Abstract
Ecological speciation has been the subject of intense research in evolutionary biology but the genetic basis of the actual mechanism driving reproductive isolation has rarely been identified. The extreme polymorphism of the major histocompatibility complex (MHC), probably maintained by parasite-mediated selection, has been proposed as a potential driver of population divergence. We performed an integrative field and experimental study using three-spined stickleback river and lake ecotypes. We characterized their parasite load and variation at MHC class II loci. Fish from lakes and rivers harbor contrasting parasite communities and populations possess different MHC allele pools that could be the result of a combined action of genetic drift and parasite-mediated selection. We show that individual MHC class II diversity varies among populations and is lower in river ecotypes. Our results suggest the action of homogenizing selection within habitat type and diverging selection between habitat types. Finally, reproductive isolation was suggested by experimental evidence: in a flow channel design females preferred assortatively the odor of their sympatric male. This demonstrates the role of olfactory cues in maintaining reproductive isolation between diverging fish ecotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Various theories have been proposed to explain speciation (Coyne and Orr 2004). While speciation modes based on neutral evolutionary processes are widely accepted (Maynard Smith 1966; Mayr 1942), the idea that ecology can play a major role in species formation has recently re-emerged (Bolnick and Fitzpatrick 2007; Dieckmann et al. 2004; Hendry 2009; Schluter and Rambaut 1996; van Doorn et al. 2009). Ecological speciation is defined as the process by which reproductive isolation between populations evolves from adaptation to different environments even in sympatry.
To date, ecological speciation has typically been examined on the basis of morphological traits such as beaks of Darwin’s finches (Grant and Grant 1982), which reflect adaptations to food regime and have been shown to be associated with mating preference (Huber et al. 2007). Other instances of ecological speciation relate to lizards (Losos et al. 1998) and whitefish (Harrod et al. 2010). In contrast to morphological adaptation, local adaptations related to parasite pressure have been neglected, although parasites are ubiquitous (Kuris et al. 2008; Poulin 1996) and impose strong selection pressure on all free-living organisms (Hamilton and Zuk 1982).
Differences in the abiotic and biotic conditions associated with different macro- and micro-habitats such as oxygen concentrations or community structure, e.g., presence of competitors or predators, are likely to influence the presence and abundance of both obligatory intermediate hosts of the parasites (Halmetoja et al. 2000), and the parasites themselves. These differences may therefore result in the development and maintenance of contrasting parasite communities (Thompson 1994), which then ultimately should lead to contrasting immunogenetic adaptations in the respective host populations.
A major challenge in elucidating the role of parasite pressure in ecological speciation is to determine which genes underlie phenotypic variation in the host. In this regard, the identification of the major histocompatibility complex (MHC) in vertebrates as a key component of the adaptive immune system to respond to pathogen infection was a breakthrough. The MHC is a large cluster of genes, which encode antigen presenting molecules and therefore play a key role in molecular self/non-self discrimination and in the activation of adaptive immune response (Janeway et al. 2005). MHC genes are the most polymorphic genes found in vertebrates: this high intra- and inter-individual polymorphism is thought to be maintained by negative frequency-dependent selection, heterozygote advantage and habitat heterogeneity mediated by parasites (for review see Apanius et al. 1997; Milinski 2006; Spurgin and Richardson 2010). Among those mechanisms, habitat heterogeneity has only recently been the focus of research. A number of studies on MHC variation in neighboring populations proposed that habitat heterogeneity shapes MHC composition, probably due to contrasting parasites communities (Alcaide et al. 2008; Babik et al. 2008; Blais et al. 2007; Ekblom et al. 2007).
In addition to pathogen resistance, MHC genes can also help maintaining a pre-mating barrier because of their crucial role in mate choice (Reusch et al. 2001a; Richardson et al. 2005; Schwensow et al. 2008; Yamazaki et al. 1976). This raises the question of what would happen to migrants or hybrids of two locally adapted ecotypes that are not able to sufficiently resist parasitism in a foreign habitat to which they are not adapted. For instance, maladapted migrants or hybrids might be unable to find a mating partner due to assortative mating that would reduce the risk of disruption of locally adapted genes for parasite-resistance (Eizaguirre et al. 2009b). Therefore, the pleiotropic role of MHC genes in resistance to parasites and mate choice could reduce gene flow and thus enhance the process of divergence between two locally adapted ecotypes, ultimately leading to speciation (Blais et al. 2007; Eizaguirre et al. 2009a; Summers et al. 2003). Theoretical models have indicated that when mate choice is based on ecologically important traits, divergence of that trait can accelerate allopatric speciation and facilitate sympatric speciation (Maynard Smith 1966). However, identification of such a “magic trait” (Gavrilets 2004) involved in both assortative mating and adaptation have rarely been reported (e.g., Jiggins et al. 2001; Podos 2001) and, except in cichlids with their polymorphic long wavelength-sensitve opsin gene (Seehausen et al. 2008), the genetic basis of magic traits is largely unknown. Due to their (i) extreme polymorphism (probably maintained by balancing selection); (ii) capacity to respond to environmental variation (parasites); and (iii) role in mate choice, MHC genes have a great potential in initiating the first steps of speciation and are ideal candidates to investigate ecology-mediated reproductive isolation.
The three-spined stickleback, Gasterosteus aculeatus, serves as prime example for rapid genetic divergence (Gibson 2005). Throughout the northern hemisphere, it is thought to have colonized freshwater habitats from adjacent estuarine or marine refuges following glacial retreat (reviewed in McPhail 1994). Over this short time period (ca. 12,000 years), ancestral marine genotypes rapidly differentiated into several ecotypes, which differ markedly in numerous morphological traits, behavior and ecology (e.g., Berner et al. 2008; Boughman 2001; Kitano et al. 2007; Marchinko and Schluter 2007) as a result of divergent selection (Schluter 1996) .
Female mate choice has been intensively investigated in the three-spined stickleback (Aeschlimann et al. 2003; Bakker 1993; Boughman 2001; Kitano et al. 2007; McLennan and McPhail 1990; Milinski and Bakker 1990). Particularly, olfactory recognition was shown to be able to contribute to reproductive isolation between sympatric benthic and limnetic sticklebacks from Paxton Lake (British Columbia, Canada) suggesting a potential role of MHC genes (Rafferty and Boughman 2006). Females assess their potential partners’ MHC make-up via perception of corresponding peptide ligands in odor signals (Milinski et al. 2005).
Reusch et al. (2001b), investigated sixteen stickleback populations in Northern Germany, and revealed the dominant role of habitat type in genetic divergence during the post glacial recolonization. Since there are no major obstacles (e.g., large dams or waterfalls) between lakes and rivers in this area, they regarded gene flow as possible among these populations. The genetic distance between ancestral marine populations and both lake or river populations was very similar, suggesting the occurrence of a single colonization event. Moreover, Reusch et al. (2001b) proposed that habitat-specific selection on phenotypic traits may have resulted in mating barriers and reduced gene flow between freshwater habitats. Here we hypothesize that MHC genes could represent the target of selection by contrasting parasites communities and as a consequence would also affect the pattern of mate choice in lake and river fish. Thus, we studied in the field whether some potential ingredients for this scenario can be found. (i) Are fish from lakes and rivers infected by different parasites? (ii) Have fish habitat-specific MHC allele pools? (iii) In the lab we tested experimentally whether female mate choice favors sympatric males, which may help maintaining population divergence. Based on previous studies (Reusch et al. 2001b; Wegner et al. 2003), we chose two characteristic lake and two river populations respectively in which we investigated parasite load, individual MHC diversity and MHC allele pools at the population level.
Methods
Fish sampling and dissections
We sampled three-spined sticklebacks (Gasterosteus aculeatus) from two small, slow-flowing rivers (Malenter Au (MA, N = 72), 54°12′16.19″N, 10°33′32.93″E and Schwale (SCW, N = 24) 54°0′24.47″N, 10°9′2.04″E) and two lakes (Großer Plöner See (GPS, N = 72), 54°9′21.16″N, 10°25′50.14″E, and Vierer See (VS, N = 29) 54°7′31.94″N, 10°26′39.55″E) from already described systems in Northern Germany (Kalbe et al. 2002; Rauch et al. 2006; Reusch et al. 2001b; Wegner et al. 2003). GPS and MA correspond to the water bodies called ASC and SÖH, respectively in the map of Reusch et al. 2001a. Young of the year fish were sampled using minnow traps and hand nets during winter 2007 (from January to March) in order to assure that fish were from the same age-cohort. Fish were killed by an overdose of MS222 and all external and internal macroparasites and ciliates were determined to the lowest taxonomic level possible following Kalbe et al. (2002). Briefly, all ectoparasites from the skin were counted as well as macroparasites in the gills, eyes, lateral fin muscles, liver, urinary bladder, head kidney, kidney and gut. Fish mass (±0.1 mg) as well as standard length (±1 mm) were recorded.
Microsatellite typing and MHC diversity
DNA extractions from tail fin tissue were performed using the Invisorb DNA Tissue HTS 96 Kit/R (Invitek, Germany) on an automated platform (Tecan, Switzerland) following the manufacturer’s protocol. All fish were typed for 9 microsatellites combined in 2 different PCR multiplex protocols (Kalbe et al. 2009). We used these markers to calculate an individual heterozygosity index (Coulson et al. 1998) and for population structure analysis.
The MHC class IIB diversity was determined by reference strand-mediated conformation analysis (RSCA) according to Lenz et al. (2009). This included amplification of the exon 2 of MHC class IIB genes, which encodes for the immunologically relevant peptide-binding groove of the MHC molecule. Cautious measures were taken to avoid the production of PCR artefacts during genotyping (Lenz and Becker 2008). The exon 2 of the MHC IIB genes in this species has previously been suggested to be under parasite-mediated positive selection as revealed by a high ratio of nonsynonymous over synonymous substitutions (dN/dS, Reusch et al. 2004). For reasons of simplicity, we refer to different sequence variants as alleles, although they may originate from different duplicated loci.
Olfactory-based mate choice
Further fish were collected between June and July 2006 and 2007 for mate choice tests from the river “Malenter Au” and the lake “Großer Plöner See”, two of the focal populations of this study. After capture, fish were held singly in 16 l tanks in summer conditions (day light cycle 16:8 h, 18°C water temperature), and males were provided with artificial nesting material (sand and nylon threads). All experimental tests were conducted between 10 days and 6 weeks post-collection.
For the olfactory mate-choice tests, a randomly-picked gravid female was placed in the choice chamber of a flow channel that was fed by two water columns, to each of which stimulus water (1L during 600 s) was continuously added, under conditions of laminar flow as described previously (Reusch et al. 2001b; Aeschlimann et al. 2003; Milinski et al. 2005). Fish were able to freely investigate the composition of water in the two halves of the chamber for two periods of 300 s each, with spatial reversal of water sources at halftime. Their choice in the chamber was video-recorded from above. There were lines drawn on the screen of the monitor, by which it was divided in front and back quarters. From the record we measured the time the female’s tip of the nose spent in each front quarter. If the two sources were equally attractive, the fish should spend an equal period of time with each source of stimulus. Odor preference as determined in the flow channel set-up reliably predicts mate choice (supplementary information of Milinski et al. 2005).
As a male’s nest is a reservoir of both MHC derived odor and a ‘male signaling factor’ that validates the MHC signal (Milinski et al. 2005, 2010; Sommerfeld et al. 2008), we took the stimulus water directly from above the nest and only from males that maintained (glued) their nest on the day of the test (see Milinski et al. 2005). On one side of the flow channel, stimulus water originated from a male from the same habitat as the test female, and on the other side from a male originating from the foreign habitat (sympatric male vs. allopatric male). All tests were conducted blind with respect to the origin of the water sources and were recorded with a digital processing video camera (Panasonic GP-KR222).
For these tests each pair of males was always tested sequentially with a female from the lake habitat and with a female from the river habitat. Each fish was used only in one quadruplet. With this procedure we obtained 9 quadruplets (i.e., 36 fish, 18 tests). Due to a limited pool of experimental animals, in two additional cases the quadruplets could not be completed: one pair of males could only be tested with a lake female and another pair of males was only tested with a river female, this resulted in a total of 20 trials. Tests were regarded as valid only if the females spawned within 24 h after the trial, which guarantees that the female had been choosy.
Statistical analyses
Except if stipulated, statistical analyses were conducted using the R statistical package (v. 2.5.0) for Windows XP and Primer v6 (Clarke and Gorley 2006). Normality of distributions and variance homoscedasticity were verified and tests conducted accordingly.
Population genetics: microsatellite analysis
Deviation from Hardy–Weinberg equilibrium was tested using the Markov Chain permutation test implemented in GENEPOP (Raymond and Rousset 1995). For each locus, we examined the resulting P-values both before and after sequential bonferroni correction for multiple tests. To examine whether river–lake sticklebacks formed genetically differentiated populations, we used STRUCTURE 2.3.1 (Pritchard et al. 2000). We used the admixture and independent allele model of the software without prior population information. We calculated the probability of there being one to four populations. We repeated each run 6 times with 100.000 equilibration steps followed by a run length of 100.000 Markov Chain Monte Carlo simulations. To determine the number of populations (K) most likely to explain the genetic data, we averaged the log-likelihoods across six runs and calculated ΔK following Evanno et al. (2005).
Then the most likely number of K clusters was used as groups for an analysis of molecular variance (AMOVA) performed in Arlequin 3.1. (Excoffier et al. 2005) to partition variance.
Parasite data
We first calculated the Shannon index for each fish, which accounts for both the diversity and the evenness of the parasite species present. The use of this index permits parasite load on fish to be compared according to their habitat of origin. We then performed a nested analysis of variance (ANOVA) with population of origin nested into habitat type as independent variables on individual Shannon index.
Difference in parasite communities among populations and between habitats was further tested with a nested analysis of similarity (ANOSIM) on a Bray-Curtis similarity matrix based on the square-root transformed abundance of different parasite species as variables. The relative abundance of each parasite species was compared using a permutation-based percentage test (SIMPER, Clarke and Gorley 2006). The parasites that contributed most (explaining up to 90% of the total difference) were then log transformed and used in a multivariate analysis of variance with the detected MHC alleles and habitat type as predictors (see MHC analysis). The same analyses were also performed splitting the parasite taxa into those transmitted either actively or trophically.
MHC analysis
Firstly, transformations of individual MHC diversity values did not improve the data’s approximation to a normal distribution, hence we conducted non-parametric tests. Number of individual MHC alleles was correlated to individual heterozygosity index and microsatellite diversity using spearman correlations. We used a Mann–Whitney test to compare individual MHC diversity between populations within habitat type and between habitat types.
Secondly, in order to estimate divergence between habitat types in MHC class II B genes, we used an ANOSIM. MHC alleles were coded as binomial (presence/absence) variables. In addition, a SIMPER test was used to estimate the contribution of each individual haplotype to the total pool. Indeed alleles occurred in fixed haplotypes (i.e., variants are found in common combinations) as determined by RSCA typing (Lenz et al. 2009) and represent the statistical unit. Following the SIMPER test, if a haplotype contributed significantly to the difference between MHC pools, we predicted this specific haplotype to be antagonistically associated to parasite load in the different habitats. To test this, we performed a multivariate analysis of variance with all parasites present in both lake and river habitat types (8 parasites species) as dependent variables and the specific haplotype and habitat type as predictors following Bonneaud et al. (2006).
Thirdly, we used the haplotype information in order to perform an analysis of molecular variance (AMOVA, Arlequin 3.1) with a single locus. This method accounts for the actual zygosity and for haplotype frequencies.
Olfactory-based mate choice
We used a three-step approach to analyze female mating decision. Firstly, with an exact binomial test, we investigated whether females more often chose males from the same habitat, which we refer to as female choice. Secondly, we investigated the time spent in each front quarter of the flow channel. For the analysis, time from both 5 min measurements for each male odor source were summed and considered as a single variable (we thus controlled for side effects). Because the total time spent in the choice area (the two front quarters of the flow channel chamber) varied between tests, we calculated the proportion of time that females spent on each front quarter of the flow channel (Rafferty and Boughman 2006). These proportions were then used for statistical tests. The data was checked for normality with a Shapiro–Wilk test (W = 0.9603, P = 0.549). Based on this result we performed parametric tests. To determine if females preferred sympatric male odor, we tested with a paired t-test if the proportion of time that females spent on the sympatric male side was greater than the random expectation of 0.5. Such deviation from the random expectation would show that females are able to discriminate between sympatric and allopatric odors and prefer sympatric odors.
Results
Population genetics: microsatellite analysis
Hardy–Weinberg equilibrium was tested for each locus and each population and none of the tests showed significant departure from equilibrium. Observed and expected heterozygosity, the number of alleles per locus and F is are reported in Suppl. Doc. 1. Wright statistics showed stronger genetic differentiation between the two habitat types (Mean F st (±SE) = 0.159 ± 0.0177, P < 0.001) than between populations within habitat type (river: F st = 0.034, P = 0.047; lake: F st = −0.0003, P = 0.689; mean = 0.019 ± 0.022). Individual heterozygosity did not vary among populations (F 3,196 = 0.909, P = 0.439) nor between habitat types (F 1,196 = 0.827, P = 0.365).
Using an individual-based cluster analysis (using the software STRUCTURE), we found two clusters as the most likely partitions, following habitat type: rivers vs. lakes (see Fig. 1; Suppl. Doc. 2). An AMOVA revealed that habitat type contributed approximately three times as much to the total genetic variance as the effect of between-population differentiation within habitats (Table 1a).
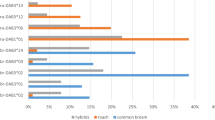
a Prevalence of parasite species by capture location (ordered for an increase in parasite prevalence, first in lake (black bars/black frames) then in river (grey bars/grey frames) fish). b Individual parasite load calculated as standardized Shannon index (H′ = −sum(Pi*log(Pi)) where Pi is the abundance of parasite i, means SE are reported). Underlined are parasite species that actively infect the fish while the others are trophically transmitted
Parasite load
We recorded 17 different parasite taxa from 6 different groups: Protozoa (Trichodina sp. and Apiosoma sp.), Monogenea (Gyrodactylus sp.), Digenea (Diplostomum sp., Apatemon cobitidis, Cyathocotyle prussica, Echinochasmus sp., Phyllodistomum folium), Cestoda (Valipora campylancristrota, Paradilepis scolecina, Proteocephalus filicollis, Diphyllobothrium sp.), Nematoda (Anguillicoloides crassus, Camallanus lacustris, Contracaecum sp., Raphidascaris acus) and glochidia, the parasitic larval stage of freshwater mussels (Mollusca) (see Fig. 1a).
Fish from the lakes harbored a higher parasite load than fish from the rivers (F 1,196 = 56.2, P < 0.001; Fig. 1b) and there was no difference between populations of the same habitat type, i.e., river or lake (F 2,195 = 0.05, P = 0.633; Fig. 1b). Furthermore, a community analysis revealed that fish from the two habitats harbored different parasite communities (Global R between habitat types = 0.48, P < 0.0001, Global R among populations = 0.316, P < 0.0001), with an average dissimilarity of 67.7% between the two habitats (Fig. 1b). Out of the 17 parasite taxa, only 8 (Trichodina sp., Apiosoma sp., Diplostomum sp., Cyathocotyle prussica, Echinochasmus sp., Gyrodactylus sp., Paradilepis scolecina and Anguillicoloides crassus) contributed to more than 90% of the difference (see Suppl. Table 3). Between habitat types, i.e., river and lake, differences remained when parasites were split into actively infecting vs. trophically transmitted parasites (trophically transmitted, R = 0.33, P = 0.0001; actively infecting, Global R = 0.83, P < 0.0001). Interestingly, trophically transmitted parasites did not differ significantly between populations of the same habitat type (between rivers, R = 0.05, P = 0.072; between lakes, R = 0.02, P = 0.098) while actively infecting parasites showed significant differences between populations of the same habitat type (between rivers, R = 0.89, P < 0.0001; between lakes, R = 0.40, P < 0.0001).
MHC analysis
We detected 36 alleles in total, with 19 specific to lake fish and 9 to river whilst 8 were shared by fish from both habitat types. The shared alleles displayed obvious frequency differences between habitats. On the total pools, allele frequencies ranged from 1 to 40% in lake and from 1 to 50.7% in the river populations (Fig. 2a). Individual MHC diversity was neither correlated with individual heterozygosity index based on microsatellites (Rho = 0.04, P = 0.667) nor with individual microsatellite diversity (Rho = 0.005, P = 0.960).
The individual number of MHC class IIB alleles did not vary between populations within habitat type (Mann–Whitney test, Lake: W = 606.5, P = 0.979; River: W = 727, P = 0.488) but varied between fish from different habitat types, with fish from lake carrying on average a higher number than fish from river (mean (±SE) lake fish = 3.7 ± 0.1; mean river fish = 3.1 ± 0.1; Mann–Whitney test, W = 3,880.5, P = 0.005; Fig. 2b)
Moreover, the pool of MHC alleles differed between the populations (ANOSIM, Global R = 0.44, P = 0.0001) with higher variance given between habitat types (mean R ± SD = 0.368 ± 0.06, P < 0.001) than between populations within habitat type (mean R = 0.11 ± 0.101, P < 0.01). A SIMPER test revealed that the average dissimilarity between habitats was 96.2% and that 18 alleles contributed to 90% of this difference. Among those alleles, one haplotype (composed of three alleles So05, SC103 and So11 Genbank accession numbers: DQ016402, DQ016404, AJ230191, respectively) contributed for 22.68% of the difference.
We predicted that if a haplotype contributed for a substantial part of the difference between river and lake allele pools, this haplotype should have antagonistic effect with regards to parasite resistance. The multivariate analysis of variance model showed that this haplotype was associated with increased resistance against Gyrodactylus sp. in the river habitats and a slightly increased susceptibility in the lake (F 1,197 = 4.33, P = 0.041; Fig. 3).
Interestingly the AMOVA showed a similar pattern as the one from the microsatellites where habitat type explained three times more variation than populations within habitat type (Table 1b).
Olfactory-based mate choice
We tested whether females preferred the odor of sympatric over allopatric males. Consistent with our hypothesis, females significantly chose the odor from males originating from their own habitat (Exact binomial test, N = 17/20, P = 0.003; Fig. 4). Moreover, we found that focal females spent a significantly higher proportion of time on the side of the sympatric male than expected (Paired t-test, expected value of 0.5, t = 3.546, df = 19, P = 0.002).
Female choice for sympatric and allopatric male odor. Time spent in each of the two front quarters of the flow channel (in seconds) during the olfactory mate choice experiment. Although statistics were conducted on proportions, for better visualization we depicted the actual time spent by the females in front quarters of the flow channel. Means and SE are shown. * P < 0.05
Splitting the dataset by female habitat of origin, our results revealed that lake females consistently preferred the lake male side (Exact binomial test, N = 9/10, P = 0.021) and spent as well a significantly higher proportion of time on that side than expected under random preference (Paired t-test, expected value of 0.5, t = 2.646, df = 9, P = 0.027. River females did not choose more often the river side than the lake side (Exact binomial test, N = 8/10, P = 0.109). However, river females demonstrated a preference for the river males’ side (Paired t-test, expected value of 0.5, t = 2.359, df = 9, P = 0.043).
Furthermore, using the 18 tests where females from lake and river habitats were each confronted with the odor from a river and a lake male, we demonstrated that females spent significantly more time on the side of the sympatric male than the allopatric male (Paired t-test, t = 3.516, df = 17, P = 0.003).
Discussion
Originating from marine refuges three-spined sticklebacks in Northern Germany have colonized rivers and lakes after the ice-age. Previous work (Reusch et al. 2001b) has shown limited gene flow between habitat types potentially due to local adaptation and suggested the existence of two distinct ecotypes. A crucial difference between rivers and lakes consists in the habitat-specific parasite fauna (Kalbe et al. 2002; Wegner et al. 2003). If adaptation of stickleback immunogenetics (MHC) to these different parasitic environments has occurred, northern German sticklebacks might be in a process of ecological speciation, which includes reproductive isolation between river and lake populations. The present study undertook to verify necessary prerequisites of this process comparing the two types of habitats.
We thus aimed at determining the potentially habitat-specific MHC allele pools and compared them in relation to the respective natural parasite fauna for two populations per habitat type.
We confirmed that fish from lakes and fish from rivers form two different clusters, where habitat type explains more of the genetic variance than population origin. Furthermore the analysis of molecular variance performed on both neutral and adaptive markers demonstrated the same variance partitioning. This population structuring could arise from genetic drift acting on the divergence of MHC allele pools between river and lake populations. Several hypotheses could explain this pattern such as habitat-specific demographic effects or habitat specific contrasting ecological selective pressures or a synergistic action of both. Potentially, these populations have been diverging in these different habitats ever since the ancestral marine population has colonized lakes and rivers.
Nonetheless, despite the potential role of neutral evolution in contributing to the observed patterns of population structuring and divergent MHC allele pools, several lines of evidence for MHC local adaptation also support the role of selection acting on the MHC genes.
As parasites are the prime selective agent for the evolution of the MHC, differences in this genomic region in fish exposed to divergent parasite communities are to be expected (Blais et al. 2007; Eizaguirre et al. 2009a; Ekblom et al. 2007; Summers et al. 2003). Accordingly, we showed for the first time that fish from lakes that harbored a higher and more diverse parasite load, also displayed a higher individual MHC diversity, i.e., a higher number of different MHC alleles, than fish from rivers. This supports the hypothesis that stickleback MHC genes display copy number variation (Lenz et al. 2009; Sato et al. 1998), but more importantly, it indicates the potential selective mechanism leading to adaptive differences in MHC gene numbers. While the duplication and deletion of genes over evolutionary time is an expected phenomenon in the MHC (Nei et al. 1997), the selective mechanism was expected to affect only MHC quality. However, selection could also work quantitatively: a more diverse parasite community selects for a larger number of different alleles (and therefore loci) in order to initiate immune responses against a larger diversity of parasites (Eizaguirre and Lenz in press). At the molecular level, the T cell repertoire might be at the root of the maintenance of a relatively low individual MHC diversity since its diversity is crucially dependent on the number of MHC molecules, and thus on the number of expressed MHC genes. During negative selection, T-cells, whose receptors react too strongly with self-peptide–MHC complexes, are eliminated. Because self/non-self discrimination is mediated by MHC molecules, recognition of self peptides as foreign would lead to the activation of the immune system and thus to auto-immune diseases (Germain 1994). Therefore, the balance between natural selection (parasite pressure) and negative selection theoretically leads to an optimal number of different MHC molecules (Nowak et al. 1992; Woelfing et al. 2009) or to an optimal MHC heterozygosity (De Boer and Perelson 1993). How the risk of T cell depletion varies across habitat types, however, still remains to be investigated (Woelfing et al. 2009). Although the total number of alleles present within a population could be the result of a smaller effective population size in river populations, it is unlikely that this affects the number of loci and thus selection may be invoked to explain the observed pattern of variation in individual diversity.
We further found that under contrasting parasite pressures, the populations displayed different and little overlapping habitat-specific pools of MHC alleles, both in constitution and in frequencies. This supports the hypothesis that MHC divergence may have evolved as a consequence of immunogenetic local adaptation to habitat-specific parasite communities. Differences in MHC allele pools between populations have already been proposed while associated data on contrasting parasite communities is scarce (but see, Blais et al. 2007; Bonneaud et al. 2006). As mentioned previously, this pattern could theoretically arise from genetic drift, however, the likelihood that both rivers (as both lakes) evolved by chance independently in the same direction, seems rather low without invoking habitat-specific homogenizing selection (Frazer and Neff 2010). Thus, under a “one event of colonization hypothesis”, (Reusch et al. 2001b), a plausible scenario might be: genetic drift has acted on populations’ divergence but similar parasite pressure slowed down divergence at MHC genes within habitat type. However, between habitat types, parasites increase the pace of MHC divergence.
The process of MHC local adaptation is indicated further by the role of a specific haplotype, which was associated with reduced Gyrodactylus sp. load in river populations whereas it was associated with slightly increased Gyrodactylus sp. load in lake populations. This result is consistent with a previous study that demonstrated that in our system Gyrodactylus sp. influences reproduction: males carrying a specific MHC genotype conferring resistance against this parasite were more chosen by females and achieved a higher reproductive success (Eizaguirre et al. 2009b). A previous survey revealed that the majority of Gyrodactylus sp. found on lake sticklebacks in our area are G. gasterostei, and it is known that Gyrodactylus species are relatively host species-specific compared to other parasites (Raeymaekers et al. 2008). The three-spined stickleback is the only host on which this parasite feeds and reproduces. Several studies have reported the impact of Gyrodactylus species on host immune system activation (Buchmann and Lindenstrøm 2002; Lindenstrom et al. 2004; Collins et al. 2007). Gyrodactylus infection has also been correlated with carotenoid input—the source of various male secondary sexual trait colourations (Kolluru et al. 2006). Antagonistic matching allele effects as the one reported in our study are predicted by and consistent with contrasting parasite-mediated selection. Such arms races favoring (i.e., through higher resistance) specific alleles in one habitat and deselecting (i.e., through higher susceptibility) them in others represent a mechanism by which different allele pools evolve. Ultimately it has been proposed that these habitat specific arms races may facilitate the formation of distinct species through the pleiotropic role of MHC genes in immunity and mate choice (Eizaguirre et al. 2009a).
The habitat specific population structuring is puzzling and we can only speculate why fish seldom migrate between lake and river habitats. Since contact zones between the two ecotypes exist, low migration rates could be the consequence of reduced fitness in either foreign habitat. Berner et al. (2009) demonstrated a shift in fish morphology and foraging behavior accross lake-stream transitions in North America, which indicates adaptation and could explain a part of the observed lack of migration in our system. Similarly, using a mark-transplant-recapture experiment on morphologically divergent parapatric populations, Bolnick et al. (2009) showed that 90% of lake and stream sticklebacks returned to their native habitat, reducing migration between habitats by 76%, thus increasing reproductive isolation. For the Northern Germany drainage systems, Scharsack et al. (2007) proposed that parasite-mediated selection could account for reduced gene flow and reduced between habitat types due to lower growth of lake fish in river and higher parasite load of river fish in lake. A previous study attempted to highlight the role of parasites in local immunogenetics adaptation, however, most probably because of a too short time of exposure, results were not conclusive (Rauch et al. 2006). Further experimental tests are needed to quantify the impact of parasites on the two ecotypes.
Adaptation to different habitats may also be reflected by mate choice, if choosing a locally adapted mate would lead to more viable offspring. We found experimental evidence for olfactory mediated reproductive isolation between three-spined sticklebacks from river and lake habitats. Recently, Milinski et al. (2010) demonstrated that the MHC signal needs to be validated by an additional male odor cue, probably signaling the species identity of the sender. It is therefore possible that non MHC olfactory cues such as the validation factor were involved in female mate choice for sympatric males. Further work will show which part of the odor signal allows females to prefer sympatric males.
Assortative mating according to morphological characters has been reported in a few studies (e.g., beaks of Darwin’s finches Huber et al. 2007). McKinnon et al. (2004), Boughman et al. (2005) and Vines and Schluter (2006) have shown limnetic/benthic and stream-lake pairs of sticklebacks, respectively, to be reproductively isolated with fish size being important. Recent work has shown limnetic/benthic sticklebacks to be exposed to contrasting parasite communities (Maccoll 2009). From our results, we would thus predict different MHC allele pools in limnetic and benthic morphs, which could have facilitated species divergence. Although not investigated, it is possible that MHC-associated olfactory cues have also played a role in these reported cases. Rafferty and Boughman (2006) later on hypothesized a potential role of MHC in their study by showing assortative mating based on olfactory cues in sympatric benthic/limnetic stickleback species, but did not investigate genetics. While shoaling preferences possibly due to odor cues resulting from diet can be reversed by fish translocation for short time periods (Ward et al. 2007), our findings suggest a different basis for female mating preference: as the fish had been acclimatized in the lab for at least 10 days and all fish were fed the same diet. Thus, our results are in line with assortative mating for locally adapted males. Interestingly, as seen in various studies (reviewed in Eizaguirre and Lenz in press) we identified asymmetrical differences in the strength of female preference, with river fish being slightly less discriminative than lake fish. Based on our hypothesis, this could be explained by the overlapping parasite species and/or a lower parasite load in river fish.
In summary, the differences of three-spined stickleback river and lake ecotypes in (i) parasite load, (ii) habitat-specific MHC allele pools and individual diversity that potentially evolved in response to contrasting parasite communities, (iii) antagonistic effects of alleles/haplotypes in parasite resistance between fish from different habitat types and (iv) olfactory mate choice for sympatric males are all in line with a hypothetical scenario of parasite-mediated local adaptation, which could ultimately lead to ecological speciation. Our approach, using ecological and molecular tools, provides one of the first data sets elucidating the potential extent of MHC local adaptation to two distinct and habitat-specific parasite communities. This study therefore contributes to our growing understanding of the potential role of parasites in driving divergent MHC pools. However, it remains difficult to disentangle the complex processes of ecological adaptation and neutral evolution. Final conclusions on the role of parasites in ecological speciation require an experimental approach which is hardly feasible.
References
Aeschlimann PB, Häberli MA, Reusch TBH, Boehm T, Milinski M (2003) Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. Behav Ecol Sociobiol 54:119–126
Alcaide M, Edwards SV, Negro JJ, Serrano D, Tella JL (2008) Extensive polymorphism and geographical variation at a positively selected MHC class IIB gene of the lesser kestrel (Falco naumanni). Mol Ecol 17:2652–2665
Apanius V, Penn D, Slev PR, Ruff LR, Potts WK (1997) The nature of selection on the major histocompatibility complex. Crit Rev Immunol 17:179–224
Babik W, Pabijan M, Radwan J (2008) Contrasting patterns of variation in MHC loci in the Alpine newt. Mol Ecol 17:2339–2355
Bakker TCM (1993) Positive genetic correlation between female preference and preferred male ornament in sticklebacks. Nature 363:255–257
Berner D, Adams DC, Grandchamp AC, Hendry AP (2008) Natural selection drives patterns of lake/stream divergence in stickleback foraging morphology. J Evol Biol 21:1653–1665
Berner D, Grandchamp AC, Hendry AP (2009) Variable progress toward ecological speciation in parapatry: stickleback across eight lake-stream transitions. Evolution 63:1740–1753
Blais J, Rico C, van Oosterhout C, Cable J, Turner GF, Bernatchez L (2007) MHC adaptive divergence between closely related and sympatric african cichlids. PLoS ONE 2:e734
Bolnick DI, Fitzpatrick BM (2007) Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst 38:459–487
Bolnick DI, Snowberg LK, Patenia C, Stutz WE, Ingram T, Lau OL (2009) Phenotype-dependent native habitat preference facilitates divergence between parapatric lake and stream stickleback. Evolution 63:2004–2016
Bonneaud C, Perez-Tris J, Federici P, Chastel O, Sorci G (2006) Major histocompatibilty alleles associated with local resistance to malaria in a passerine. Evolution 60:383–389
Boughman JW (2001) Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature 411:944–948
Boughman JW, Rundle HD, Schluter D (2005) Parallel evolution of sexual isolation in sticklebacks. Evolution 59:361–373
Buchmann K, Lindenstrom T (2002) Interactions between monogenean parasites and their fish hosts. Int J Parasitol 32:309–319
Clarke KR, Gorley RN (2006) Primer v6: user manual/tutorial. PRIMER-E, Plymouth
Collins CA, Olstad K, Sterud E et al (2007) Isolation of a novel fish thymidylate kinase gene, upregulated in Atlantic salmon (Salmo salar L.) following infection with the monogenean parasite Gyrodactylus salaris. Fish Shellfish Immunol 23:793–807
Coulson TN, Pemberton JM, Albon SD, Beaumont M, Marshall TC, Slate J, Guinness FE, Clutton-Brock TH (1998) Microsatellites reveal heterosis in red deer. Proc R Soc Lond Ser B Biol Sci 265:489–495
Coyne JA, Orr HA (2004) In speciation. Sinauer, Sunderland
De Boer RJ, Perelson AS (1993) How diverse should the immune system be? Proc R Soc Lond B Biol Sci 252:171–175
Dieckmann U, Doebeli M, Metz J, Tautz D (2004) Adaptive speciation. In: Dieckmann U, Doebeli M, Metz J, Tautz D (eds) Adaptive speciation. Cambridge Studies in Adaptive Dynamics, vol 3
Eizaguirre C, Lenz TL (in press) Dynamics and consequences of parasite-mediated local adaptation: a future for MHC studies? J Fish Biol
Eizaguirre C, Lenz TL, Traulsen A, Milinski M (2009a) Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol Lett 12:5–12
Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M (2009b) MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol Ecol 18:3316–3329
Ekblom R, Saether SA, Jacobsson PAR, Fiske P, Sahlman T, Grahn M, Kalas JA, Hoglund J (2007) Spatial pattern of MHC class II variation in the great snipe (Gallinago media). Mol Ecol 16:1439–1451
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Frazer BA, Neff BD (2010) Parasite mediated homogenizing selection at the MHC in guppies. Genetica (in press)
Gavrilets S (2004) Fitness landscapes and the origin of species. Princeton University Press, Princeton
Germain RN (1994) Mhc-dependent antigen-processing and peptide presentation—providing ligands for T-lymphocyte activation. Cell 76:287–299
Gibson G (2005) Evolution: the synthesis and evolution of a supermodel. Science 307:1890–1891
Grant BR, Grant PR (1982) Niche shifts and competition in Darwin’s finches: Geospiza conirostris and congeners. Evolution 36:637–657
Halmetoja A, Valtonen ET, Koskenniemi E (2000) Perch (Perca fluviatilis L.) parasites reflect ecosystem conditions: a comparison of a natural lake and two acidic reservoirs in Finland. Int J Parasit 30:1437–1444
Hamilton WD, Zuk M (1982) Heritable true fitness and bright birds: a role for parasites? Science 218:384–387
Harrod C, Mallela J, Kahilainen KK (2010) Phenotype-environment correlations in a putative whitefish adaptive radiation. J Anim Ecol doi: 10.1111/j.1365-2656.2010.01702.x
Hendry AP (2009) Ecological speciation! Or the lack thereof? Can J Fish Aquat Sci 66:1383–1398
Huber SK, De Leon LF, Hendry AP, Bermingham E, Podos J (2007) Reproductive isolation of sympatric morphs in a population of Darwin’s finches. Proc R Soc B Biol Sci 274:1709–1714
Janeway CA, Travers P, Walport M, Sclomchik MJ (2005) Immunobiology: the immune system in health and disease. Garland Science Publishing, New York
Jiggins CD, Naisbit RE, Coe RL, Mallet J (2001) Reproductive isolation caused by colour pattern mimicry. Nature 411:302–305
Kalbe M, Wegner KM, Reusch TBH (2002) Dispersion patterns of parasites in 0+ year three-spined sticklebacks: a cross population comparison. J Fish Biol 60:1529–1542
Kalbe M, Eizaguirre C, Dankert I, Reusch TBH, Sommerfeld RD, Wegner KM, Milinski M (2009) Lifetime reproductive success is maximized with optimal MHC diversity. Proc R Soc Lond Ser B Biol Sci 276:925–934
Kitano JUN, Mori S, Peichel CL (2007) Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linnean Soc 91:671–685
Kolluru GR, Grether GF, South SH et al (2006) The effects of carotenoid and food availability on resistance to a naturally occurring parasite (Gyrodactylus turnbulli) in guppies (Poecilia reticulata). Biol J Linn Soc 89:301–309
Kuris AM, Hechinger RF, Shaw JC, Whitney KL, Aguirre-Macedo L, Boch CA, Dobson AP, Dunham EJ, Fredensborg BL, Huspeni TC, Lorda J, Mababa L, Mancini FT, Mora AB, Pickering M, Talhouk NL, Torchin ME, Lafferty KD (2008) Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature 454:515–518
Lenz TL, Becker S (2008) Simple approach to reduce PCR artefact formation leads to reliable genotyping of MHC and other highly polymorphic loci—implications for evolutionary analysis. Gene 427:17–123
Lenz TL, Eizaguirre C, Becker S, Reusch TBH (2009) RSCA genotyping of MHC for high-throughput evolutionary studies in the model organism three-spined stickleback Gasterosteus aculeatus. BMC Evol Biol 9:57
Lindenstrom T, Secombes CJ, Buchmann K (2004) Expression of immune response genes in rainbow trout skin induced by Gyrodactylus derjavini infections. Vet Immunol Immunopathol 97:137–148
Losos JB, Jackman TR, Larson A, Queiroz Kd, Rodriguez-Schettino L (1998) Contingency and determinisn in replicated adaptive radiations of island lizards. Science 279:2115–2118
Maccoll ADC (2009) Parasites may contribute to ‘magic trait’ evolution in the adaptive radiation of three-spined sticklebacks, Gasterosteus aculeatus (Gasterosteiformes: Gasterosteidae). Biol J Linnean Soc 96:425–433
Marchinko KB, Schluter D (2007) Parallel evolution by correlated response: lateral plate reduction in threespine stickleback. Evolution 61:1084–1090
Maynard Smith J (1966) Sympatric speciation. Am Nat 100:637–650
Mayr E (1942) Systematics and the origin of species. Columbia University Press, New York
McKinnon JS, Mori S, Blackman BK, David L, Kingsley DM, Jamieson L, Chou J, Schluter D (2004) Evidence for ecology’s role in speciation. Nature 429:294–298
McLennan DA, McPhail JD (1990) Experimental investigations of the evolutionary significance of sexually dimorphic nuptial coloration in Gasterosteus-Aculeatus (L)—the relationship between male color and female behavior. Can J Zool Revue Canadienne de Zoologie 68:482–492
McPhail JD (1994) Speciation and the evolution of reproductive isolation in the sticklebacks (Gasterosteus) of south western British Columbia. In: Bell MA, Foster SA (eds) The evolutionary biology of the threespine stickleback. Oxford University Press, Oxford, pp 399–437
Milinski M (2006) The major histocompatibility complex, sexual selection, and mate choice. Annu Rev Ecol Evol Syst 37:159–186
Milinski M, Bakker TCM (1990) Female sticklebacks use male coloration in mate choice and hence avoid parasitized males. Nature 344:330–333
Milinski M, Griffiths S, Wegner KM, Reusch TBH, Haas-Assenbaum A, Boehm T (2005) Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci U S A 102:4414–4418
Milinski M, Griffiths SnW, Reusch TBH, Boehm T (2010) Costly major histocompatibility complex signals produced only by reproductively active males, but not females, must be validated by a ‘maleness signal’ in three-spined sticklebacks. Proc R Soc B Biol Sci 227:391–398
Nei M, Gu X, Sitnikova T (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A 94:7799–7806
Nowak MA, Tarczyhornoch K, Austyn JM (1992) The optimal number of major histocompatibility complex-molecules in an individual. Proc Natl Acad Sci U S A 89:10896–10899
Podos J (2001) Correlated evolution of morphology and vocal signal structure in Darwin’s finches. Nature 409:185
Poulin R (1996) How many parasite species are there: are we close to answers? Int J Parasit 26:1127–1129
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raeymaekers JAM, Huyse T, Maelfait H, Hellemans B, Volckaert FAM (2008) Community structure, population structure and topographical specialisation of Gyrodactylus (Monogenea) ectoparasites living on sympatric stickleback species. Folia Biol 55:187–196
Rafferty NE, Boughman JW (2006) Olfactory mate recognition in a sympatric species pair of three-spined sticklebacks. Behav Ecol 17:965–970
Rauch G, Kalbe M, Reusch TBH (2006) Relative importance of MHC and genetic background for parasite load in a field experiment. Evol Ecol Res 8:373–386
Raymond M, Rousset F (1995) Genepop (version-1.2)—population-genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reusch TBH, Haberli MA, Aeschlimann PB, Milinski M (2001a) Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 414:300–302
Reusch TBH, Wegner KM, Kalbe M (2001b) Rapid genetic divergence in postglacial populations of threespine stickleback: the role of habitat type, drainage and geographical proximity. Mol Ecol 10:2435–2445
Reusch TBH, Schaschl H, Wegner KM (2004) Recent duplication and inter-locus gene conversion in major histocompatibility class II genes in a teleost, the three-spined stickleback. Immunogenetics 56:427–437
Richardson DS, Komdeur J, Burke T, von Schantz T (2005) MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc R Soc Lond Ser B Biol Sci 272:759–767
Sato A, Figueroa F, O’hUigin C, Steck N, Klein J (1998) Cloning of major histocompatibility complex Mhc genes from threespine stickleback, Gasterosteus aculeatus. Mol Mar Biol Biotechnol V7:221–231
Scharsack JP, Kalbe M, Harrod C, Rauch G (2007) Habitat-specific adaptation of immune responses of stickleback (Gasterosteus aculeatus) lake and river ecotypes. Proc R Soc Lond B Biol Sci 274:1523–1532
Schluter D (1996) Ecological speciation in postglacial fishes. Philos Trans R Soc Lond B Biol Sci 351:807–814
Schluter D, Rambaut A (1996) Ecological speciation in postglacial fishes. Philos Trans R Soc B Biol Sci 351:807–814
Schwensow N, Eberle M, Sommer S (2008) Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc R Soc Lond Ser B Biol Sci 275:555–564
Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R, van der Sluijs I, Schneider MV, Maan ME, Tachida H, Imai H, Okada N (2008) Speciation through sensory drive in cichlid fish. Nature 455:U620–U623
Sommerfeld RD, Boehm T, Milinski M (2008) Desynchronising male and female reproductive seasonality: dynamics of male MHC-independent olfactory attractiveness in sticklebacks. Ethol Ecol Evol 20:325–336
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc R Soc B Biol Sci 277:979–988
Summers K, McKeon S, Sellars J, Keusenkothen M, Morris J, Gloeckner D, Pressley C, Price B, Snow H (2003) Parasitic exploitation as an engine of diversity. Biol Rev 78:639–675
Thompson JN (1994) In the coevolutionary process. University of Chicago Press, Chicago
van Doorn GS, Edelaar P, Weissing FJ (2009) On the origin of species by natural and sexual selection. Science 326:1704–1707
Vines T, Schluter D (2006) Strong assortative mating between allopatric sticklebacks as a by-product of adaptation to different environments. Proc R Soc Lond Ser B Biol Sci 273:911–916
Ward AJW, Webster MM, Hart PJB (2007) Social recognition in wild fish populations. Proc R Soc B Biol Sci 274:1071–1077
Wegner KM, Reusch TBH, Kalbe M (2003) Multiple parasites are driving major histocompatibility comple polymorphism in the wild. J Evol Biol 16:224–232
Woelfing B, Traulsen A, Milinski M, Boehm T (2009) Does intra-individual MHC diversity keep a golden mean? Phil Trans R Soc B Biol Sci 364:117–128
Yamazaki K, Boyse EA, Mike V, Thaler HT, Mathieson BJ, Abbott J, Boyse J, Zayas ZA, Thomas L (1976) Control of mating preferences in mice by genes in the major histocompatibility complex. J Exp Med 144:1324–1335
Acknowledgments
We thank G. Augustin and D. Martens for their help with maintaining the fish, I. Schultz, H. Buhtz, E. Blohm-Sievers, S. Dembeck and K. Brzezek for lab assistance, and D. Benesh, A. Traulsen, A. Nolte, J. Meunier, C. Peichel for their helpful comments on an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eizaguirre, C., Lenz, T.L., Sommerfeld, R.D. et al. Parasite diversity, patterns of MHC II variation and olfactory based mate choice in diverging three-spined stickleback ecotypes. Evol Ecol 25, 605–622 (2011). https://doi.org/10.1007/s10682-010-9424-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-010-9424-z