results were the same as RNA-Seq (Fig. 8A); in OIV vs T, the mRNA expression levels of amh, dmrt1, cyp17a, cyp11a, cyp19a, sox9b, and dmrt2 were increased; other genes were reduced, showing the same trend as RNA-Seq (Fig. 8B); in OII vs T, the mRNA expression levels of the sox3, sox4, sox11, and sox17 were reduced, while other genes of these were increased, which were the same as RNA-Seq (Fig. 8C). The above results showed similar patterns of mRNA abundance in RNA-Seq and qRT-PCR analysis, demonstrating the reliability of the RNA-Seq results in Pearlscale angelfish.
Moreover, we verified the relative expression of these genes during the gonadal development (OII, OT, OIV, and T); amh, sox9b, dmrt1, dmrt2, cyp11a, cyp17a, and cyp19a were significantly expressed in testes; and the expression of amh, sox9b, dmrt2, and dmrt1 was significantly increased during the hermaphroditic period (OT), and finally reached the highest level in pure testis after sex reversal (Fig. 9); the cyp11a, cyp17a, and cyp19a genes showed no such trend (Fig. 10). Sox3, sox4, and hsd3b7 were significantly expressed in ovary at the perinucleolus stage (OII) (Fig. 11), but sox11 and sox17 were significantly expressed in ovary at the yolk vesicle stage (OIV) (Fig. 12); the same thing is that these genes were significantly reduced during hermaphroditic period (OT) (Figs. 11 and 12).
Discussion
The reversal of sex reflects the particularity and diversity of reproductive strategies in fish. The order of gonadal differentiation is different in the fish of the natural reversal. Pearlscale angelfish is a kind of protogynous hermaphrodite fish with a natural sexual reversion. As we can see from the results of the paraffin section, in the process of gonadal development, Pearlscale angelfish differentiated into ovaries at first (Fig. 1A, B), and then, male germ cells gradually differentiated from the ovary of the largest individual female fish, forming facultative gonads which coexisted of the male and female germ cells (Fig. 1C, D), and finally completely differentiated into pure testis (Fig. 1E). The difference in the order of gonadal differentiation reflects species specificity and may be related to the reproductive strategy of fish. For most fish, the growth rates and individual sizes of male and female fish showed obvious differences, and some fish have sexual reversal in their life history, such as protogyny in Monopterus albus (Chan and Phillips 2010), Dentex dentex (Chatzifotis et al., 2004), Esox lucius (Senay et al., 2017), and many kinds of Epinephelus (Xiao et al., 2020; Wu et al., 2020b), and another protandry such as yellowtail clownfish (Amphiprion clarkii) (Nakamura et al. 2015), Acanthopagrus schlegeli (Chang et al., 1994), and various kinds of clownfish (Casas et al., 2020; Zhang et al., 2020a, 2020c). Although Nile tilapia (Paul-Prasanth et al., 2014) and zebrafish (Danio rerio) (Takatsu et al. 2013) also have the phenomenon of female prematuration, male and female gonads coexist in the larval stage. Then, the ovary developed at first, and some of the females in the ovary at the perinucleolus stage degradation and masculine then turn into a functional male fish. There were no oocytes in the sperms of the male medaka after sex reversal (Hayasaka et al., 2019); females in the protandrous Amphiprion also have only ovarian tissue after sex reversal (Nakamura et al., 2015); and there was no obvious testis in the gonads of the mature females after sex reversal in blackhead seabream (Acanthopagrus schlegelii) (Chang et al., 1994) and yellowfin seabreams (Acanthopagrus latus) (Abou-Seedo et al., 2003; Li et al., 2020). The coexistence of bisexual cells was not found in pure testis and ovary of the Pearlscale angelfish (Fig. 1A, B, E), which may be the reason for its one-way reversal from female to male.
In this study, RNA-Seq techniques were used for transcriptome analysis; at the same time, many genes related to gonadal development was identified and screened for understanding their corresponding functions from a biological perspective. Some of these genes are involved in gonadal development or sexual differentiation and are likely key genes for sexual reversal in Pearlscale angelfish. As reported, a unique new gene was screened from the gonadal tissue of rainbow trout by RNA-Seq technology, which was only expressed in the testis. Further studies showed that this unique gene was located on Y chromosome and tightly connected to the sex locus, which could be concluded as a necessary condition for the initiation of testis differentiation (Yano et al., 2012). Therefore, our study is also aimed at screening sex-related genes to provide a large number of sex-biased genes for future research. After sequencing, the total of 142, 589 unigenes was assembled in our study. The N50 value of the assembled unigenes was 2263 bp (Table 2), which was longer than those assembled in other teleost fish, such as Lateolabrax japonicus (1,710 bp) (Zhao et al., 2018), Oncorhynchus mykiss (758 bp) (Hao et al., 2011), Scophthalmus maximus (756 bp) (Salem et al. 2010), and Misgurnus anguillicaudatus (611 bp) (Long et al., 2013). From the results of all-unigene function annotation, we can see that 26.28% of unigenes had significant similarity to L. crocea (8019) (Fig. 4), which indicated that gene homology was conserved between Pearlscale angelfish and L. crocea, and that Pearlscale angelfish was more closely related to L. crocea, both of which belonged to the order Perciformes and no enough comparable fish. For most fish, the growth rates and individual sizes of male and female fish showed obvious differences. Pearlscale angelfish is a kind of protogynous hermaphrodite fish with a natural sexual reversion, whose growth, development, and reproduction are also very important. To identify DEGs showing significant differences in expression in the ovaries and testes of Pearlscale angelfish, the comparisons between the OII vs OIV, OIV vs T, and OII vs T were carried out. Furthermore, all-unigene identified above was clustered by GO and KEGG for screening reproduction and growth-related genes that are specifically expressed in OII, OIV, and T. The results of the transcriptomes of gonads of flounder showed that the sex-related biological pathways are elucidated, such as gonadal steroid generation and estrogen signaling pathway, providing further insights into sex determination and gonadal development (Fan et al., 2014). The key expression genes were identified by RNA-Seq in gonad of southern bluefin tuna (Thunnus maccoyii), as well as the specific expression patterns in male and female gonadal cells were studied for further identifying the key genes which involved in the reproductive molecular pathway, especially in gonadal germ cell development (Bar et al., 2016).
As a special organ of female or male fish, ovary or testis plays an important role in the growth and reproduction of fish, which is responsible for the production of gametes, the formation of sex hormones, and catalysis (Sarà et al., 2012). Steroids are a kind of small and hydrophobic hormones, which can be combined with the specific receptors through the cell membrane by osmosis, such as estrogen receptor (ER) and androgen (Sandra and Norma 2010). Then, the steroids combination goes into the nucleus and causes certain physiological reaction in turn, such as germ cell proliferation, gonad development, and gender differentiation (Devlin and Nagahama 2002). After comparing with the result of KEGG pathway in OIV vs OII, OIV vs T, and OII vs T, the top three KEGG pathway were different, which may be related to the activation of different signaling pathways during gonadal development; thus, there are differences. In the present study, 29 genes were identified to be involved in the ovarian steroidogenesis pathway, 15 genes were as the male-biased genes, and 12 genes were as the female-biased genes (see Fig. S6 and Table S1 in Supplementary). In this study, 3-hydroxyl steroid dehydrogenase (hsd3b) was identified as a mature ovary-bias expressed gene and indicates that it plays an important role in the synthetic estrogen and may be involved in the ovarian steroid generated pathway of Pearlscale angelfish, as reported hsd3b7 in Takifugu rubripes (Zou et al,. 2020) and P. olivaceus (Yan et al., 2018). Transcriptome of undifferentiated gland (PG), juvenile ovary (OJ), and adult ovary (OA) of the Yellow River carp (Cyprinus carpio) was studied; in the DEGs between PG and OJ, some upstream regulators of gonadal development, such as Cyp19a and sox9, were strongly expressed in PG, while some oocyte-specific genes, such as Nobox, BMP15, and Zp2, were up-regulated in OJ; in the DEGs between OJ and OA, many oocyte physiological function-related genes were up-regulated in OA, such as Fern-1 and foxl2 (Jia et al., 2017). From the results of DEGs in RNA-Seq in our study, some male sex-determining genes, such as amh, dmrt2, dmrt1, sox9b, cyp17a, and cyp19a, the expression of these genes in testes were significantly increased, while the expression of some female-bias genes in ovaries was significantly increased, such as sox3, sox4, and hsd3b7. The interaction of these genes may affect gender change and then reverse to spermatozoa under the influence of biological factors such as environments during the transition from female to male. It is suggested that the sexual reversion in Pearlscale angelfish may be subject to complex controls by multiple agents via pathways, not only the ovarian steroid generated pathway, estrogen signaling pathway, but also the steroid hormone biosynthesis, the autophagy, apoptosis pathways, and others.
In order to more accurately elucidate their relationship to gender determination and differentiation, and to understand their characteristics and molecular mechanisms of sexual reversal in Pearlscale angelfish, we verified the relative expression of these twelve genes during the gonadal development (OII, OT, OIV, and T). The expression of the amh, sox9b, dmrt2, and dmrt1 was low in the OII and OIV, while they were significantly increased during the hermaphroditic period (OT), and finally reached the highest level in pure testis after sex reversal (Fig. 9), and sox3, sox4, hsd3b7, sox11, and sox17 were significantly reduced during the hermaphroditic period (OT) (Figs. 11 and 12). These genes may play an important role in the process of sex reversal. As reported by it, the expression of dmrt1 in the gonads of male and female fish showed sexual dimorphism at different stages by in situ hybridization and RT-qPCR, indicating that dmrt1 played an important role in the determination and differentiation of the sex of largemouth bass (Yan et al., 2019). In addition, amh, Cyp19a, and dmrt1 interact during gonadal development and sexual differentiation (Chen et al., 2019a; Liu et al., 2020; Wu et al., 2020a). Dmrt1 and sox9 are specifically expressed in male gonads, and the expressions of dmrt1 and sox9 in early gonads are necessary for testicular differentiation (Raghuveer et al., 2011). This is consistent with our research, so it is further speculated that sox9b and dmrt1 play an important role in male gonadal maturation. The gonadal transcription of the spotted scat (Scatophagus argus) showed that a total of 136, 561 unigenes were obtained, the expression of sox9b was up-regulated in the testes, and sox3 was up-regulated in the ovaries (He et al., 2019), which was consistent with the results of our study. By combining PacBio isoform sequencing (Iso-Seq) and Illumina short-read RNA-Seq method, 60 early gamete-related genes were identified and screened; among them, the amh and Gsdf expressed higher in the testes, and the Cyp19a and foxl2 expressed higher in the ovaries in Amur sturgeon (Zhang et al., 2020b). Which was consistent with the results of our study, according to the changing trend of their expression levels at different stages of the gonad, it is speculated that the expression levels of sox3, sox4, hsd3b7, and other genes are decreased, which activates the expressions of amh, sox9b, dmrt2, dmrt1, and other genes, thus causing the atrophying and disappearance of oocytes and promoting the generation of male germ cells at different stages. And cyp11a, cyp17a, and cyp19a genes were not significantly changed during the hermaphroditic period (OT) (Fig. 10), but were highly expressed in the testis, suggesting that these genes may not be the key genes involved in the process of sexual reversal, but related genes in the process of testis development. In this study, the results showed similar patterns of mRNA abundance in RNA-Seq analysis and qRT-PCR, demonstrating the reliability of the RNA-Seq results in Pearlscale angelfish. The sex-related genes selected in this study may provide important clues for future studies on gonadal development and sex-determining mechanism in Pearlscale angelfish.
Conclusions
The transcriptomes of the ovary at the perinucleolus stage (OII), ovary at the yolk vesicle stage (OIV), and testis (T), respectively, have screened the gonadal differential expression genes (DEGs) and analyzed the expression of these genes in gonads of different sexes. At the same time, the results verified by RT-qPCR were consistent with the variation trend of transcriptomics data. Among the results, the expression of the amh, sox9b, dmrt2, and dmrt1 were low in the OII and OIV, while they were significantly increased during the ovotestis in the hermaphroditic period (OT), and finally reached the highest level in pure testis after sex reversal, and sox3, sox4, hsd3b7, sox11, and sox17 were significantly reduced during the hermaphroditic period (OT). The interaction of these genes may affect gender change and then reverse to spermatozoa under the influence of biological factors such as environments during the transition from female to male. However, the interaction details of the sexual reversion with the other regulatory pathways, such as ovarian steroid generated pathway, estrogen signaling pathway, steroid hormone biosynthesis, the autophagy, apoptosis pathways, and so on, are worth being further investigated. These results provide a data source to study the regulatory mechanisms and molecular characteristics of reproduction in Pearlscale angelfish. The results also present basic support for sex and reproduction control of Pearlscale angelfish in the aquaculture industry and provide a theoretical basis for fish developmental biology.
Data availability
BioSample accessions: SAMN17817928, SAMN17817929, SAMN17817930, SAMN17817931, SAMN17817932, SAMN17817933, SAMN17817934, SAMN17817935, SAMN17817936 Temporary SubmissionID: SUB9037523. Release date: 2022–02-01, or with the release of linked data, https://www.ncbi.nlm.nih.gov/biosample/17817929, https://www.ncbi.nlm.nih.gov/biosample/17817929, https://www.ncbi.nlm.nih.gov/biosample/17817930, https://www.ncbi.nlm.nih.gov/biosample/17817931, https://www.ncbi.nlm.nih.gov/biosample/17817932, https://www.ncbi.nlm.nih.gov/biosample/17817933, https://www.ncbi.nlm.nih.gov/biosample/17817934, https://www.ncbi.nlm.nih.gov/biosample/17817935, https://www.ncbi.nlm.nih.gov/biosample/17817936.
Code availability
Not applicable.
References
Abou-Seedo FS, Dadzie S, Al-Kanaan KA (2003) Sexuality, sex change and maturation patterns in the yellowfin seabream, Acanthopagrus latus (Teleostei : Sparidae) (Houttuyn, 1782). J Appl Ichthyol 19(2):65–73. https://doi.org/10.1046/j.1439-0426.2003.00355.x
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI BLAST: a new generation of protein database search programs. FASEB Journal, 25(17): 3389–3402. http://blast.ncbi.nlm.nih.gov/Blast.cgi
Bar I, Cummins S, Elizur A (2016) Transcriptome analysis reveals differentially expressed genes associated with germ cell and gonad development in the southern bluefin tuna (Thunnus maccoyii). BMC Genomics 17:217. https://doi.org/10.1186/s12864-016-2397-8
Barros NHC, de Souza AA, Peebles EB, Chellappa S (2017) Dynamics of sex reversal in the marbled swamp eel (Synbranchus marmoratus Bloch 1795), a diandric hermaphrodite from Marechal Dutra reservoir northeastern Brazil. Journal of Applied Ichthyology 33(3) 443 449https://doi.org/10.1111/jai.13273
Casas L, Saenz-Agudelo P, Irigoien X (2020) High-throughput sequencing and linkage mapping of a clownfish genome provide insights on the distribution of molecular players involved in sex change. Sci Rep 8:4073. https://doi.org/10.1038/s41598-018-22282-0
Chan STH, Phillips JG (2010) The structure of the gonad during natural sex reversal in Monopterus albus (Pisces: Teleostei). J Zool 151(1):129–141. https://doi.org/10.1111/j.1469-7998.1967.tb02868.x
Chang CF, Lee MF, Chen GR (1994) Estradiol-17β associated with the sex reversal in protandrous black porgy, Acanthopagrus schlegeli. J Exp Zool 268(1):53–58. https://doi.org/10.1002/jez.1402680107
Chao QH, Shen FF, Xue YD, Wu JK, Zhang JL (2020) Cbx2, a PcG family gene, plays a regulatory role in medaka gonadal development. Int J Mol Sci 21(4):1288. https://doi.org/10.3390/ijms21041288
Chatzifotis S, Muje P, Pavlidis M, Agren J, Paalavuo M, Molsa H (2004) Evolution of tissue composition and serum metabolites during gonadal development in the common dentex (Dentex dentex). Aquaculture 236(1–4):557–573. https://doi.org/10.1016/j.aquaculture.2003.12.004
Chen CY, Tsai YJ, Chang CF (2019a) The roles of cyp19ala and dmrt1 during gonadal sex differentiation and sex change in orange-spotted grouper, Epinepheluscoioides. J Mar Sci Technol 27(3):282–291. https://doi.org/10.6119/JMST.201906_27(3).0011
Chen J, Xiao L, Peng C, Ye ZF, Wang DD, Yang YQ, Zhang HF, Zhao M, Li SS, Lin HR, Zhang Y (2019b) Socially controlled male-to-female sex reversal in the protogynous orange-spotted grouper, Epinephelus coioides. J Fish Biol 94(3):414–421. https://doi.org/10.1111/jfb.13911
Devlin RH, Nagahama Y (2002) Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208(3–4):191–364. https://doi.org/10.1016/S0044-8486(02)00057-1
Dibattista JD, Gaither MR, Hobbs JPA, Rocha LA, Bowen BW (2016) Angelfishes, paper tigers, and the devilish taxonomy of the Centropyge flavissima complex. J Hered 107(7):647–653. https://doi.org/10.1093/jhered/esw062
Fan ZF, You F, Wang LJ, Weng SD, Wu ZH, Hu JW, Zou, YX, Tan XG, Zhang PJ (2014) Gonadal transcriptome analysis of male and female olive flounder (Paralichthys olivaceus). Biomed Research International, 291067https://doi.org/10.1155/2014/291067
Fernandez-Silva I, Henderson JB, Rocha LA, Simison WB (2018) Whole-genome assembly of the coral reef pearlscale pygmy angelfish (Centropyge vrolikii). Sci Rep 8(1):1489. https://doi.org/10.1038/s41598-018-19430-x
Finn RD, Mistry J, Tate J, Coggill P (2010) Pfam: the protein families database. Nucleic Acids Research 38:D211–D222. https://doi.org/10.1093/nar/gkp985
Forner-Piquer I, Fakriadis I, Mylonas CC, Piscitelli F, Di-Marzo V, Maradonna F, Calduch-Giner J, Perez-Sanchez J, Carnevali O (2019) Effects of dietary bisphenol A on the reproductive function of gilthead sea bream (Sparus aurata) testes. Int J Mol Sci 20(20):5003. https://doi.org/10.3390/ijms20205003
Gao JN, Li PZ, Zhang W, Wang ZG, Wang XB, Zhang QQ (2015) Molecular cloning, promoter analysis and expression profiles of the sox3 gene in Japanese flounder, Paralichthys olivaceus. Int J Mol Sci 16(11):27931–27944. https://doi.org/10.3390/ijms161126079
Gust N (2004) Variation in the population biology of protogynous coral reef fishes over tens of kilometres. Can J Fish Aquat Sci 61(2):205–218. https://doi.org/10.1139/f03-160
Hao DC, Ge GB, Xiao PG, Zhang YY, Yang L (2011) The first insight into the tissue specific taxus transcriptome via Illumina second generation sequencing. PLoS ONE 6(6):e21220. https://doi.org/10.1371/journal.pone.0021220
Hayasaka O, Takeuchi Y, Shiozaki K, Anraku K, Kotani T (2019) Green light irradiation during sex differentiation induces female-to-male sex reversal in the medaka Oryzias latipes. Sci Rep 9:2383. https://doi.org/10.1038/s41598-019-38908-w
He FX, Jiang DN, Huang YQ, Mustapha UF, Yang W, Cui XF, Tian CX, Chen HP, Shi HJ, Deng SP, Li GL, Zhu CH (2019) Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish Physiol Biochem 45(6):1963–1980. https://doi.org/10.1007/s10695-019-00693-8
Jia Y, Nan P, Zhang WW, Wang F, Zhang RH, Liang TT, Ji XL, Du QY, Chang ZJ (2017) Transcriptome analysis of three critical periods of ovarian development in Yellow River carp (Cyprinus carpio). Theriogenology 105:15–26. https://doi.org/10.1016/j.theriogenology.2017.08.027
Jin SB, Zhang Y, Dong XL, Xi QK, Song D, Fu HT, Sun DJ (2015) Comparative transcriptome analysis of testes and ovaries for the discovery of novel genes from Amur sturgeon (Acipenser schrenckii). Genet Mol Res 14(4):18913–18927. https://doi.org/10.4238/2015.December.28.40
Lande R, Seehausen O, Alphen J (2001) Mechanisms of rapid sympatric speciation by sex reversal and sexual selection in cichlid fish. Genetica 112–113(1):435–443. https://doi.org/10.1023/A:1013379521338
Liu W, Zhang H, Xiang YX, Jia KT, Luo MF, Yi MS (2020) A novel germline and somatic cell expression of two sexual differentiation genes, dmrt1 and foxl2 in marbled goby (Oxyeleotris marmorata). Aquaculture 516:734619. https://doi.org/10.1016/j.aquaculture.2019.734619
Li SZ, Lin GM, Fang WY, Huang PL, Gao D, Huang J, Xie JG, Lu JG (2020) Gonadal transcriptome analysis of sex-related genes in the protandrous yellowfin seabream (Acanthopagrus latus). Front Genet 11:709. https://doi.org/10.3389/fgene.2020.00709
Long Y, Li Q, Zhou BL, Song GL, Li T, Cui Z (2013) De Novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PLoS ONE 8(2):e56998. https://doi.org/10.1371/journal.pone.0056998
Maxfield JM, Cole KS (2019) Structural changes in the ovotestis of the bidirectional hermaphrodite, the blue-banded goby (Lythrypnus dalli), during transition from ova production to sperm production. Environ Biol Fishes 102(11):1393–1404. https://doi.org/10.1007/s10641-019-00914-2
Michael T (2007) Reversed sexual dimorphism and courtship by females in the Topaz cichlid, Archocentrus myrnae (Cichlidae, Teleostei), from Costa Rica. Southwest Nat 52(3):371–377. https://doi.org/10.1894/0038-4909(2007)52[371:RSDACB]2.0.CO;2
Nakamura M, Miura S, Nozu R, Kobayashi Y (2015) Opposite-directional sex change in functional female protandrous anemonefish, Amphiprion clarkii: effect of aromatase inhibitor on the ovarian tissue. Zool Lett 1(30):1–5. https://doi.org/10.1186/s40851-015-0027-y
Nakashima Y, Sakai Y, Karino K, Kuwamura T (2000) Female-female spawning and sex change in a haremic coral-reef fish Labroides Dimidiatus. Zool Sci 17(7):967–970. https://doi.org/10.2108/zsj.17.967
Paul-Prasanth B, Bhandari RK, Kobayashi T, Horiguchi R, Kobayashi Y, Nakamoto M, Shibata Y, Sakai F, Nakamura M, Nagahama Y (2014) Estrogen oversees the maintenance of the female genetic program in terminally differentiated gonochorists. Sci Rep 3:2862. https://doi.org/10.1038/srep03787
Qian X, Ba Y, Zhuang QF, Zhong GF (2014) RNA-Seq technology and its application in fish transcriptomics. Omics J Int Biol 18(2):98–110. https://doi.org/10.1089/omi.2013.0110
Qu M, Ding SX, Schartl M, Adolfi MC (2020) Spatial and temporal expression pattern of sex-related genes in ovo-testis of the self-fertilizing mangrove killifish (Kryptolebias marmoratus). Gene 742:144581. https://doi.org/10.1016/j.gene.2020.144581
Raghuveer K, Senthilkumaran B, Sudhakumari CC, Sridevi P, Rajakumar A, Singh R, Murugananthkumar R, Majumdar KC (2011) Dimorphic expression of various transcription factor and steroidogenic enzyme genes during gonadal ontogeny in the air-breathing catfish, Clarias Gariepinus. Sexual Development 5(4):213–223. https://doi.org/10.1159/000328823
Sakai Y, Karino K, Kuwamura T, Nakashima Y, Maruo Y (2003) Sexually dichromatic protogynous angelfish Centropyge ferrugata (Pomacanthidae) males can change back to females. Zoolog Sci 20(5):627–633. https://doi.org/10.2108/zsj.20.627
Salem M, Rexroad CE, Wang JN, Thorgaard GH, Yao JB (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11(1):564. https://doi.org/10.1186/1471-2164-11-564
Sandra GE, Norma MM (2010) Sexual determination and differentiation in teleost fish. Rev Fish Biol Fisheries 20(1):101–121. https://doi.org/10.1007/s11160-009-9123-4
Sarà G, Reid G, Rinaldi A, Palmeri V, Troell M, Kooijman S (2012) Growth and reproductive simulation of candidate shellfish species at fish cages in the Southern Mediterranean: dynamic Energy Budget (DEB) modelling for integrated multi-trophic aquaculture. Aquaculture 324:259–266. https://doi.org/10.1016/j.aquaculture.2011.10.042
Senay C, Harvey-Lavoie S, Macnaughton C, Bourque G (2017) Morphological differentiation in northern pike (Esox lucius): the influence of environmental conditions and sex on body shape. Canadian Journal of Zoology, 95(6): https://doi.org/10.1139/cjz-2016-0159
Shao CW, Li QY, Chen SL, Zhang P, Lian JM, Hu QM, Sun B, Jin LJ, Liu SS, Wang ZJ (2014) Epigenetic modification and inheritance in sexual reversal of fish. Genome Res 24(4):604–615. https://doi.org/10.1101/gr.162172.113
Takatsu K, Miyaoku K, Roy SR, Murono Y, Sago T, Itagaki H, Nakamura M, Tokumoto T (2013) Induction of female-to-male sex change in adult zebrafish by aromatase inhibitor treatment. Sci Rep 3:3400. https://doi.org/10.1038/srep03400
Tian F, Liu SJ, Shi JQ, Qi HF, Zhao K, Xie BS (2019) Transcriptomic profiling reveals molecular regulation of seasonal reproduction in Tibetan highland fish Gymnocypris Przewalskii. BMC Genomics 20(1):2. https://doi.org/10.1186/s12864-018-5358-6
Trukhina AV, Lukina NA, Wackerow-Kouzova ND, Smirnov AF (2013) The variety of vertebrate mechanisms of sex determination. BioMed Research International, 587460. https://doi.org/10.1155/2013/587460
Wang X, Liu Q, Xiao Y, Yang Y, Wang Y, Song Z, You F, An H, Li J (2017) High temperature causes masculinization of genetically female olive flounder Paralichthys olivaceus accompanied by primordial germ cell proliferation detention. Aquaculture 479:808–816. https://doi.org/10.1016/j.aquaculture.2017.07.019
Wang WX, Liang SS, Zou YX, Wu ZH, Wang LJ, Liu Y, You F (2020) Amh dominant expression in sertoli cells during the testicular differentiation and development stages in the olive flounder Paralichthys olivaceus. Gene 755:144906. https://doi.org/10.1016/j.gene.2020.144906
Wei L, Li X, Li M, Tang Y, Wei J, Wang D (2019) Dmrt1 directly regulates the transcription of the testis-biased sox9b gene in Nile tilapia (Oreochromis niloticus). Gene 687:109–115. https://doi.org/10.1016/j.gene.2018.11.016
Wu K, Song WY, Zhang ZW, Ge W (2020a) Disruption of dmrt1 rescues the all-male phenotype of the cyp19a1a mutant in zebrafish a novel insight into the roles of aromatase/estrogens in gonadal differentiation and early folliculogenesis. Development, 147(4): dev182758. https://dev.biologists.org/content/147/4/dev182758.supplemental
Wu X, Yang Y, Zhong CY, Guo Y, Wei TY, Li SS, Lin HR, Liu XC (2020b) Integration of ATAC-seq and RNA-seq unravels chromatin accessibility during sex reversal in orange-spotted grouper (Epinephelus coioides). Int J Mol Sci 21(8):2800. https://doi.org/10.3390/ijms21082800
Xiao L, Guo Y, Wang DD, Zhao M, Hou X, Li SH, Lin HR, Zhang Y (2020) Beta-hydroxysteroid dehydrogenase genes in orange-spotted grouper (Epinephelus coioides): genome-wide identification and expression analysis during sex reversal. Front Genet 11:161. https://doi.org/10.3389/fgene.2020.00161
Yan HW, Shen XF, Cui X, Wu YM, Wang LS, Zhang L, Liu Q, Jiang YS (2018) Identification of genes involved in gonadal sex differentiation and the dimorphic expression pattern in Takifugu rubripes gonad at the early stage of sex differentiation. Fish Physiol Biochem 44(5):1275–1290. https://doi.org/10.1007/s10695-018-0519-8
Yan NN, Hu J, Li J, Dong JJ, Sun CF, Ye X (2019) Genomic organization and sexually dimorphic expression of the dmrt1 gene in largemouth bass (Micropterussalmoides). Comp Biochem Physiol Mol Biol 234:68–77. https://doi.org/10.1016/j.cbpb.2019.05.005
Yang Y, Liu Q, Xiao Y, Xu S, Wang X, Yang J, Li J (2020) Effects of environmental stress (sex steroids and heat) during sex differentiation in Japanese flounder (Paralichthys olivaceus): insight from germ cell proliferation and gsdf-amh-cyp19a1a expression. Aquaculture 515:734536. https://doi.org/10.1016/j.aquaculture.2019.734536
Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, Bouchez O, Fostier A, Guiguen Y (2012) An immune-related gene evolved into the master sex-determining gene in rainbow trout Oncorhynchus Mykiss. Curr Biol 22(15):1423–1428. https://doi.org/10.1016/j.cub.2012.05.045
Yoshinaga N, Shiraishi E, Yamamoto T, Iguchi T, Abe SI, Kitano T (2004) Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem Biophys Res Commun 322(2):508–513. https://doi.org/10.1016/j.bbrc.2004.07.162
Yu HY, Du XX, Li XJ, Qu JB, Zhu H, Zhang QQ, Wang XB (2018) Genome-wide identification and transcriptome-based expression analysis of sox gene family in the Japanese flounder Paralichthysolivaceus. J Oceanol Limnol 36(5):1731–1745. https://doi.org/10.1007/s00343-018-7216-4
Yue H, Li C, Du H, Zhang S, Wei Q (2015) Sequencing and de novo assembly of the gonadal transcriptome of the endangered Chinese sturgeon (Acipenser sinensis). PLoS ONE 10(6):e0127332. https://doi.org/10.1371/journal.pone.0127332
Zhang H, Zhang YY, Guo Y, Zhang X, Wang Q, Liu XC, Lin HR (2020a) Kiss2 but not kiss1 is involved in the regulation of social stress on the gonad development in yellowtail clownfish Amphiprion Clarkii. Gen Comp Endocrinol 298:113551. https://doi.org/10.1016/j.ygcen.2020.113551
Zhang XJ, Zhou JB, Li LM, Huang WZ, Ahmad HI, Li HM, Jiang HY, Chen JP (2020b) Full-length transcriptome sequencing and comparative transcriptomic analysis to uncover genes involved in early gametogenesis in the gonads of Amur sturgeon (Acipenser schrenckii). Front Zool 17(1):11. https://doi.org/10.1186/s12983-020-00355-z
Zhang YY, Zhang H, Wang J, Zhang X, Bu SY, Liu XC, Wang Q, Lin HR (2020c) Molecular characterization and expression patterns of glucocorticoid receptor (GR) genes in protandrous hermaphroditic yellowtail clownfish Amphiprion Clarkii. GENE 745:144651. https://doi.org/10.1016/j.gene.2020.144651
Zhao C, Wang P, Qiu L (2018) RNA-Seq-based transcriptome analysis of reproduction- and growth-related genes in Lateolabrax japonicus ovaries at four different ages. Mol Biol Rep 45(6):2213–2225. https://doi.org/10.1007/s11033-018-4383-5
Zou CC, Wang LJ, Zou YX, Wu ZH, Wang WX, Liang SS, Wang L, You F (2020) Characteristics and sex dimorphism of 17 beta-hydroxysteroid dehydrogenase family genes in the olive flounder Paralichthys olivaceus. J Steroid Biochem Mol Biol 199:105597. https://doi.org/10.1016/j.jsbmb.2020.105597
Funding
This project was supported by the Natural Science Foundation of Fujian Province (2018J01451); the Fujian Engineering Research Center of Aquatic Breeding and Healthy Aquaculture (DF201905); the Fujian University Students Innovation and Entrepreneurship Training Program project (201910390047); and the Jimei University Students Innovation and entrepreneurship training program project (2020xj053; 2020xj059).
Author information
Authors and Affiliations
Contributions
Zhong ZW and Wang SH carried out the experiments, performed the statistical analyses, and drafted the manuscript. Ao LL, Zhao LP, and Ma SW participated in protocol development and data analysis. Jiang YH and Wang YL designed and supervised the experiments, analyzed the data, and critically edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All experimental protocols involved in this study were approved by the Regulations for the Administration of Affairs Concerning Experimental Animals for the Science and Technology Bureau of China. Experiments involving Pearlscale angelfish were approved by the Animal Research and Ethics Committee of Fisheries College Jimei University.
Consent to participate
All authors have discussed the study procedures and have been satisfied with the relevant questions, and all have agreed to participate in the study.
Consent for publication
All authors have read, reviewed, and revised this article, and all agree to publish this article.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, Z., Ao, L., Wang, Y. et al. Comparison of differential expression genes in ovaries and testes of Pearlscale angelfish Centropyge vrolikii based on RNA-Seq analysis. Fish Physiol Biochem 47, 1565–1583 (2021). https://doi.org/10.1007/s10695-021-00977-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-021-00977-y