Abstract
A new collection of introgression lines (ILs) have been developed using the Spanish cultivar ‘Piel de Sapo’ (PS, subsp. melo, group ibericus) as the recurrent parent, and ‘Queen’s pocket melon’ (DUD, subsp. melo, group dudaim PI 273438) as donor. Genome-wide markers assisted selection has been proceed in several backcross generations to obtain a set of 16 ILs. An average of 1.4 introgressions/plant representing altogether about 62% of the DUD genome, and with an average of recovery per IL of 93.9% of the PS genetic background, has been accomplished in the selected ILs after several backcrosses and selfings. QTLs for abscission layer, external aroma, rind and flesh firmness, rind netting, fruit weight and shape, and sugar content have been identified on this set of ILs some of them corresponding to genomic regions previously described or with interesting candidate genes, but, also, new allelic diversity has been identified. Thus, this set of ILs may be useful to exploit new underexploited genetic variability from DUD. Based on GBS results for five of this ILs, further steps of backcrossing will be necessary to clean up the small introgressions detected in some of them. Novel phenotypes like small PS melons or aromatic and medium-climacteric PS melons were obtained, which might become pre-breeding lines with potential commercial interest.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melon (Cucumis melo L.) is a cucurbit species with high agronomic and economic importance worldwide, with almost 32 million tons produced worldwide (FAO 2017), and exportation values of more than 1.6 billion U.S. dollars (FAO 2016), Due to this economic impact, the development of new and more adapted cultivars is required to satisfy consumer producer and market demands. The first step in a breeding program is to find useful genetic variability. The exploitation of exotic genotypes related to crops is a strategy to increase the variability pool available for breeders.
Cucumis melo presents a high level of genetic variability due to at least two domestication events (Endl et al. 2018), that have led to a large diversification regarding to seed, vegetative and flowering traits, fruit size and shape, flesh and rind color, ripening behavior, resistances, etc. (Dantas et al. 2015; Esteras et al. 2013, 2018; Lázaro et al. 2017; López et al. 2015; Nunes et al. 2017; Pitrat 2017; Sabato et al. 2015). This species has been traditionally subdivided into two subspecies, subsp. melo and agrestis, and recently, Pitrat (2017) has described 19 groups: agrestis, kachri, chito, tibish, acidulus, momordica, conomon, makuwa, chinensis, flexuosus, chate, dudaim, chandalak, indicus, ameri, cassaba, ibericus, inodorus, and cantalupensis. All this huge diversity can be potentially employed for breeding commercial cultivars, using different strategies, although the high diversity on fruit traits among cultivar types preclude the transfer of genetic variability through different types.
Although high production and disease/pest resistance continue being important breeding objectives, nowadays the development of cultivars with high quality standards is the main goal in melon breeding programs in response to the current consumer’s demand. Melon fruit quality is a complex feature that involves morphological traits (fruit shape, size, rind and flesh color) and also organoleptic and nutritional properties such as sweetness, content in vitamins and antioxidants, and aroma (Burger et al. 2006; Gur et al. 2016). Most of these quality traits show a quantitative variation, as a consequence of a polygenic genetic control (Quantitative Trait Loci, QTLs) (Amanullah et al. 2018; Argyris et al. 2017; Galpaz et al. 2018; Gur et al. 2016; Pereira et al. 2018; Perpiñá et al. 2016), and also influenced by environmental factors. Therefore, their breeding is complex and requires the generation of specific populations. Introgression Lines (ILs) are appropriate populations to study QTLs, especially when dealing with exotic variability (Eshed and Zamir 1995; Zamir 2001), allowing a rapid and straightforward association for traits to specific genomic regions and accelerating the development of new commercial cultivars with favourable QTLs. With that purpose, the use of exotic or wild genotypes as donors of new alleles, is the most used strategy to exploit the extant diversity through the generation of IL libraries. These are collections of lines, obtained by backcrossing after the first F1 cross, each one containing a chromosome fragment of a selected donor genotype within an elite genetic background. Finally, the whole collection represents the donor’s complete genome.
To date, two IL collections have been reported in melon, both using genotypes with contrasting phenotypes belonging to the two subspecies. The first one used a Spanish cultivar type ‘Piel de Sapo’ (PS, subsp. melo, group ibericus) as a recurrent parent and the Korean accession PI 161375 (SC, Songwan Charmi) (subsp. agrestis, group chinensis) as a donor parent (Eduardo et al. 2005). The second one was developed from the cross of a Charentais ‘Vedrantais’ type (subsp. melo, group cantalupensis), as the recurrent parent, and the Japanese accession PI 420176 (Ginsen makuwa) (subsp. agrestis, group makuwa) (Perpiñá et al. 2016). Other two IL collections, reciprocal between ‘Vedrantais’ and ‘Piel de Sapo’ genotypes, will be reported soon (Pereira 2018).
Apart from the incorporation of the new variability found in exotic genotypes, these ILs collections also allowed the detection of new QTLs in the species (Eduardo et al. 2007; Perpiñá et al. 2016). In the last few years, using other strategies such as different mapping populations derived from other exotic melons like flexuosus, momordica and agrestis types (Amanullah et al. 2018; Díaz et al. 2017; Galpaz et al. 2018; Ramamurthy and Waters 2015), or even germplasm collections through GWA (genome-wide association analysis) (Gur et al. 2017; Nimmakayala et al. 2016), more than 250 common and new QTLs have been reported for the species regarding fruit quality traits. Moreover, Pereira et al. (2018) recently developed a RIL population derived from the two most important commercial cultivars in Europe, a ‘Vedrantais’ and a ‘Piel de Sapo’, detecting 33 QTLs mainly associated to fruit quality.
The large collections of molecular markers currently available in melon as the result of several sequencing projects (Blanca et al. 2011, 2012; Pavan et al. 2017), as well as saturated genetic maps anchored to the genome sequence like the ones reported by Garcia-Mas et al. (2012) and Argyris et al. (2015), and the consensus maps by Díaz et al. (2011, 2015), are genomic tools which highly improve the efficiency in the development of IL populations through marker assisted selection (MAS), since their use reduces significantly the number of backcross generations to obtain ILs with single donor introgressions (Perpiñá et al. 2016). High Resolution Melting, medium-throughput platforms like Agena Bioscience iPLEX ® Gold MassARRAY, or Genotyping-by-Sequencing (GBS) are available genotyping technologies, which are currently easily affordable, and that can be very useful during ILs development in the different moments of the process.
In this context, we present the development of a new IL collection in melon using a dudaim melon as new exotic donor with interesting quality traits to introduce new variation in the most important non-climacteric melon background in the market (‘Piel de Sapo’). Dudaim melons are cultivated from Turkey to Afghanistan. They firstly were reported in ninth century by its medical properties and fragrance in Persia (Paris et al. 2012). In fact, dudaim fruits have a very unique fragrant, exotic, and musky smell (Aubert and Pitrat 2006; Esteras et al. 2018). Also, they are characterized by their small reddish-yellow fruits with ochre stripes and smooth surface, round or slightly oval shaped, and with white, mealy flesh. They are early maturing and with dehiscent peduncle.
Materials and methods
Plant material
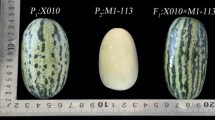
The two genotypes employed in the generation of the IL population were the Spanish cultivar ‘Piel de Sapo’ (PS) (subsp. melo, group ibericus) as the recurrent parent, and ‘Queen’s pocket melon’ (DUD) (a selection by selfing from the accession PI 273438, subsp. melo, group dudaim, of the NPGS GRIN collection) as the exotic donor parent (Fig. 1). Both accessions were multiplied and conserved at the Genebank of the Institute for the Conservation and Breeding of Agrobiodiversity (COMAV-UPV) in Valencia (Spain) and were previously phenotyped, along with other accessions from the COMAV melon core collection, to confirm their classification in the corresponding horticultural group (Esteras et al. 2018; Leida et al. 2015).
DUD is an exotic Asian variety, mainly used with ornamental purposes. It produces small-size, round, no sutured and white-fleshed fruits, that are highly aromatic and climacteric with low sugar content. In contrast, PS bears fruits which are big and oval. They are white-fleshed, sweet, without aroma, and non-climacteric.
Breeding scheme
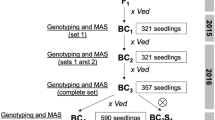
The F1 generation derived from the cross between PS and DUD was backcrossed to the recurrent PS parent to generate the BC1 population. Fourteen BC1 plants were then backcrossed with the recurrent parent thus producing 14 BC2 families. A total of 409 BC2 plants (28–30 plants per BC2 family) were genotyped at the seedling stage using Agena Bioscience iPLEX® Gold MassARRAY with a set of SNPs evenly distributed throughout the melon genome (see details below). According to these results, a subset of plants was selected, those having the highest proportion of the recurrent (PS) genome while containing DUD introgressions in order to cover the entire donor genome. The selected BC2 plants were grown in the greenhouse and backcrossed to construct the BC3 population.
The same genotyping platform was employed to select the best BC3 individuals (among a total of 332) according to genotype, using the same criteria that in BC2 to produce the following generations. Eighty-two BC3 plants, carrying three or fewer DUD introgressions, were selfed, and their offsprings (BC3S1 and BC3S2) were genotyped using the SNPs located in the corresponding target introgressions by High Resolution Melting (HRM) (Vossen et al. 2009) in order to finally select plants with single/double homozygous introgressions. Subsequently, the selected BC3S1 and BC3S2 plants were genotyped using the Agena Bioscience array designed in previous generations. Moreover, in order to remove a few remaining heterozygous markers and provide seeds for future characterization assays, selected plants of the BC3S2 generation were subjected to further selfing processes (BC3S3 and BC3S4).
In this work, a first set of 16 ILs (developed in BC3S3 and BC3S4 generations) with single or double homozygous introgressions was agronomically characterized. This is a medium-sized IL population, with one or two lines representing each DUD chromosome.
Markers and genotyping methods
Genomic DNA was extracted from young leaves following the method described by Doyle and Doyle (1990) with minor modifications. Final concentration was adjusted to 10 ng/μl for the Agena Bioscience array and 30 ng/μl for the HRM genotyping (Perpiñá et al. 2016).
Markers included in the Agena Bioscience array performed in this study were selected from the SNP set used by Perpiñá et al. (2016). The selected SNPs were polymorphic between PS and DUD, and evenly distributed in chromosomes, based on the first genetic map anchored to the melon genome by Garcia-Mas et al. (2012) (Online resource 1). This Agena Bioscience array was employed to genotype the full BC2 and BC3 populations, the selected BC3S1 and BC3S2, and the final set of ILs selected for phenotyping (BC3S3 and BC3S4). The Agena Bioscience genotyping was carried out at the Epigenetics and Genotyping laboratory at the Central Research Unit of the Faculty of Medicine (UCIM) belonging to the Universitat de València (Spain).
A total of 89 SNPs, out of the 107 employed in the full Agena Bioscience platform, were adapted for HRM analysis. HRM genotyping was used to accelerate the selection and fixation of target introgressions during the construction of the IL population in several specific offsprings. HRM reactions were performed using the LightCycler 480 with the Roche LightCycler 480 High Resolution Melting Master kit.
Five ILs with the most interesting phenotypes regarding climacteric ripening, fruit shape and yellowish rind, out of the 16 ILs selected, were also genotyped by GBS (DUD_3-1; DUD_4-2; DUD_5-1; DU_6-1; DUD_6-2). Genomic DNA extraction was carried out employing DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and about 400 ng of each one were sent to LGC Genomics GmbH (Berlin, Germany) for the GBS analysis. Libraries were constructed using ApeKI, and 2.5–2.9 million filtered reads per sample were obtained with an average size of 70 bp. Quality reads were mapped against the last version of the melon genome (genome assembly v.3.6.1, Melonomics) and SNP calling was performed using freebayes selecting SNPs with at least 3 reads. Polymorphic SNPs between parentals and monomorphic within them were filtered to have a set of useful markers in our lines.
Agronomic evaluation and traits measured
The selected plants in each generation were transplanted to the greenhouse for seed obtention. Generations BC3S3 and BC3S4 were also phenotyped. A total of 22 BC2, 82 BC3, 99 BC3S1 and 126 BC3S2 plants were grown from 2011 to 2015, during the spring–summer growing season at the greenhouse facilities of the Polytechnic University of Valencia (Valencia, Spain) and the Fundacion Cajamar in Paiporta (Valencia, Spain). Sixteen ILs were selected from the BC3S3 and BC3S4 populations, and evaluated in two trials conducted in 2 years, 2016 and 2017, in the facilities of the Fundacion Cajamar in Paiporta (Paip16 and Paip17). Each assay included six to eight plants of each IL in a fully randomized design with five to ten plants of each parental line (PS and DUD). Pollination was ensured with insects. Temperature was controlled with cooler and automatic window aperture (with a temperature range of 10–25/10–20 and of 25–37/18–35, minimum and maximum during the whole growing season for Paip2016/2017, respectively). Plants were grown in substrate bags (70% coconut fiber and 30% coconut chips) with fertirrigation.
Plants were phenotyped for flowering-related traits: the number of male and female flowers 15 and 30 days after the opening of the first flower on each plant was counted (NMaF15, NFeF15, NMaF30 and NFeF30). Two fruits per plant were set and characterized at full maturity with the following traits: fruit weight (FW in grams, with digital scale), fruit length and diameter (FL, FD in mm, with graduated rule), fruit shape index (FS, as the ratio of fruit length to fruit diameter), cavity width (CW, as the ratio of the width of the seminal cavity to the fruit diameter), flesh and rind thickness (Fth, Rth in mm, with electronic digital caliper, I.C.T, S.L., La Rioja, Spain), rind and flesh firmness (RF, FF, measured as kg/cm2 with a fruit pressure tester, FT 327, with a plunger diameter of 8 mm, Alfonsine, Italy), pH on a scale from 1 to 14 (1 = pH acid, 14 = pH basic), formation of abscission layer, the occurrence of rind netting (AL and NET, scored visually as 0, absent and 1, present), and external aroma of the whole fruit (AR assessed by olfaction, without opening the fruit, as 0, absent and 1, present); flesh color measured with a CR-400 colorimeter (Konica Minolta, Inc., Tokyo, Japan) (coordinates Hunter Lab. L* express luminosity (L = 0 black and L = 1 white), a* expresses the color direction between red (positive) and green (negative) and b* expresses the color direction between yellow (positive) and blue (negative) (FCHl, FCa, FCb), and also rind color (evaluated visually, as parentals present a clearly distinguishable rind color). Soluble solids concentration (SSC) was measured as °Brix from drops of juice with a hand-held “Pocket” refractometer (PAL-α, Atago CO., LTD, Tokyo, Japan). Flesh samples were taken from the same regions in the equatorial slice of the fruit and used for firmness, flesh color, ºBrix and pH measurements.
QTL detection in introgression lines
With the aim of checking the normality of every trait distribution, the Kolmogorov–Smirnov test was carried out using Statgraphics Centurion XVII.II (Online resource 2).
An analysis of variance (ANOVA) was performed with ILs data for trials Paip16 and Paip17 to examine the effects of genotype, environment and genotype x environment interaction. In addition, the mean of every IL in each environment (Paip16 and Paip17) was compared to the control PS mean using the Dunnett’s contrast with Type-I error a ≤ 0.05 (Dunnett 1955). QTLs for each trait in the DUD introgression were considered to exist in those lines with means significantly different from PS in the two environments.
Results and discussion
Development of the several IL generations
The genome of BC2 plants showed an average of 8.7 introgressions/plant and 81.0% of the PS background genome (ranging from 68.7 to 88.3%). The DUD genome was fully represented among the BC2 plants. In the next generation, selected BC3 plants presented an average of 5.9 introgressions/plant and 89.4% of the PS background genome (range 81.1–97.2%). The early selection in a large number of plants facilitated the recovery of the recurrent genetic background, and the reduction of the number of introgressions per plant (to about 3) in one backcross generation. Several rounds of selfing (BC3S1, BC3S2, BC3S3, BC3S4) were performed to fix introgressions and to generate enough seeds for further assays. A first collection of 16 ILs (Online resource 3) was obtained. This IL collection has an average of 1.4 introgressions/IL (3.2 introgressions/IL with single-marker introgressions), representing more than 60% of the DUD genome with 93.9% of the PS background genome (range 86.9–96.3%) (Online resource 3). The average size of the introgressions, not considering single ones, was 8.42 Mbp.
Five ILs (DUD_3-1; DUD_4-2; DUD_5-1; DU_6-1; DUD_6-2) were further genotyped by GBS. A total of 3479 SNPs resulted informative for these 5 ILs and the parent genotypes (Online resource 4A). The introgressions defined previously with the Agena Bioscience assay (Online resource 3) were confirmed, but also some additional small introgressions were detected (Online resource 4B), showing a percentage of PS genome recovery slightly lower than the previously calculated with Agena Bioscience assay (88.4%). All this suggests that at the final steps in the process of IL development, a more exhaustive genotyping with thousands of markers is necessary.
Parent phenotypes
The two parents showed contrasting phenotypes in a number of traits related to flowering time, fruit morphology, fruit ripening behavior and traits related to organoleptic and nutritive value, such as sugars content and aroma. Online resource 5 depicts the mean values and standard deviations, along with ANOVA results for means comparison of both parents, PS and DUD, for each studied trait in the two trials in which they were phenotyped along with the ILs.
PS fruits were significantly heavier [average fruit weight (FW) 1665.7 g] than DUD fruits (213.8 g). PS fruits were more elongated with lower percentages of seminal cavity (FS 1.7 vs. 1.0, and CW 0.39 vs. 0.53, respectively for PS and DUD) (Fig. 1). Additionally, DUD presented an abscission layer at the time of ripening (AL) and the occurrence of external aroma in mature fruits (AR), while these two traits were absent in PS. Accordingly with their ripening behavior, fruit flesh at time of maturity was firmer in PS [2.00 vs. 0.79 kg/cm2 in DUD, for flesh firmness (FF)].
While PS is a sweet melon, DUD is a low sugar melon, showing a significant difference of the values of soluble solids content (SSC) in the fruits (13.8° vs. 8.4° brix degrees for PS and DUD, respectively) (Online resource 5).
Differences in other traits, such as rind thickness (RTh, 3.4 mm vs. 1.7 mm in PS and DUD, respectively), rind firmness (RF, 13.0 vs. 4.8 kg/cm2, in PS and DUD, respectively), and rind netting (Net, PS presents low values in netting, while in DUD is absent) were also observed (Online resource 5). Regarding other traits related to flesh quality, both fruits, PS and DUD were white fleshed, with similar values of luminosity Hl and b* parameter (FCHl 61.9 and FCb 10.5 vs. FCHl 62.1 and FCb 7.3, in PS and DUD, respectively), although the greenish-white fleshed PS fruit had significantly lower values of the colorimeter parameter a* (FCa − 2.5) compared to the white fleshed DUD fruit (FCa − 0.6) (Online resource 5).
PS and DUD also showed differences in male and female flowering, being DUD more early flowering (with an average of 0.8 vs. 3.3 male flowers (NMaF15), 0.4 vs. 3.2 female flowers (NFeF15), 0.3 vs. 2.5 male flowers (NMaF30), and 0.4 vs. 2.8 female flowers (NFeF30), for PS and DUD respectively).
Introgression line analysis
The QTLs detected in this set of 16 ILs, representing altogether 62.6% of the DUD genome, in the two environments using the Dunnet test are summarized in Table 1 (with complete information in Online resource 6) and Figs. 2, 3, 4, 5 and 6. These QTLs are described with detail in the next section.
Comparison of the means of the set of ILs with the mean of the recurrent parent (PS) using the Dunnett’s test. The means and standard errors for AL = presence or absence of abscission layer, AR = presence or absence of external aroma, RF = rind firmness in kg/cm2 and FF = flesh firmness in kg/cm2 are shown for each trial (gray bars for Paip16, white bars for Paip17). Bars with an asterisk show significantly different (p < 0.05) IL and PS means
Comparison of the means of the set of ILs with the mean of the recurrent parent (PS) using the Dunnett’s test. The means and standard errors for FW = fruit weight in grams, FS = fruit shape as the ratio between fruit length and fruit diameter, FL = fruit length in mm, FD = fruit diameter in mm, CW = cavity width (as the ratio between the width of the seminal cavity and the fruit diameter) and Fth = flesh thickness in mm are shown for each trial (gray bars for Paip16, white bars for Paip17). Bars with an asterisk show significantly different (p < 0.05) IL and PS means
Comparison of the means of the set of ILs with the mean of the recurrent parent (PS) using the Dunnett’s test. The means and standard errors for Rth = rind thickness in mm and Net = presence or absence of rind netting, are shown for each trial (gray bars for Paip16, white bars for Paip17). Bars with an asterisk show significantly different (p < 0.05) IL and PS means
Comparison of the means of the set of ILs with the mean of the recurrent parent (PS) using the Dunnett’s test. The means and standard errors for Hunter coordinates, FCHl = flesh color luminosity, FCa = flesh color a* parameter and FCb = flesh color b* parameter, are shown for each trial (gray bars for Paip16, white bars for Paip17). Bars with an asterisk show significantly different (p < 0.05) IL and PS means
Comparison of the means of the set of ILs with the mean of the recurrent parent (PS) using the Dunnett’s test. The means and standard errors for SSC = soluble solids content in Brix degree and pH = degree of acidity, are shown for each trial (gray bars for Paip16, white bars for Paip17). Bars with an asterisk show significantly different (p < 0.05) IL and PS means
Flowering and ripening behavior
Although DUD presents a significant early flowering in comparison to PS (Online resource 5), no IL from this set showed significant differences in the number of male and female flowers 15 and 30 days after the opening of the first flower.
As previously said, significant differences between parentals for ripening-associated traits like the presence of abscission layer (AL), occurrence of aroma (AR), and flesh and rind firmness (FF and RF) were detected in the ANOVA analysis (Online resource 5). The presence of the abscission layer had a strong genotype effect (56.9%) and almost no environmental and no interaction G × E (0.7 and 1.8% respectively) (Online resource 7). As previously reported in other studies (Bernillon et al. 2013), aroma also had an important genotype effect (23.4%) with moderate interaction G × E (5.6%) (Online resource 7). Similarly, rind and flesh firmness had a notable genotype effect (19.4–24.3% of total variation), with no environmental effect for FF and low effect for RF (0.8%) and moderate interaction G × E (7.0–9.8% of total variation).
The abscission layer was observed in the fruits of the lines DUD_3-1 and DUD_6-2 (Fig. 7a) with significant differences detected for these ILs in comparison to PS based on ANOVA and Dunnett’s test (Online resource 5, Fig. 2). DUD_3-1 has a main introgression on chromosome 3 (spanning 25.9 Mbp), only partially shared with DUD_8-1 (Online resources 3 and 4), and a minor introgression on chromosome 9 (0.55 Mbp). Additional small introgressions, ranging from 0.01 to 0.14 Mbp, were also detected after GBS analysis on chromosomes 4, 5, and 10 (Table 1, Online resources 4 A, B and 6). QTLs controlling melon abscission layer were reported for first time by Périn et al. (2002), who identified the genes Al-3 and Al-4 in chromosomes 8 and 9, respectively. Moreno et al. (2008) and Vegas et al. (2013) reported a QTL (ETHQB3.5) associated to ethylene production in fruits, in a region of chromosome 3, spanning from 22 to 30 Mbp. The introgression in DUD_3-1 includes partly this region (Online resource 4). ETHQB3.5 alleles of the chinensis SC induce climacteric behavior in the non-climacteric ‘Piel de Sapo’ background, with a similar behavior to DUD_3-1, supporting the presence of a QTL for ripening in this introgression. CmACS5 (MELO3C010779, 30,906,213–30,908,525 bp in chromosome 3) was proposed as candidate for ETHQB3.5, although it can be discarded as the responsible of the phenotype observed in DUD_3-1, since this gene is out of the IL DUD_3-1 introgression (located 199 Kbp–26.1 Mbp) and other IL, DUD_5-1, contains a DUD introgression including CmACS5 (30–31.5 Mbp), and does not develop abscission layer (Online resource 4).
Pictures showing characteristic phenotypes of some ILs in comparison to PS. a Mature fruits of parental PS without abscission layer and of line DUD_3-1 and DUD_6-2 with abscission layer. b Variation of fruit morphology in DUD_4-2 and DUD_5-3 likely caused by the DUD introgressions in chromosomes 4, 6 or 12 in the genetic background of PS affecting the FS and FL. c Effect of the DUD introgressions in the line DUD_1-2 produces increase the CW (18.1%) and reduction the Rth (15.7%) and Fth (24.1%) in comparison with the parental PS. d Mature fruits of parental PS (netted) and of lines DUD_5-1 and DUD_5-2 with more intense netting. e Mature fruits of parental PS and DUD_6-1 showing the effect of yellowish rind
DUD_6-2, also displaying abscission layer, carries two main introgressions according to Agena Bioscience assay, on chromosomes 1 (spanning 2.6 Mbp) and 6 (spanning 17.7 and 2.9 Mbp) (Online resources 3 and 4). Additional small introgressions have been found using GBS on chromosomes 3, 4, 5, 7, and 10 (from 0.01 to 0.65 Mbp in length; Online resource 4). This line, DUD_6-2, was the only IL that showed consistently significant differences with respect to PS in other ripening related traits since it presents external aroma (AR), and a loss of flesh and rind firmness (FF, RF) (59.5 and 17.4%, respectively) in both years (Fig. 2, Table 1, Online resource 6). Based on GBS genotyping, the main DUD introgression of chromosome 6 in DUD_6-2 is located from 14.5 to 32.7 Mbp and from 34.8 to 37.7 Mbp. In this region is located the previously reported QTL ETHQV6.3 also involved in climacteric ripening of the non-climacteric ‘Piel de Sapo’ with chinensis SC introgressions (Moreno et al. 2008; Vegas et al. 2013). The gene CmNAC-NOR (MELO3C016540, 27,663,292–27,665,351 bp; Online resource 8), which encodes a NAC transcription factor involved in the regulation of climacteric ripening, has been identified as the responsible of the altered ripening controlled by ETHQV6.3 (Ríos et al. 2017). Although this gene is located within the introgression of DUD_6-2, also the non-climacteric IL DUD_5-1 contains a DUD introgression in this region (27.4–27.9 Mbp) showing no effects (Online resource 4). The DUD_6.2 introgression in chromosome 6 contains other four genes annotated as NAC domain proteins (MELO3C014922, 20,900,809–20,902,991, MELO3C016444 29,775,058–29,777,132, MELO3C013971, 35,529,924–35,533,985, MELO3C014141, 37,363,939–37,367,039) (Online resource 8) that could also contribute to the climacteric phenotype.
Interestingly, other NAC domain-containing proteins such as MELO3C019845 and MELO3C019954 are located in the genomic region of chromosome 3 from DUD_3-1 (Online resource 8). Also, a recent study using a ‘Vedrantais’ IL with a chromosome 10 introgression from a makuwa melon (PI 420176) (Perpiñá et al. 2017), suggested a NAC/NAM transcription factor (MELO3C012390) as a candidate gene responsible for the delayed ripening process and the absence of abscission layer and external aroma in the climacteric genetic background of ‘Vedrantais’, highlighting again the close relation between NAC genes and the climacteric behavior.
Our results, along with previous studies, suggest that the genetic basis of fruit ripening in melon is complex and variety-specific. The presence of NAC factors in most of the main introgressions coming from different exotic genetic backgrounds, suggests that common genetic mechanisms may be involved in ripening regulation through the action of different transcriptions factors of the NAC family located in different genomic regions and that can induce climacteric ripening in non-climacteric background or delay ripening in climacteric-backgrounds, leading to a wide range of climacteric behavior that can be useful for melon breeding.
DUD_3-1 and DUD_6-2 also showed differences in ripening related traits among them. DUD_3-1 forms abscission layer, likely induced by the QTL al.3 defined by this line, but fruits are not aromatic and do not suffer flesh softening, whereas DUD_6-2 forms abscission layer and develops aromatic fruit with an enhanced flesh softening process. Although the genomic region in chromosomes 6 defined by DUD_6-2 is the one more consistently supported by previous studies of QTLs related to climacteric ripening and flesh firmness (Monforte et al. 2004; Paris et al. 2008; Ríos et al. 2017; Vegas et al. 2013), this line has an additional major introgression in chromosome 1, where other studies have reported QTLs related to flesh softness. For instance, Galpaz et al. (2018) detected a QTL for flesh firmness, but this is located 30 Mbp away from our rf.1 and ff.1 (Table 1, Online resource 6). Another QTL involved in ethylene production, eth1.1, was reported in chromosome 1 (Moreno et al. 2008; Périn et al. 2002), being suggested the ethylene receptor CmERS1 (MELO3C015961) as a candidate gene. Although this gene locates outside DUD_6-2 introgression (at 32 Mbp), several ethylene-related genes like MELO3C024268 and MELO3C024315, two ethylene-responsive transcription factors, were located in the QTL region of chromosome 1 defined by DUD_6-2 IL. Additionally, other genes involved in volatile metabolism like MELO3C029261 or MELO3C024348 (a malonate-CoA ligase-like protein and a lipoxygenase, enzymes involved in the pathway of phenylpropanoid-derived compound metabolisms) collocated with ar.1 (Online resource 8). Galpaz et al. (2018) reported more than 140 QTLs, mainly in chromosomes 6, 8 and 11, corresponding to many volatiles detected differentially in their RILs derived from the momordica PI 414723 and the reticulatus Dulce. Only some QTLs in chromosome 6 such as the ones for S-methyl propanethioate or total sesquiterpenes collocate with ar.6. The stronger impact of QTL al.6 compared to al.3 on the ripening process, or the combined impact of al.6 with al.1 in IL DUD 6-2, along with effects from other minor introgressions, could account for these differences.
Segregation of the additional introgressions detected with GBS in these ILs will allow verifying that the QTLs are in the chromosome 3 and 6 introgressions. Aroma is one of the most important quality traits for the consumer, whereas flesh softness is an undesired trait. Both lines might be a prebreeding material useful to develop Piel de sapo varieties with improved fruit quality and new characteristics.
Fruit morphology
The fruit morphology studied in the present report includes fruit weight (FW), length (FL), diameter (FD), shape (FS) and cavity (CW), traits which showed significant differences among PS and DUD (Online resource 5). As expected, these traits showed a strong genotype effect (12.7–47.4%) and low interaction G × E (3.0–9.9%). CW and FS also displayed a significant environmental effect (Online resource 7). In addition, some of these traits are closely related. For instance, FS was positively correlated with FL (r = 0.65, mean Paip16 and Paip17), and FW presented a strong correlation with FL and FD (r = 0.80 and r = 0.91, respectively) (Online resource 9). However, FS and FW showed low correlation (r = 0.19) (Online resource 9), similar to the results reported by Perpiñá et al. (2016).
DUD_1-1, DUD_1-2, DUD_4-2, DUD_5-1, DUD_5-3, DUD_6-2 showed a significant reduction of FS from 14.4 to 30.9% compared with PS in both trials (Fig. 3, Table 1, Online resource 6). In fact, many of these lines also showed a FL reduction from 20.0 to 38.8% with respect to PS parental (with exception of DUD_1-1 and DUD_6-2). These lines define 6 QTLs in fs.1.1, fs.1.2, fs.4, fs.5, fs.6 and fs.12 (Table 1).
Regarding fruit shape, Monforte et al. (2014) reported several meta-QTLs derived from different studies and populations. In chromosome 1, FSMQ1 was described with subsp. agrestis allele leading to elongated fruits. Our fs.1.1 and fs.1.2 defined by DUD_1-1, DUD_1-2 and DUD_6-2 collocate in this region, with alleles from DUD decreasing the shape index (rounder fruits; 22.5, 14.4, 24.9%, respectively; Table 1). In fact, the line DUD_1-2 also defined fl.1, displaying a significant decrease in fruit length (22.3%).
The introgression on chromosome 4 of DUD_4-2 defined fs.4 and fl.4 (Table 1, Online resource 6). This IL presented fruits with a significant decrease in fruit shape index (30.9%) and also in length (38.8%). No meta-QTL involved in fruit shape was reported by Monforte et al. (2014) in this chromosome. However, QTLs involved in fruit length have been reported by Díaz et al. (2015) in that region of chromosome 4 in their consensus map (FLQC4.3, FLQX4.1), and Ramamurthy and Waters (2015) found that the snake melon allele for QTL FS2 (same region as fs.4) increased the fruit length, while the cantaloupe alleles decreased the trait. Díaz et al. (2017) detected flqt4.1, faqt4.1, and fdqt4.1 in a region partly collocalized with fs.4, in a population derived from wild Cucumis trigonus (Trigonus, now reclassified as Cucumis melo subspecies agrestis) crossed to ‘Piel de sapo’, with the PS alleles increasing length fruit and size, which is coherent with our results for that region.
Wu et al. (2018) presented strong evidences that members of Ovate Family Protein (OFP) gene family are involved in the determination of organ shape in different plant species having a critical role in regulating cell division patterns early in the development of organs. Consistently, Díaz et al. (2017) indicated that the QTLs described in chromosome 4 included some candidate genes belonging to the OFP family (CmOFP4 corresponding to MELO3C009113, 34,135,042–34,136,208 bp; and CmOFP5, corresponding to MELO3C009514, 31,566,369–31,567,286 bp), along with other genes like MELO3C013074 (17,152,898–17,154,990 bp), a calmodulin-binding family protein (involved in orientation during cell division; Huang et al. 2013; Wu et al. 2011, 2015). Although the region of fs.4 did not include any of these OFP candidates, it included the calmodulin-binding family protein MELO3C013074.
Interestingly, the lines DUD_4-2, DUD_5-1, DUD_5-3 and DUD_6-2, which showed rounder shape in comparison to PS, include OFP genes in their DUD introgressions of chromosomes 1, 5 and 12, based on the list of OFPs reported by Monforte et al. (2014) (Online resource 8). For example, DUD_4.2 shares with DUD_5-3 an introgression in chromosome 12. In this region maps the Transcription repressor OFP6 (MELO3C025581, 12,990,776–12,991,288), that can be a candidate to explain fruit shape variation in these lines (Fig. 7b). Apart from OFP6, a probable candidate for this QTL is the pentamerous locus (p), which controls the number of carpels (Monforte et al. 2014) and has been mapped in this region on chromosome 12 (16.4–22.4 Mbp; Díaz et al. 2011). As an increase in the number of carpels has been associated to longer diameters and rounder fruits in tomato (Rodriguez et al. 2011), alleles from accessions with 5 carpels (not the usual 3) such as the chinensis SC and dudaim melons may be lead also to rounder shapes. A meta-QTL involved in fruit shape (FSQM12) was reported in chromosome 12 (Monforte et al. 2014). Our fs.12 collocates with this meta-QTL whose conomon alleles lead to rounder fruits, similarly to DUD alleles.
Moreover, based on the DUD introgressions in this line DUD_5-3, putative QTLs in chromosomes 5 and 6 (fs.5, fl.5, fs.6, fl.6), and not only in chromosome 12, were defined (Table 1, Online resource 6). With respect to the introgression on chromosome 6 of DUD_5-3 (fs.6), it is shared with DUD_1-1, DUD_5-1, and DUD_6-2 with the same effects on FS. Recently, Pereira et al. (2018) detected FSQU6.1 and FSQU6.2 (QTLs spanning 20.7–31.9 Mbp and 31.6–36.4 Mbp, respectively) on chromosome 6 in which ‘Vedrantais’ alleles, like DUD ones, lead to rounder shapes in contrast to more elongated PS.
Wu et al. (2018), using NILs from the cross between the Indian momordica PI 124112 and ‘Piel de Sapo’ as recurrent parent, identified a gene of the ovate family (OFP1 transcription repressor, MELO3C025206) on chromosome 8, as the responsible for the decrease of the length, producing round shape if the momordica allele is present. This gene is not represented in our final ILs collection, but the results described above suggest that other OFP-related genes, may also cause differences in fruit shape. Derivation of additional ILs from BC3 and new BC4 is needed to cover this genomic region and confirm if DUD alleles for this gene in a PS background have similar effects on fruit shape.
Regarding fruit size, only DUD_1-2 and DUD_2-1 showed significant differences with respect to PS in both environments (Fig. 3), with a reduction in weight of 36.5 and 33.9%, respectively. The former IL not only presents smaller fruits but also rounder ones, as indicated previously (fl.1, fs.1.1). Previous studies using PS also reported a QTL located in the same region (FWQC1.4, Eduardo et al. 2007) with PS alleles decreasing fruits weight. In addition, fw.1 collocates with cw.1, a QTL involved in the variation of seed cavity width, and fth.1, involved in flesh thickness (Table 1, Online resource 6). Only this IL DUD_1-2 showed significant effects increasing seed cavity and reducing flesh thickness in both environments (Figs. 3, 7c, Table 1) compared to PS (17.2 and − 24.3%, respectively).
In contrast, DUD_2-1 only presented a difference in weight, as both length and diameter were also significantly lower than PS without an effect on shape (Fig. 3, Online resource 5). Thus, fw.2, fl.2 and fd.2 collocate in a large region on chromosome 2 (from 170 Kbp to 15.8 Mbp). Previous studies reported QTLs involved in fruit length associated to size and in shape in this region of chromosome 2. For instance, Zalapa et al. (2007) detected a QTL that collocates with our fw.2 (FWQI2.1, Díaz et al. 2015), and Harel-Beja et al. (2010), using RILs derived from the momordica PI 414723 and the reticulatus Dulce, reported flqn2.1 (0.7–23 Mbp, with Dulce reducing the length) and fsh2.1 (0.8–20.0 Mbp). In our PS background, DUD alleles lead to a reduction in length (19.8%, Table 1) like Dulce alleles in flqn2.1. Subsequently, a QTL for weight including the same region was detected using ILs derived from makuwa and ‘Vedrantais’, although in this case makuwa alleles increased fruit weight, the opposite of dudaim. Recently, Díaz et al. (2017), using the F2 from PS and the wild Trigonus, detected flqt2.1 in an overlapping region to our fw.2 [1.4–2.1 Mbp, region of andromonoecious a gene (CmACS7; MELO3C015444: 1,709,035–1,711,473 bp)], with PS alleles decreasing fruit length, similar to our DUD reported effect. Monforte et al. (2014) reported a meta-QTL for weight in that region (FWMQ2) co-segregating with a gene. Previously, Monforte et al. (2005) suggested that sex expression affects fruit size and shape since the presence of stamens prevent the normal longitudinal growth leading to a reduced elongation of the ovary. Since DUD and PS melons are andromonoecious and carry the non-functional allele for CmACS7 (a), this pleiotropic effect does not explain the observed phenotype caused by this DUD introgression in chromosome 2. Therefore, we suggest that maybe other gene is involved in this trait in our population, or simply that fruit length and andromonoecious phenotype only co-segregates without a cause-effect relation as previously thought.
The small size of melons may be a desired attribute in current markets. Thus both DUD_1-2 and DUD_2-1 can be promising pre-breeding lines, and especially DUD_2-1, since flesh thickness is not affected.
Rind thickness and netting
The rind characteristics, such as the thickness and netting are important in the processes of resistance to storage and transport (shelf life). These two traits, Rth and Net, displayed a negative correlation of r = −0.17 (Online resource 9). Moderate heritability has been reported for them (Perpiñá et al. 2016). We found important G effects (17.7–44.9%), non-significant E effects, and moderate G X E interaction (Online resource 7).
Based on ANOVA, parentals showed significant differences for Rth in Paip16 and Paip17, while for Net the differences were only significant in one environment (Online resource 5). Four ILs presented significant differences with PS in both trials for Rth (DUD_4-2, DUD_5-1, DUD_6-3 and DUD_7-1) and two for Net (DUD_5-1 and DUD_5-2) (Fig. 4). For rind thickness these lines presented an average of 2.2, 2.3, 2.5 and 2.6 mm respectively versus 3.4 in PS (Online resource 5). Putative genomic regions responsible to the decrease in these lines were identified (rth.3, rth.4, rth.6 and rth.7) (Table 1, Online resource 6), although DUD_4-2 and DUD_5-1 carries some other introgressions based on GBS analysis (from 0.02 to 11.2 Mbp long) that will be necessary to study.
One region in chromosome 5 seemed to control netting density (Table 1, Online resource 6). Although DUD is a non-netted type (likely due to a genetic background effect), DUD alleles in this region spanning 2 Mbp lead to netting in DUD_5-1 and DUD_5-2 fruits (Fig. 7d), with an increase of 451.3 and 379.5% with respect to PS, which is also non-netted, respectively. Perpiñá et al. (2016) identified QTLs associated with this trait on chromosomes 5, 6 and 7, with no correspondence with the reported herein. Also, Paris et al. (2008) using a cross with the cantaloupe Top Mark, reported PNQJ5.1 in chromosome 5, which is located 1Mbp away from our net.5.
Rind and flesh color and sugars content
Despite the differences in rind color between parentals (dark green in PS and reddish-yellow with ochre stripes in DUD; Fig. 1) only one IL, DUD_6-1, presented a yellowish rind (Fig. 7e). It is well known that the rind color in melon changes during the ripening process due to the loss of the green pigment chlorophyll and the synthesis of other pigments such as carotenoids and flavonoids. Monforte et al. (2004) detected in chromosome 10 a QTL involved in this trait, which is not associated to the climacteric behavior. Subsequently, Feder et al. (2015) identified a Kelch domain-containing F-box protein coding gene (CmKFB, MELO3C011980: 3,475,283–3,476,416 bp) in that region as the causal factor of the yellow rind phenotype. This gene negatively regulates the accumulation of naringenin chalcone, a yellow flavonoid, and affects to downstream flavonoid pathway. Based on the GBS genotyping, DUD_6-1 has two main introgressions in chromosome 10 (4.2–6.2 Mbp, and 19–26.5 Mbp) and small ones in 1, 2, 4, 5, 6, 7, 8 and 11 (spanning small regions from 0.02 to 1.68 Mbp) (Online resource 4). However, CmKFB is outside the DUD introgressions in chromosome 10, therefore other genes must be controlling this trait in this dudaim derived population. For example, in the chromosome 10 introgression of DUD_6-1 is located MELO3C018335 (19,402,491–19,403,056) that catalyzes the 3′,5′-hydroxylation of naringenin and other flavonoids and could be a good candidate for this genotype.
PS and DUD have similar white flesh, with similar values of FCHl and FCb, and differences in FCa (greenish flesh in PS fruit) (Online resource 5). ILs did not show high variability for these traits (Fig. 5). For these color parameters a considerable genotype effect (10.9, 23.8 and 26.1%, for FCHl, FCb and FCa, respectively) and interaction G X E (6.6, 14.8 and 15.6%, respectively) were found, while low or no environmental effect was estimated (1.5% for FCa) (Online resource 7). Only some lines showed significant effects based on Dunnett’s test for FCa and FCb, but only in one environment, none showing a consistent effect in both assays (Fig. 5).
Regarding nutritional content, sweetness is one of the most appreciated traits. Sugar content in melon has a complex genetic control (Gur et al. 2016), so a large number of QTLs related to this trait have been identified in different chromosomes to date (Argyris et al. 2017; Díaz et al. 2015; Pereira et al. 2018). Parentals showed differences in SSC (13.8 ºBrix for sweet PS vs. 8.18 ºBrix for non-sweet DUD). Although a significant environmental effect (3.1%) and G × E (6.9%) was estimated, also a moderate genotype effect existed for this trait (16.9%) (Online resource 7). Only DUD_2-1, had a consistent effect in both environments (average 10.9° Brix vs. 13.8° Brix in PS) with a significant decrease in °Brix (19.2–23.1%) (Table 1, Fig. 6). This IL defined QTL ssc.2, and as expected, DUD alleles led to a reduction in the soluble solid content (Table 1). Harel-Beja et al. (2010) reported some QTLs in that region of chromosome 2 (sscqn2.1, sscqc2.2), with Dulce allele increasing SSC. Although only in one location, Argyris et al. (2017), using NILs derived from SC, also detected QTLs in this chromosome for SSC and sucrose in the region 0–1.4 Mbp overlapping ssc.2. SC alleles led to a decrease of the sugar content relative to PS (more than 25%). Despite the fact that RIL population by Pereira et al. (2018) was also developed from PS, no QTL for SSC was detected in common with our population. Many other studies have mapped sugar-related traits also in other chromosomes, however, some QTLs seem to be conserved across distinct germplasm suggesting that common genetic mechanisms regulate sugar content (Argyris et al. 2017), especially in populations involving non-sweet Oriental alleles (from DUD, SC, Trigonus).
Differences between parentals for flesh acidity (pH) only were detected in one environment (Paip17) and only DUD_6-2 was slightly more acid than PS demonstrating there is low genetic variability for this trait in this population (Fig. 6).
Conclusions
The set of introgression lines presented in this work represents the first IL set developed with the aim to exploit and study dudaim genomic variability into a ‘Piel de Sapo’ genetic background. Although the whole dudaim genome is not represented in these 16 ILs, interesting variability for several fruit quality traits such as climacteric ripening accompanied or not of external aroma, fruit shape and weight, development of thinner and netted rind, and different rind color not associated to climacteric ripening has been found. Therefore, some of these ILs could be considered prebreeding lines for future improvement of PS commercial types with new characteristics. Associated to these traits, several QTLs have been defined by these ILs, in some cases reinforcing previously reported QTLs and discussing the location of some candidate genes in these genomic regions, and in other cases suggesting new candidates and regions involved. The need of an in depth genotyping using GBS methodologies in the final steps of IL library construction has been also suggested.
References
Amanullah S, Liu S, Gao P, Zhu Z, Zhu Q, Fan C, Luan F (2018) QTL mapping for melon (Cucumis melo L.) fruit traits by assembling and utilization of novel SNPs based CAPS markers. Sci Hortic 236:18–29. https://doi.org/10.1016/j.scienta.2018.02.041
Argyris JM, Ruiz-Herrera A, Madriz-Masis P, Sanseverino W, Morata J, Pujol M, Ramos-Onsins SE, Garcia-Mas J (2015) Use of targeted SNP selection for an improved anchoring of the melon (Cucumis melo L.) scaffold genome assembly. BMC Genom 16:1–14. https://doi.org/10.1186/s12864-014-1196-3
Argyris JM, Díaz A, Ruggieri V, Fernández M, Jahrmana T, Gibon Y, Picó B, Martin-Hernández AM, Monforte AJ, Garcia-Mas J (2017) QTL analyses in multiple populations employed for the fine mapping and identification of candidate genes at a locus affecting sugar accumulation in melon (Cucumis melo L.). Front Plant Sci 8:1–20. https://doi.org/10.3389/fpls.2017.01679
Aubert C, Pitrat M (2006) Volatile compounds in the skin and pulp of Queen Anne’s pocket melon. J Agric Food Chem 54:8177–8182. https://doi.org/10.1021/jf061415s
Bernillon S, Biais B, Deborde C, Maucourt M, Cabasson C, Gibon Y, Hansen TH, Husted S, de Vos RCH, Mumm R, Jonker H, Ward JL, Miller SJ, Baker JM, Burger J, Tadmor Y, Beale MH, Schjoerring JK, Schaffer AA, Rolin D, Hall RD, Moing A (2013) Metabolomic and elemental profiling of melon fruit quality as affected by genotype and environment. Metabolomics 9:57–77
Blanca J, Cañizares J, Ziarsolo P, Esteras C, Mir G, Nuez F, Garcia-Mas J, Picó B (2011) Melon Transcriptome characterization: simple sequence repeats and single nucleotide polymorphisms discovery for high throughput genotyping across the species. Plant Genome 4(2):118–131. https://doi.org/10.3835/plantgenome2011.01.0003
Blanca J, Esteras C, Ziarsolo P, Pérez D, Fernández-Pedrosa V, Collado C, de Pablos RR, Ballester A, Roig C, Cañizares J, Picó B (2012) Transcriptome sequencing for SNP discovery across Cucumis melo. BMC Genom 13:1–18. https://doi.org/10.1186/1471-2164-13-280
Burger Y, Sa’ar U, Paris HS, Lewinsohn E, Katzir N, Tadmor Y, Schaffer AA (2006) Genetic variability for value fruit quality traits in Cucumis melo. Isr J Plant Sci 54:233–242. https://doi.org/10.1560/ijps
Dantas ACA, Araujo IS, Esteras C, Nunes GHS, Pico MB (2015) Diversity of melon accessions from northeastern Brazil and their relationships with germplasms of diverse origins. J Am Soc Hortic Sci 10:505–517. https://doi.org/10.21273/JASHS.140.5.504
Díaz A, Fergany M, Formisano G et al (2011) A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol 11:111. https://doi.org/10.1186/1471-2229-11-111
Díaz A, Forment J, Argyris JM, Fukino N, Tzuri G, Harel-Beja R, Katzir N, Garcia-Mas J, Monforte AJ (2015) Anchoring the consensus ICuGI genetic map to the melon (Cucumis melo L.) genome. Mol Breed 35:1–7. https://doi.org/10.1007/s11032-015-0381-7
Díaz A, Martín-Hernández AM, Dolcet-Sanjuan R, Garces-Claver A, Álvarez JM, Garcia-Mas J, Picó B, Monforte AJ (2017) Quantitative trait loci analysis of melon (Cucumis melo L.) domestication-related traits. Theor Appl Genet 130:1837–1856. https://doi.org/10.1007/s00122-017-2928-y
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(13–15):58
Dunnett C (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121
Eduardo I, Arús P, Monforte AJ (2005) Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor Appl Genet 112:139–148. https://doi.org/10.1007/s00122-005-0116-y
Eduardo I, Arús P, Monforte AJ, Obando J, Fernández-trujillo JP, Martínez JA, Alarcón AL, Álvarez JM, van der Knaap E (2007) Estimating the genetic architecture of fruit quality traits in melon using a genomic library of near isogenic lines. J Am Soc Hortic Sci 132:80–89. https://doi.org/10.21273/JASHS.132.1.80
Endl J, Achigan-Dako EG, Pandey AK, Monforte AJ, Picó B, Schaeffer H (2018) Repeated domestication of melon (Cucumis melo) in Africa and Asia and a new close relative from India. Am J Bot 105:1662–1671. https://doi.org/10.1002/ajb2.1172
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Esteras C, Formisano G, Roig C, Díaz A, Blanca J, Garcia-Mas J, Gómez-Guillamón ML, López-Sesé AI, Lázaro A, Monforte AJ, Picó B (2013) SNP genotyping in melons: genetic variation, population structure, and linkage disequilibrium. Theor Appl Genet 126:1285–1303. https://doi.org/10.1007/s00122-013-2053-5
Esteras C, Rambla JL, Sánchez G, López-Gresa MP, González-Mas MC, Fernández-Trujillo JP, Bellés JM, Granell A, Picó B (2018) Fruit flesh volatile and carotenoid profile analysis within the Cucumis melo L. species reveals unexploited variability for future genetic breeding. J Sci Food Agric 98:3915–3925. https://doi.org/10.1002/jsfa.8909
FAO (FAOSTAT) (2016) http://www.fao.org/faostat/en/#data. Accessed 30 Jan 2019
FAO (FAOSTAT) (2017) http://www.fao.org/faostat/en/#data. Accessed 30 Jan 2019
Feder A, Burger J, Gao S et al (2015) A Kelch domain-containing F-box coding gene negatively regulates flavonoid accumulation in Cucumis melo L. Plant Physiol 169:1714–1726. https://doi.org/10.1104/pp.15.01008
Galpaz N, Gonda I, Shem-Tov D et al (2018) Deciphering genetic factors that determine melon fruit-quality traits using RNA-Seq-based high-resolution QTL and eQTL mapping. Plant J 94:169–191. https://doi.org/10.1111/tpj.13838
Garcia-Mas J, Benjak A, Sanseverino W et al (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci 109:11872–11877. https://doi.org/10.1073/pnas.1205415109
Gur A, Gonda I, Portnoy V et al (2016) Genomic aspects of melon fruit quality. In: Grumet R, Katzir N, García-Mas J (eds) Genetics and genomics of cucurbitaceae. Springer, Cham, pp 377–408
Gur A, Tzuri G, Meir A, Sa’ar U, Portnoy V, Katzir N, Schaffer AA, Li L, Burger J, Tadmor Y (2017) Genome-wide linkage-disequilibrium mapping to the candidate gene level in melon (Cucumis melo). Sci Rep 7:1–13. https://doi.org/10.1038/s41598-017-09987-4
Harel-Beja R, Tzuri G, Portnoy V et al (2010) A genetic map of melon highly enriched with fruit quality QTLs and EST markers, including sugar and carotenoid metabolism genes. Theor Appl Genet 121:511–533. https://doi.org/10.1007/s00122-010-1327-4
Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E (2013) Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genom 288:111–129. https://doi.org/10.1007/s00438-013-0733-0
Lázaro A, Fernandez IC, Borrero MJ, Cabello F, Lopez-Sese A, Gómez-Guillamon M, Picó B (2017) Agromorphological genetic diversity of spanish traditional melons. Genet Resour Crop Evol 7(64):1687–1706. https://doi.org/10.1007/s10722-016-0466-0
Leida C, Moser C, Esteras C, Sulpice R, Lunn JE, de Langen F, Monforte AJ, Picó B (2015) Variability of candidate genes, genetic structure and association with sugar accumulation and climacteric behavior in a broad germplasm collection of melon (Cucumis melo L.). BMC Genet 16:1–17. https://doi.org/10.1186/s12863-015-0183-2
López C, Ferriol M, Picó MB (2015) Mechanical transmission of tomato leaf curl New Delhi virus to cucurbit germplasm: selection of tolerance sources in Cucumis melo. Euphytica 204:679–691. https://doi.org/10.1007/s10681-016-1816-x
Monforte AJ, Oliver M, Gonzalo MJ, Alvarez JM, Dolcet-Sanjuan R, Arús P (2004) Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.). Theor Appl Genet 108:750–758. https://doi.org/10.1007/s00122-003-1483-x
Monforte AJ, Iban E, Silvia A, Pere A (2005) Inheritance mode of fruit traits in melon: heterosis for fruit shape and its correlation with genetic distance. Euphytica 144:31–38. https://doi.org/10.1007/s10681-005-0201-y
Monforte AJ, Díaz A, Caño-Delgado A, Van Der Knaap E (2014) The genetic basis of fruit morphology in horticultural crops: lessons from tomato and melon. J Exp Bot 65:4625–4637. https://doi.org/10.1093/jxb/eru017
Moreno E, Obando JM, Dos-Santos N, Fernández-Trujillo JP, Monforte AJ, Garcia-Mas J (2008) Candidate genes and QTLs for fruit ripening and softening in melon. Theor Appl Genet 116:589–602. https://doi.org/10.1007/s00122-007-0694-y
Nimmakayala P, Tomason YR, Abburi VL et al (2016) Genome-wide differentiation of various melon horticultural groups for use in GWAS for fruit firmness and construction of a high resolution genetic map. Front Plant Sci 7:1–15. https://doi.org/10.3389/fpls.2016.01437
Nunes EWLP, Esteras C, Ricarte AO et al (2017) Brazilian melon landraces resistant to Podosphaera xanthii are unique germplasm resources. Ann Appl Biol 171:214–228. https://doi.org/10.1111/aab.12370
Paris MK, Zalapa JE, McCreight JD, Staub JE (2008) Genetic dissection of fruit quality components in melon (Cucumis melo L.) using a RIL population derived from exotic x elite US Western Shipping germplasm. Mol Breed 22:405–419. https://doi.org/10.1007/s11032-008-9185-3
Paris HS, Amar Z, Lev E (2012) Medieval history of the Duda’im melon (Cucumis melo, Cucurbitaceae). Econ Bot 66(3):276–284
Pavan S, Marcotrigiano AR, Ciani E, Mazzeo R, Zonno V, Ruggieri V, Lotti C, Ricciardi L (2017) Genotyping-by-sequencing of a melon (Cucumis melo L.) germplasm collection from a secondary center of diversity highlights patterns of genetic variation and genomic features of different gene pools. BMC Genom 18:1–10. https://doi.org/10.1186/s12864-016-3429-0
Pereira L (2018) Genetic dissection of fruit quality and ripening traits in melon. Barcelona (doctoral thesis), Universitat Autònoma de Barcelona. Barcelona, Spain. Retrieved from https://ddd.uab.cat/record/201529. Accessed 10 April 2019.
Pereira L, Ruggieri V, Pérez S, Alexiou KG, Fernández M, Jahrmann T, Pujol M, Garcia-Mas J (2018) QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol 18:1–17. https://doi.org/10.1186/s12870-018-1537-5
Périn C, Gomez-jimenez M, Hagen L, Dogimont C, Pech JC, Latché A, Pitrat M, Lelièvre JM (2002) Molecular and genetic characterization of a non-climacteric phenotype in melon reveals two loci conferring altered ethylene response in fruit. Plant Pathol 129:300–309. https://doi.org/10.1104/pp.010613
Perpiñá G, Esteras C, Gibon Y, Monforte AJ, Picó B (2016) A new genomic library of melon introgression lines in a cantaloupe genetic background for dissecting desirable agronomical traits. BMC Plant Biol 16:1–21. https://doi.org/10.1186/s12870-016-0842-0
Perpiñá G, Cebolla-Cornejo J, Esteras C, Monforte AJ, Picó B (2017) “MAK-10”: a long shelf-life charentais breeding line developed by introgression of a genomic region from Makuwa melon. HortScience 52:1633–1638. https://doi.org/10.21273/HORTSCI12068-17
Pitrat M (2017) Melon genetic resources: phenotypic diversity and horticultural taxonomy. In: Grumet R, Katzir N, Garcia-Mas J (eds) Genetics and genomics of the Cucurbitaceae. Springer, New York, pp 25–60
Ramamurthy RK, Waters BM (2015) Identification of fruit quality and morphology QTLs in melon (Cucumis melo) using a population derived from flexuosus and cantalupensis botanical groups. Euphytica 204:163–177. https://doi.org/10.1007/s10681-015-1361-z
Ríos P, Argyris J, Vegas J, Leida C, Kenigswald M, Tzuri G, Troadec C, Bendahmane A, Katzir N, Picó B, Monforte AJ, Garcia-Mas J (2017) ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J 91:671–683. https://doi.org/10.1111/tpj.13596
Rodriguez GR, Muños S, Anderson C, Sim S-C, Michel A, Causse M, Gardener BBM, Francis D, van der Knaap E (2011) Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol 156:275–285. https://doi.org/10.1104/pp.110.167577
Sabato D, Esteras C, Grillo O, Pico B, Bacchetta G (2015) Seed morpho-colourimetric analysis as complementary method to molecular characterization of melon diversity. Sci Hortic 192:441–452. https://doi.org/10.1016/j.scienta.2015.06.006
Vegas J, Garcia-Mas J, Monforte AJ (2013) Interaction between QTLs induces an advance in ethylene biosynthesis during melon fruit ripening. Theor Appl Genet 126:1531–1544. https://doi.org/10.1007/s00122-013-2071-3
Vossen RHAM, Aten E, Roos A, Den Dunnen JT (2009) High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30:860–866. https://doi.org/10.1002/humu.21019
Wu J, Chang Z, Wu Q, Zhan H, Xie S (2011) Molecular diversity of Chinese Cucurbita moschata germplasm collections detected by AFLP markers. Sci Hortic 128:7–13. https://doi.org/10.1016/j.scienta.2010.12.006
Wu S, Clevenger JP, Sun L, Visa S, Kamiya Y, Jikumaru Y, Blakeslee J, van der Knapp E (2015) The control of tomato fruit elongation orchestrated by sun, ovate and fs8.1 in a wild relative of tomato. Plant Sci 238:95–104. https://doi.org/10.1016/j.plantsci.2015.05.019
Wu S, Zhang B, Keyhaninejad N et al (2018) A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat Commun 9:1–12. https://doi.org/10.1038/s41467-018-07216-8
Zalapa JE, Staub JE, McCreight JD, Chung SM, Cuevas H (2007) Detection of QTL for yield-related traits using recombinant inbred lines derived from exotic and elite US Western Shipping melon germplasm. Theor Appl Genet 114:1185–1201. https://doi.org/10.1007/s00122-007-0510-8
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989. https://doi.org/10.1038/35103590
Acknowledgements
This work was supported by the Spanish Ministerio de Educación through an ERA-NET Plant Genomics project (MELRIP: GEN2006-27773-C2-2-E), by the Spanish Ministerio de Economía de Empresa through a Plant KBBE Project (SAFQIM: PIM2010PKB-00691), by the Spanish Ministerio de Economía y Competitividad AGL2015-64625-C2-2-R, and by the Spanish Ministerio de Ciencia, Innovación y Universidades AGL2017-85563-C2-1-R (jointly funded by FEDER). This work was also partly supported by the Programa de Valorización y Recursos Conjuntos de I + D + i de VLC/CAMPUS funded by the Ministerio de Educación, Cultura y Deporte as part of the Programa Campus de Excelencia Internacional.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
. SNPs employed in the construction of the IL population. This set was selected from those previously mapped and employed to anchor the first version of the melon genome (Garcia-Mas et al. 2012). Also, positions in the new genetic map by Argyris et al. (2015) are indicated, as well as the physical position for genome assembly in the version v.3.5.1 and the most recent v.3.6.1, available at Melonomics. Information about the SNP and flanking sequence is included for all the markers used in the Agena Bioscience assay, and the primers for those that were adapted to the HRM genotyping procedure are also provided. Markers that could not be accurately called in the Agena Bioscience assay are marked as failed markers (f) (XLSX 31 kb)
Online resource 2
. Evaluation of the normality of every trait distribution for Paip16 and Paip17 populations with the Shapiro–Wilk and Kolmogorov–Smirnov tests using Statgraphics Centurion XVII.II (XLSX 17 kb)
Online resource 3.
Graphical genotype of the 16 ILs selected. Location in chromosomes including genetic position according to maps by Garcia-Mas et al. (2012) and Argyris et al. (2015), and physical position in melon genome assembly v.3.6.1, for the genetic background markers is indicated. Violet boxes indicate DUD homozygous introgressions, green boxes indicate the PS genetic background, yellow boxes indicate DUD heterozygosis and red boxes correspond to no calls (XLSX 23 kb)
Online resource 4.
GBS results. A. Genotype data for the 5 selected ILs and parents PS and DUD genotyped by GBS. B. Summary of introgressions found in the 5 selected ILs genotyped using GBS technology (only introgressions longer than 5Kbp) (XLSX 525 kb)
Online resource 5.
Mean value, standard deviation and standard error of the mean, along with ANOVA results for means comparison of both parents, PS and DUD, is shown for each trait in the two trials in which they were phenotyped along with the ILs. The asterisk (*) indicate significant difference with respect to the parent PS (XLSX 29 kb)
Online resource 6.
QTLs identified by the Dunnet’s Test in two environments in the IL collection. Abbreviated trait name, QTL name, chromosome, QTL position (cM) and flanking markers, DUD effect relative to the PS parental abçnd expressed in percentage (%) with positive/negative effects, IL carrying the introgression. Introgression position (cM) and flanking markers in the IL, and information about other introgressions in these ILs (XLSX 17 kb)
Online resource 7.
Percentage of variance explained for every trait for genotype, environment and the interaction (XLSX 12 kb)
Online resource 8
. Genes located in the introgressions detected in the set of 16 ILs related to the traits analyzed (XLSX 23 kb)
Online resource 9.
Correlation coefficient (p < 0.05) for the traits analyzed in both trials Paip16 and Paip17 (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Castro, G., Perpiñá, G., Monforte, A.J. et al. New melon introgression lines in a Piel de Sapo genetic background with desirable agronomical traits from dudaim melons. Euphytica 215, 169 (2019). https://doi.org/10.1007/s10681-019-2479-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2479-1