Abstract
Developing host resistance is an effective measure to minimize the yield losses caused by biotic stresses in crop plants. Generation of genomic resources greatly facilitates the development of resistant plants. Microsatellite markers speed up the selection procedure and introgression of resistant alleles in rice breeding programme. Candidate gene based SSR (cgSSR) markers are preferred over genomic SSR due to their tighter linkage with the trait governing loci. This study describes the identification and analysis of cgSSR markers from biotic stress responsive genes of rice. Among the selected 308 different biotic stress responsive genes of rice, 176 gene sequences were found to harbour a total of 364 SSR loci. Tri-nucleotide motif was found to be the most abundant (51.09%), followed by di- (45.05%) and tetra-nucleotide (3.84%). Intron and CDS are the two locations where most of the cgSSR loci were found, followed by 5′UTR and 3′UTR. In order to validate, polymorphism survey was done in 25 Oryza sativa genotypes using 35 cgSSR primer pairs. Among the 35 cgSSR, 27 cgSSR exhibited loci specific amplification with an average allele number of 5.66 per primer and mean PIC value of 0.226. Further, out of 27, 21 cgSSRs were found to be cross transferable to the wild species belonging to the Sativa complex; while 19 were found to be transferable to the species belonging to the Officinalis complex. The novel biotic stress-responsive cgSSR markers developed here could be used in marker-assisted introgression and pyramiding of resistant allele into elite rice cultivars from other rice germplasm as well as from wild relatives of rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L) is the second most important cereal crop after corn (Zea maize L.) in terms of production and is considered the principal food crop of the developing world (Awika, 2011). The current rate of rice yield increase is estimated at 1%, which is far behind the estimated required rate of 2.4% to double global production by 2050 in order to feed the world population (Ray et al. 2013). Moreover, the trend of declining agricultural land area and occurrence vis-á-vis resurgence of several diseases and pests of rice are the two major limiting factors to achieving world food security.
Despite of coexistence of biotic stress and plant species since their evolution, the dynamics are changing continuously and as a result managing disease and insect-pests still remain a major challenge for rice farmers around the world. Oerke (2006) reported a yield loss of about 37% exclusively due to pest infestation in rice. Yellow stem borer (Scirpophaga incertulas), gall midge (Orseolia oryzae) and brown planthopper (Nilaparvata lugens) are the major insect pests of rice adding to the menace of yield loss due to biotic stress. In addition, the rapidly changing global climate is playing a significant role in the dynamics of the crop-pest relationship. New pathogens are attacking crops and some erstwhile minor pests are also creating havoc. False smut, earlier considered as a minor disease has emerged as a major disease in almost all rice cultivating regions of the world in recent times (Nessa et al. 2015). Large areas under monoculture and the varietal uniformity caused due to the introduction of high yielding varieties have facilitated the evolution of otherwise minor pests and pathogens to major ones. Sheath blight is one such rapidly emerging disease that has become the second most significant biotic stress (after blast) of rice causing considerable yield losses after the adoption of high yielding varieties (Molla et al. 2016).
Developing genomic resources to facilitate rapid and precise selection of genotypes by plant breeders shall be highly beneficial for rapid development of resistant/tolerant varieties and combating the economic risk associated with emerging pests and diseases. Molecular marker, which were initially developed mostly to establish identity of individuals, has now emerged as one of the most enriched and useful genomic resources having tremendous impact on today’s rice breeding. It greatly facilitates and fast tracks the identification and selection procedure of target loci containing genotypes. DNA based markers are of two types, viz., nucleic acid hybridization based markers and polymerase chain reaction (PCR) based markers. PCR based markers outplayed hybridization based markers since PCR based markers need much less template DNA, are more cost-effective, and are easy to handle. Simple sequence repeat (SSR) or microsatellite is the most popular PCR based marker nowadays owing to their abundance throughout the genome, multi-allelic nature, co-dominant segregation pattern, and high reproducibility across laboratories. With the availability of whole genome sequence of rice and fast-growing knowledge of bioinformatics it has become possible to further fine-tune the technology to make it more precise and efficient. Conventional genomic SSR markers have uncertainty of linkage with the transcribed regions of the genome, while candidate gene based SSR markers (cgSSR) have better possibility of linkage with the trait governing loci (Molla et al. 2015). CgSSR markers developed from candidate gene sequence are regarded as highly valuable because of their suitability in functional diversity analysis, applicability as anchor markers for comparative mapping and evolutionary studies and of course, high transferability among related species (Varshney et al. 2002).
Since wild species are reservoir of favorable alleles of genes governing resistance to several significant biotic and abiotic stresses, markers cross transferable among related species are very useful in gene/QTL introgression studies. Among five different species complexes of Oryza, the Sativa and Officinalis complexes are most frequently used for introgression of biotic stress tolerance genes to cultivated rice. O. brachyantha, containing FF genome, is a potential donor for resistance to yellow stem borer. With the advancement in genomic resources, novel genes/QTLs for different biotic and abiotic stress tolerance/resistance may be anticipated to be identified and mined from these wild species for further utilization in breeding programs. Availability of SSR markers within those favorable genes will be highly beneficial for their direct use in marker assisted introgression, selection and breeding. Although there has been an increasing trend to identify genic SSRs from EST sequences, transcriptomes, and deep sequencing data (Thumilan et al. 2016; Feng et al. 2016; Sun et al. 2016), very few studies reported the development of cgSSRs related to defined traits (Sharma and Chauhan 2008; Babu et al. 2012; Molla et al. 2015). In a previous report, a set of salinity stress responsive cgSSR markers was identified and their ability to distinguish rice genotypes on the basis of salinity response was validated (Molla et al. 2015). However, there is not a single study describing the development of cgSSR from rice biotic stress responsive genes.
In the present study, we made an effort to develop rice biotic stress responsive candidate genes based novel microsatellite markers (cgSSRs) and to validate them in cultivated, as well as, in wild gene pool.
Materials and methods
Rice disease responsive genes and SSR mining
All biotic stresses of rice including diseases and pests were taken into account in our study for which stress responsive genes have been reported. An extensive manual literature search was performed to find all reported genes responsive to different diseases and pests of rice. In a holistic approach, all identified and cloned R genes conferring resistance to a particular rice disease/pest, genes validated through either transgenic over-expression or silencing and genes exhibiting up- or down-regulation in response to rice pests and diseases were included in the study. Locus id numbers or accession numbers for all the surveyed genes were retrieved and subsequently sequence for each locus was downloaded from either MSU database (http://rice.plantbiology.msu.edu/) or NCBI Genbank database (https://www.ncbi.nlm.nih.gov/nucco), respectively. The downloaded sequences were subjected to mine SSR using the Simple Sequence Repeat Identification Tool of GRAMENE database (Temnykh et al. 2001). Di- to penta-nucleotide motifs and a minimum repetition for 5 times were set as search parameters. All rice biotic stress responsive genes harbouring SSR motifs were physically mapped on 12 rice chromosomes using Graphical GenoTypes (GGT 2.0) software (van Berloo, 2008).
Plant material
On the basis of published reports, a total of 25 Oryza sativa genotypes which showed susceptibility, tolerance and resistance to different rice diseases and pests were selected for validating the SSR markers. Details of the genotypes selected for the study are given in Table 1. The genotypes are available in the Genebank of ICAR-National Rice Research Institute (ICAR-NRRI), Cuttack and may be obtained on request through Standard Material Transfer Agreement (SMTA) as per the names provided in the list to Director, ICAR-NRRI (http://icar-nrri.in).
Isolation and purification of genomic DNA
The seeds of 25 rice genotypes were obtained from the Genebank of National Rice Research Institute, Cuttack. After taking out the seeds from medium term storage modules, they were incubated at 50 °C for 3 days. Seeds were germinated in a germination tray filled with sterilized soil. About 100 mg fresh green leaves were harvested from each plant at 5-leaf stage and immediately ground to fine powder with liquid nitrogen in morter/pastle. DNA was isolated and purified using DNeasy Plant Mini Kit (Qiagen, Germany) following manufacturer’s protocol. The quality of the isolated DNA was assessed by 0.7% agarose gel electrophoresis. DNA concentration, 260/280 and 260/230 absorption ratios were measured by a Nanodrop spectrophotometer (Thermo Scientific, USA).
Polymerase chain reaction and polyacrylamide gel electrophoresis
Primers for PCR were designed manually from the flanking sequences of the SSR motifs. The following criteria were used for primer designing: length 20–25 bp, GC percentage 40–60, melting temperature 55–60 °C, maximum Tm difference of right and left primers 2 °C and amplicon size range 100–300 bp. PCR was performed in a gradient mastercycler (Eppendorf, Germany) with 35 SSR primer pairs using GoTaq Green Master Mix (Promega, USA). Amplification was carried out in a total volume of 10 µl reaction mixture containing 5 µl of GoTaq Green Master Mix (2X), 0.5 µl of each of the forward and reverse primer (10 µM), 1 µl of genomic DNA (25 ng/µl), 3 µl of PCR grade H2O. Annealing temperature (Ta) was optimized for each primer pair using the gradient temperature in mastercycler. Optimized PCR cycle was as follows: initial 5 min at 95 °C followed by 35 cycles of 1 min at 94 °C, 30 s at Ta (variable with primer pair), 1 min at 72 °C, with a final extension at 72 °C for 7 min.
The amplified PCR products were resolved in 6% polyacrylamide gel (non denaturing) using a vertical gel electrophoresis system (Hoefer, USA). Electrophoresis was done at a constant 100 V in 0.5XTris borate EDTA (TBE) buffer system (pH-8.0). The gel was stained with ethidium bromide and photographed in gel documentation system (Syngene, Synoptics, United Kingdom).
Allele scoring and data analysis
Alleles for each SSR markers were identified and scored manually according to Molla et al. (2015). Band size (molecular weight) was determined using alpha view software (Protein Simple, USA) based on the relative migration of GeneRuler 50 bp DNA ladder (Thermo Scientific, USA). A binary data matrix was generated by scoring the presence and absence of a particular allele as 1 and 0, respectively. Polymorphism Information Content (PIC) values for each SSR marker were determined using the formula, PIC = 1 − ΣPi2 (Pi is the frequency of an allele for locus i) as described (Botstein et al. 1980). DARwin 5.0.158 software was utilized for studying phylogenetic relationship and principal coordinates analysis (PCoA) among the 25 genotypes using the developed binary data matrix (Perrier and Jacquemoud-Collet 2006). Genetic distances between the accessions were estimated by computing Euclidean distance matrix which is subsequently utilized for constructing dendrogram following the Neighbor Joining method as described earlier (Saitou and Nei 1987). Confidence of each node was inferred by bootstrapping data for 1000 replications of the original binary data matrix.
Cross transferability analysis
There are four major species complexes in the genus Oryza, viz. Sativa complex (contains AA genome), Officinalis complex (BB, CC, BBCC, CCDD and EE genomes), Ridleyi complex (HHJJ genome) and Meyeriana complex (GG genome). There is a prominent outgroup comprised of a single species O. brachyantha (FF genome). O. schlechteri (HHKK) and O. coarctata (KKLL) belong to unclassified complexes (Brar and Khush 2018). Species belonging to Sativa complex, Officinalis complex and O. brachyantha have been used most widely in breeding for biotic stress tolerance (Ray et al. 2016, 2018, Kar et al. 2018). All eight species belonging to Sativa complex (includes two ecotypes japonica and indica of O. sativa and seven other species), nine species belonging to Officinalis complex and O. brachyantha (Table 1) were chosen to evaluate the cross species transferability of those new cgSSR markers. The leaf samples of all the wild rice were collected from the ‘Oryza Garden’ facility of ICAR-NRRI, Cuttack, India where single sample of all Oryza species are maintained for taxonomic reference. DNA isolation, PCR and diversity analysis were performed as described before for 25 cultivated genotypes except that the electrophoresis done in 3% metaphor agarose gels instead of polyacrylamide gels. Our main interest in wild rice was to check the cross species amplification rather than polymorphism survey. The multiple non-specific amplicons observed in PAGE may create difficulty to find out the actual amplicons of wild species. Two different panels were used separately for genotyping. One panel (panel 1) consisted of all six wild species belong to Sativa complex, O. glaberrima, two indica and one japonica cultivars, whereas other panel (panel 2) contained 9 species belong to Officinalis complex, one indica, one japonica cultivars and O. brachyantha as an out-group.
Result
Rice biotic stress responsive genes and the nature, frequency and distribution of biotic stress responsive cgSSR
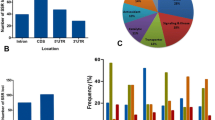
All the genes reported to be responsive to rice diseases and pests were selected and mined for SSR motifs. Fungal diseases like blast (Magnaporthe oryzae), sheath blight (Rhizoctonia solani Anastomosis Group 1-1A), brown spot (Helminthosporium oryzae), sheath rot (Sarocladium oryzae) and false smut (Ustilaginoidea virens), bacterial diseases like bacterial leaf blight (Xanthomonas oryzae oryzae) and bacterial leaf streak (Xanthomonas oryzae pv. oryzicola), virus like rice dwarf virus (RDV), rice stripe virus (RSV) and rice tungro virus, insect pests like yellow stem borer (Scirpophaga incertulas), brown plant hopper (Nilaparvata lugens), white backed plant hopper (Sogatella furcifera), gall midge (Orseolia oryzae) and aphid (Hysteroneura setariae) responsive rice genes conferring resistance/tolerance/susceptibility reported elsewhere were listed in Supplementary Table 1. Maximum numbers of genes have been reported for blast disease followed by bacterial leaf blight (BLB), rice dwarf virus (RDV) and then sheath blight. However, we could not find any study describing host genes related to sheath rot of rice. In the present study, a total of 308 gene sequences were screened for the presence of microsatellite repeats. As a result, 176 (57.14%) genes were found to contain 364 SSR loci (Supplementary Table 1). Among the motifs, most abundant class was tri-nucleotide accounting for 51.09% (186), followed by di-nucleotide (45.05%, 164) and tetra-nucleotide (3.84%, 14) (Fig. 1a). When the nature of motifs was analyzed, we found 42, 12 and 14 different kinds of tri-, di- and tetra-nucleotide repeats, respectively. The number of iterations of a given repeat unit was recorded to vary from 5 to 46. The motif CGG (Arginine), CGC (Arginine), CCG (proline) were found to be more frequent than other tri-nucleotide repeats, whereas AT and TA were most abundant among the di-nucleotide repeats (Supplementary Fig. 1). Maximum numbers of tri-nucleotide repeats were positioned in CDS (Coding DNA Sequence) (102, 54.83%), followed by 5′UTR (Untranslated region) (44, 23.65%), intron (28, 15.05%) and 3′UTR (12, 6.45), whereas di- and tetra-nucleotide repeats were mostly found in intron and least in CDS. Taking all types of repeats into consideration, highest number of SSR motifs was found to be present in intron (34.25%) followed by CDS (28.45%), 5′UTR (25.69%) and 3′UTR (11.6%) (Fig. 1b). The identified cgSSRs were found to be distributed throughout all 12 chromosomes of rice genome. With 14.08% frequency, chromosome 1 possesses the highest number of biotic stress responsive cgSSRs while chromosome 10 showed the least frequency (3.3%) of such cgSSRs (Fig. 1c). Genes containing cgSSR were mapped on twelve rice chromosomes to generate a physical map (Fig. 2).
Functional analysis of the genes containing SSR
Gene ontology analysis of the 178 genes harboring cgSSR was done to categorize them into different Gene Ontology (GO) classes (Fig. 3a). Genes were assigned to two principal categories, cellular process and metabolic process. Next two abundant classes included the genes involved in regulation of various biological processes. Response to stimulus, death and signaling GO class members are well known to play role in stress response and as expected a number of genes fall into those three GO classes. The cgSSR containing genes were also classified manually into eight different functional classes (Fig. 3b). Transcription factor, catalytic, NBS-LRR, transporter and PR genes were found to be the major classes.
Gene ontology (GO) and functional grouping of cgSSR containing genes. a Classification on the basis of GO term using agriGO v2.0 (Tian et al. 2017). Blue bars represent the GO classes of 178 biotic stress responsive genes harbouring SSR from this study and green bars are for the background GO classes of all rice genes from public databases. b Classification on the basis of specific functions. (Color figure online)
Development of cgSSR markers from biotic stress responsive genes and their validation
Primers were designed for all motifs found. For validation purpose, primers for 35 cgSSR motifs were synthesized from 30 different genes. Annealing temperature for each of these markers were standardized and allele specific amplification was performed using the genomic DNA of 25 rice genotypes to study the polymorphism. Among 35 different loci, 27 loci specific amplification were detected and 8 loci could not be amplified (Fig. 4a). A total of 153 alleles were detected along with a few rare and unique alleles. The average number of alleles was found to be 5.66 per primer. Among the polymorphic cgSSR markers, LOC_Os03g40540 (BRD1) (BLB responsive) produced highest number of alleles (15), whereas Gns1 (Blast responsive) exhibited the lowest number of alleles (2). Existence of stutter band was detected with the main bands for about 75% of the markers used. Polymorphism information content (PIC) value denotes the allelic diversity among genotypes and is a measure of a marker’s usefulness. The PIC value for cgSSR markers ranged from 0.11 to 0.40 with a computed mean of 0.226. The lowest PIC value (0.112) was obtained for the cgSSR from Gns1, whereas the cgSSR from Xa40 gene exhibited highest PIC value (0.401) (Table 2). An attempt was made to find out the relation between the SSR repeat length and PIC value. Although not strong, but a positive correlation (r = 0.3404) was found to exist between the repeat unit length and PIC value.
Representative gel profile of amplified product using biotic stress responsive cgSSR. a Amplified bands of 25 rice (O. sativa) genotypes in 6% polyacrylamide gel using BRD1 (BLB responsive) cgSSR marker, b amplified bands (using PR1A) of Panel II (one indica, one japonica, 9 species of Officinalis complex and O. brachyantha) in 3% agarose gel, c amplified bands (using BRD1) of panel 1 (two indica, one japonica and seven species belong to Sativa complex) in 3% agarose gel. M- 50 bp DNA ladder
Assessment of genetic relationship among 25 Oryza sativa genotypes using cgSSR markers
The applicability of cgSSR markers developed from biotic stress-responsive genes in rice genotyping was ascertained in 25 rice accessions. Dissimilarity analysis, factorial analysis and cluster analysis were performed to study the genetic diversity employing the data matrix generated from 27 cgSSR profiling of 25 rice genotypes. An UnWeighted neighbor-joining tree was constructed based on genetic distance matrix. The analysis revealed that the dissimilarity value among rice accessions ranges from 25 to 96%. The generated tree suggested the existence of three distinct clusters (Supplementary Fig. 2). Cluster I was the largest containing 12 genotypes, while cluster II and III contain 7 and 6 genotypes, respectively. Cluster I contained 3 sub clusters, of which IA comprised of Lemont, Rasi, ASD7, Jasmine 85, HR12 and Co39, IB contained Mudgo and PTB33, and IC comprised of 4 genotypes IRBB1, IRBB21, T12 and IR24. It is noteworthy that Mudgo and PTB33, both known for BPH resistance (Jena and Kim 2010), were grouped in the same sub cluster IB. Another two BPH tolerant genotypes Swarnalatha and Babawee exhibited similar grouping pattern in Cluster II. Similarly two popular susceptible check cultivars (HR12 and Co39) (Mahesh et al. 2016) for blast disease are clustered in the sub cluster IA.
Principal Coordinates analysis (PCoA) has been performed and it distinctly separated each genotype (Supplementary Fig. 3). In accordance with the dendrogram, the BPH tolerant genotypes Mudgo and PTB33 were in the same group. Unlike the dendrogram, another BPH tolerant genotype ASD7 also appeared in the same group with Mudgo and PTB33. In PCoA, the bacterial blight resistant IRBB1 (carrying Xa-1) and IRBB21 (carrying Xa-21) were found to be separated from the susceptible IR24. In a similar fashion, sheath blight tolerant Tetep and Tadukan (Hossain et al. 2016) were also grouped together.
Cross-species transferability of biotic stress responsive cgSSR markers
We evaluated cross transferability of the cgSSR markers, which amplified properly in different accessions of O. sativa, in all the species of Sativa and Officinalis complex beside O. brachyantha. Out of the 27 markers amplified in O. sativa, 77.77% (21) and 70.3% (19) cgSSRs were found to be transferable among the species of Sativa and Officinalis complex, respectively (Fig. 4b, c). An overall graphical representation of transferability of those cgSSR markers are represented in Fig. 5. A total of six markers could not be amplified, while 21 markers showed allele specific amplification in the Sativa complex. Out of 21 transferable markers amplified in the Sativa complex, 19 were polymorphic with a PIC value ranging from 0.165 to 0.429 (average PIC- 0.323). In the second panel comprising O. sativa (Indica and Japonica) and nine species of Officinalis complex, out of 21 cgSSR markers amplified in Sativa complex, two markers from the genes Gns1 and PR1A did not amplify in any species of Officinalis complex. Twelve markers were monomorphic out of 19 transferable cgSSR with an average PIC value of 0.294. Polymorphism of the markers in different wild species with respect to cultivated japonica and indica rice has been presented in Supplementary Table 2. The dendogram constructed from the marker profile in the two panels could not show proper phylogenetic relationship among the species (Supplementary Fig. 4 & 5).
Discussion
cgSSR markers distribution in the genomic context
Every year, rice suffers an average 37% yield loss due to various pest and diseases (Oerke, 2006). In addition, use of chemical pesticide and fungicide to control disease and pest increases the input cost and also severely deteriorate environment and farmer’s health. In order to get rid of all those problems, development of host resistance against disease and pest is the best way. Generation of more and more genomic resources is one of the essential requirements for the improvement of genetic resistance of host plant. Towards enriching genomic resources for disease and pest resistance, here we report the development of candidate gene based SSR markers (cgSSR) from pest and disease responsive genes of rice plant. Among the 308 genes, most number of genes was found to be reported for blast (167) followed by BLB (103), RDV (24) and ShB (11). This result indicates that blast and BB are the two most studied diseases of rice, whereas diseases like ShB and false smut of rice are less studied despite of their importance. Therefore, those emerging diseases are needed to be paid more attention. Although ShB is considered as the second most devastating disease of rice after blast (Molla et al. 2013), the molecular details of host–pathogen interaction for ShB are not studied as much extent as it should be. Our study reveals that Chromosome 1 and Chromosome 11 are the two which harbor most number of reported biotic stress responsive genes of rice. Chromosome 11 is believed to be rich in disease resistance gene (Rice Chromosome 11 and 12 Sequencing Consortia). In our study, SSR loci were found in 57.42% of biotic stress responsive candidate gene sequences. However, a previous study reported a lower percentage (48.18%) of salt responsive candidate genes containing microsatellite loci (Molla et al. 2015). When rice ESTs were mined for SSR loci in a previous study, only 4.7% ESTs were found to contain microsatellite motif (Kantety et al. 2002). An in silico analysis of ESTs of barley, maize, oats, rice, rye and wheat revealed the presence of microsatellite motifs in about 7–10% of totals ESTs (Varshney et al. 2002). In comparison to that, a much higher percentage of genes containing SSR were observed in our study. This might be due to the inclusion of intron sequences of candidate genes to mine SSR in our study. Among the cgSSR identified from biotic stress responsive genes, tri-nucleotide repeats were most frequent followed by di- and tetra-nucleotide repeats which is in concurrence with the previous study of salt responsive cgSSR (Molla et al. 2015). Our results also corroborate the earlier reports of tri-nucleotide repeats being the most abundant class of SSR in rice (Cho et al. 2000; Singh et al. 2010; Temnykh et al. 2001), mulberry (Thumilan et al. 2016), Dendrocalamus latiflorus (Bhandawat et al. 2016) and mungbean (Gupta et al. 2014). In contrast, di-nucleotide repeats were reported as most abundant class of genic SSR in Nelumbo nucifera (Zheng et al. 2015), rice (Parida et al. 2006) and pear buds (Yue et al. 2014). In agreement with previous studies in rice (Molla et al. 2015; Parida et al. 2009) and wheat (Yu et al. 2004), tri-nucleotide repeats were found to be located mostly in CDS (54.9%) followed by in 5′UTR (23.37%), intron (15.21) and 3′UTR (6.52) in our study. The high frequency of tri-nucleotide repeats in CDS regions might be due to selection against frameshift mutations limiting expansion of nontriplet repeats (Metzgar et al. 2000). However, in the present study, overall intron region contained the highest frequency of biotic stress responsive cgSSR followed by CDS, 5′UTR and 3′UTR which is unlike the salt responsive cgSSRs from rice where CDS was found to contain most of the repeat motifs (Molla et al. 2015). GC rich repeats like CGG (arginine coding), CGC (arginine coding), CCG (proline coding) were observed as most abundant class among tri-nucleotide repeats, whereas most abundant classes of di-nucleotide repeats were AT/TA. Abundance of arginine coding repeats was also recorded in previous studies (Molla et al. 2015; Parida et al. 2006). Genic microsatellites might play important role to control or regulate a particular trait as transcription and translation can be affected by SSR in 5′-UTRs and gain or loss of gene function via frameshift mutation can be resulted from SSR in CDS (Li et al. 2004). Similarly, SSR in 3′-UTRs can cause disruption of splicing and gene transcription, and mRNA splicing can be hampered by intronic SSR (Li et al. 2004). Therefore, the cgSSR found in the sequences of biotic stress-responsive genes can further be studied for their any possible role in controlling disease or pest response of a genotype.
cgSSR markers in genetic diversity and phylogenetic analysis
In order to validate the cgSSR markers from biotic stress responsive genes, we synthesized a set of 35 markers on the basis of the notion that polymorphism of SSR markers is directly correlated with the number of repeat units (Kayser et al. 2004). Out of the 35, 27 markers showed specific amplification when we tested them with 25 O. sativa genotypes. SSR motifs which reiterated for a minimum of 9 times have only been selected for validation purpose. The chances of obtaining polymorphism in the cgSSRs are expected to reduce significantly if the repeat motifs are reiterated lesser number of times. Those markers would not be of much practical use. Among the 27 amplified cgSSR markers, 23 markers contained dinucleotide repeat units. Dinucleotide motifs always have larger number of repeats and have a tendency to show high polymorphism (Zhang et al. 2014). Average PIC value for biotic stress responsive cgSSR primers in our study was 0.226 which is slightly lower than the previous report describing rice salt responsive cgSSR markers (Molla et al. 2015). However, SSR primers from the genomic sequences of rice (Ray et al. 2013) and Foxtail millet (Zhang et al. 2014) were reported to exhibit higher PIC values. The lower mean PIC value of biotic stress responsive cgSSRs could be attributed to the fact that genic sequences are more conserved and reveal less polymorphism than the genomic sequences (Varshney et al. 2002).
Notably, our results of clustering exhibited similar pattern of grouping of genotypes known for similar reaction against a particular disease/pest. For example, IR28, a false smut tolerant genotype (Han et al. 2015) was found to group together with another tolerant genotype Pankaj (Baruah et al. 1992) in cluster III. In a similar note, blast resistant rice variety IR64 clustered with two other blast resistant genotypes Tetep and Tadukan (Mahesh et al. 2016; Sallaud et al. 2003). However, quite a number of out-grouping or intermixing of genotypes of similar disease reaction is also observed in our study. The bacterial blight susceptible IR24 clustered with the tolerant genotypes IRBB1 and IRBB21 in Cluster I. Similarly, Jasmine 85 and Lemont, ShB tolerant and susceptible respectively (Jia et al. 2012), were found to be grouped together in Cluster I. In this regard, our result did not fully concur with the previous report on salt responsive cgSSR which showed remarkable ability to clearly distinguish salinity susceptible and tolerant rice genotypes (Molla et al. 2015). The inability of biotic stress responsive cgSSRs to clearly distinguish all susceptible genotypes from tolerant/resistant genotypes for a particular disease might be due to the inclusion of cgSSRs from all disease/pest responsive genes. The genes play positive role in a disease might play different role in other diseases. In order to get an overview, for genotyping purpose, we had selected resistant/tolerant and susceptible genotypes for many diseases (Blast, ShB, Blast, False Smut) and pests (BPH). It is obvious that no particular genotype can show similar kind of reaction (susceptibility/tolerance) against all pest/diseases. These factors probably played role for the outgrouping and intermixing of genotypes. However, this was not the case for genotyping by salt responsive cgSSR markers as the study included a single stress response, i.e. salinity (Molla et al. 2015).
cgSSR markers are highly cross-transferable
Markers that are readily transferable between and among species are called cross-species or trans-species transferrable markers and are in high demand. The degree of cross transferability of a marker is the determining factor for their suitability in comparative genome mapping, phylogenetics (Dillon et al. 2014) and gene introgression studies. Wild relatives of cultivated rice are considered as valuable to the modern breeders as they harbour valuable characteristics including several biotic stresses to improve cultivated rice (Mondal and Henry 2018). We examined the extent of transferability of the developed cgSSR markers in six and nine different wild species belong to Sativa and Officinalis complex, respectively and also in O. glaberrima and O. brachyantha. On an average, our study revealed high transferability (77.77%) of cgSSR markers to the wild members of Sativa complex, whereas a slightly lower percentage (70.37%) of transferability to the members of Officinalis complex was observed possibly due to of being other than AA genomes. The second set was constituted with nine species belonging to Officinalis complex of the genus Oryza besides O. brachyantha all of which belong to secondary and tertiary gene pool of rice and require special techniques like embryo rescue to recover the F1 progenies on crossing with O. sativa. O. sativa (both var. japonica and indica) was used as control in the second panel. Among the members of the second panel, genome sequence information is available only in case of O. punctata of Officinalis complex and O. brachyantha (Goicoechea et al. 2010). Insilico cross transferability and expected amplicon size prediction was not possible for all the members of this panel except for two. Direct checking of amplification by polymerase chain reaction was the only available option. A high percentage of transferability of cgSSRs in our study may be attributed to the more conserved nature of biotic stress responsive candidate genes across the species, even belonging to different species complexes. In a previous study, Ray et al. (2016) reported a much lower overall cross transferability (0.75%) of O. sativa RM series STMS (Sequence tagged microsatellite site) markers to O. brachyantha, a FF genome containing species. However, they also found the presence of much higher proportion (74.58%) of genic STMS markers among the cross-transferred markers (Ray et al. 2016). In a similar fashion, 16.67% of RM markers were reported to be transferable in 28 accessions of 16 Oryza species belong to Sativa and Officinalis complexes (Brondani et al. 2003). However, a higher percentage of transferability of O. sativa markers to O. rufipogon (AA), O. officinalis (CC) and O. granulata (GG) was observed in a previous study (Gao et al. 2005). For inferring the genetic relationship among the wild species using the cgSSR primers, we constructed separate dendrograms for Sativa and Officinalis complex. However, in case of both the panel I and II, constructed dendrograms did not exhibit proper phylogenetic relationship among different species of Sativa and Officinalis complex. As alleles can co-migrate as a result of orthology (identical by descent) as well as paralogy (identical in state) it is sometimes controversial to infer phylogeny using molecular markers (Brondani et al. 2003). This inability of cgSSR to show proper phylogenetic relationship may be attributed to the fact that those cgSSR were developed from continuously evolving disease responsive gene sequences. Therefore, they are more likely to establish relationship on the basis of disease response rather than phylogeny. However, it is interesting to note that in case of cgSSR of PGIP, except O. rufipogon, all the other members of Sativa complex were polymorphic with respect to O. sativa japonica and indica, while all members of Officinalis complex were monomorphic.
The utility of biotic stress-responsive cgSSR markers
As it is not possible to ensure whether the monomorphism/polymorphism for a particular SSR marker between any two individuals aroused due to “identical/dissimilar by descent” or “identical/dissimilar by type” phenomenon, we suggest to avoid these markers to draw any phylogenetic relationship of genotypes or species. However, the markers are expected to serve as useful resource for marker assisted breeding. There is scope to test the markers in rice collection(s) or mapping population(s) segregating for the specific biotic stress response. However, it must be noted that the cgSSRs reported here can’t directly be validated in any population/segregating populations showing resistance and susceptibility for a particular disease or insect. There are multiple genes for resistance to a particular disease or insect (for example Xa genes for Bacterial Blight or BPH genes for brown plant hopper resistance). A particular cgSSR marker for a particular gene can be used only in two genotypes differing for alleles of one particular resistance gene (i.e. gene differentials), while alleles of other genes for resistance to same stress should be constant. As we dealt with cgSSR for multiple genes in this study, obtaining such a large number of host-differentials and/or developing such large numbers of populations was beyond the scope of our present study as different genes from which cgSSR markers were developed have been reported by diverse research groups worldwide. However, with the information of these cgSSRs being available in public domain, it will be possible by different research groups having those populations at their disposal to validate the markers as well as use those in their resistance breeding programme. Majority of the cgSSR markers also showed amplification and polymorphism in different wild species which are traditionally known for the presence of strong alleles for genes of biotic stress resistance. The most effective dominant resistance gene for bacterial blight resistance (Xa21) identified from O. longistaminata is one such classical example (Khush et al. 1990). Similarly, large number of genes for BPH resistance have been cloned from O. officinalis complex by researchers worldwide (Ray et al. 2018). If allele mining and phenotyping can identify such favourable alleles from the wild gene pool or even cultivated gene pools for the genes from where we could find out polymorphic cgSSRs markers, they will have direct utility in tracking the inheritance of those stress-responsive genes due to their almost absolute linkage with those genes. It must be noted that most of the markers were polymorphic between the cultivated species and wild rice making them suitable for use during wide hybridization and generation advancement. With species of Sativa complex, the markers will be useful for foreground selection of target genes during development of introgression lines or subsequent transfer in multiple genotypes. For species of Officinalis complex and O. brachyantha, Monosomic Alien Addition Lines (MAALs) are also developed in the background of cultivated rice. In such cases, the 19 cross transferable cgSSR markers distributed over 8 rice chromosomes will be helpful for tracking the particular chromosomes in MAALs both during development as well as chacracterization. A similar strategy earlier have been successfully used in O. brachyantha derived MAALs (Ray et al. 2016).
SSR markers are chosen based on their monomorphic or polymorphic nature in two genotypes contrasting for a trait of interest where the markers are closely linked to the genes responsible for contrasting phenotypes. The cgSSRs being located within the gene of interest are very useful, if they are polymorphic in contrasting parents where resistance/susceptibility is due to genes within which the cgSSRs are located. As the genes chosen by us are already reported, cloned, characterized & validated for biotic stress resistance; identification of markers within those genes is expected to serve as useful resource for breeders. Despite of significant advancements in marker technologies, it is still a long way to go for finding out effective functional markers for many of the agronomically important genes. Until the functional markers for those genes are available, cgSSR remains a better alternative.
Conclusion
The novel cgSSR markers developed and validated in the study can be a valuable tool to ascertain the functional diversity at a particular locus. As the developed cgSSR markers have high potential of linkage, these markers could also be utilized in marker assisted pyramiding of disease responsive genes for rice resistance breeding. As evidenced from our study, those biotic stress responsive cgSSR are highly transferable to wild species which could serve as valuable resources to introgress resistant allele into cultivated rice (O. sativa) from wild relatives. However, we do not recommend to use these cgSSR markers for deciphering phylogeny as these are developed from the genes which confer adaptability traits under stress conditions and hence been adopted by the plants as a process of convergent evolution.
Abbreviations
- SSR:
-
Simple sequence repeat
- STMS:
-
Sequence tagged microsatellite site
- cgSSR:
-
Candidate gene based SSR
- UTR:
-
Untranslated region
- PIC:
-
Polymorphism information content
- QTL:
-
Quantitative trait loci
- EST:
-
Expressed sequence tag
- CDS:
-
Coding DNA sequence
- PCR:
-
Polymerase chain reaction
- Ta:
-
Annealing temperature
- EDTA:
-
Ethylenediaminetetraacetic acid
- PCoA:
-
Principal coordinates analysis
- BLB:
-
Bacterial leaf blight
- RDV:
-
Rice dwarf virus
- RSV:
-
Rice stripe virus
- ShB:
-
Sheath blight
- GO:
-
Gene ontology
References
Awika JM (2011) Major Cereal Grains Production and Use around the World thousands of years. Publication Date (Web): 30 Nov 2011
Babu BK, Agrawal PK, Gupta HS, Kumar A, Bhatt JC (2012) Identification of candidate gene–based SSR markers for lysine and tryptophan metabolic pathways in maize (Zea mays). Plant Breed 131:20–27
Baruah B, Senapoty D, Ali M (1992) False smut: a threat to rice growers in Assam. Indian J Mycol Plant Pathol 22:274–277
Bhandawat A, Singh G, Raina AS, Kaur J, Sharma RK (2016) Development of genic SSR marker resource from RNA-Seq data in Dendrocalamus latiflorus. J Plant Biochem Biotechnol 25:179–190
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314
Brar DS, Khush GS (2018) Wild relatives of rice: a valuable genetic resource for genomics and breeding research. In: Mondal TK, Henry RJ (eds) The Wild Oryza genomes. Springer, Cham, pp 1–25
Brondani C, Rangel PHN, Borba TCO, Brondani RPV (2003) Transferability of microsatellite and sequence tagged site markers in Oryza species. Hereditas 138:187–192
Cho YG et al (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Dillon NL, Innes DJ, Bally IS, Wright CL, Devitt LC, Dietzgen RG (2014) Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Mangifera indica L.). Diversity 6:72–87
Feng S et al (2016) Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front Genet. https://doi.org/10.3389/fgene.2016.00113
Gao L, Zhang C, Jia J (2005) Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol 52:931–940
Goicoechea JL, Ammiraju JSS, Marri PR, Chen M, Jackson S, Yu Y, Rounsley S, Wing RA (2010) The future of rice genomics: sequencing the collective Oryza genome. Rice 3(2–3):89–97
Gupta S, Bansal R, Gopalakrishna T (2014) Development and characterization of genic SSR markers for mungbean (Vigna radiata (L.) Wilczek). Euphytica 195:245–258
Han Y et al (2015) Differential expression profiling of the early response to Ustilaginoidea virens between false smut resistant and susceptible rice varieties. BMC Genom 16:955
Hossain MK, Jena KK, Bhuiyan MAR, Wickneswari R (2016) Association between QTLs and morphological traits toward sheath blight resistance in rice (Oryza sativa L.). Breed Sci 66(4):613–626
Jena KK, Kim S-M (2010) Current status of brown planthopper (BPH) resistance and genetics. Rice 3:161–171
Jia L et al (2012) Allelic analysis of sheath blight resistance with association mapping in rice. PLoS ONE 7:e32703
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kar MK, Bose LK, Cjhakraborti M., Azharudheen M., Ray S, Sarkar S, Dash SK, Reddy JN, Pani DR, Jena M, Mukherjee AK, Lenka S, Mohapatra SD, Jambhulkar NN (2018) Utilization of cultivated and wild Gene pools of rice for resistance to biotic stresses. In: Rice research for enhancing productivity, profitability and climate resilience, pp 52–72. ICAR-National Rice Research Institute, Cuttack, India. ISBN: 81-88409-04-9
Kayser M et al (2004) A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet 74:1183–1197
Khush GS, Bacalangco E, Ogawa T (1990) 18. A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet News Lett 7:121–122
Li Y-C, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Mahesh H, Shirke MD, Singh S, Rajamani A, Hittalmani S, Wang G-L, Gowda M (2016) Indica rice genome assembly, annotation and mining of blast disease resistance genes. BMC Genom 17:242
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Molla KA, Karmakar S, Chanda PK, Ghosh S, Sarkar SN, Datta SK, Datta K (2013) Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol Plant Pathol 14:910–922
Molla KA, Debnath AB, Ganie SA, Mondal TK (2015) Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L.). BMC Plant Biol 15:122
Molla KA, Karmakar S, Chanda PK, Sarkar SN, Datta SK, Datta K (2016) Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci 250:105–114
Mondal TK, Henry RJ (eds) (2018) The Wild Oryza genomes. Springer, Berlin
Nessa B et al (2015) Spatial pattern of natural spread of rice false smut (Ustilaginoidea virens) disease in fields. Am J Agric Biol Sci 10:63
Oerke E-C (2006) Crop losses to pests. J Agric Sci 144:31–43
Parida SK, Kumar KAR, Dalal V, Singh NK, Mohapatra T (2006) Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet 112:808–817
Parida SK, Dalal V, Singh AK, Singh NK, Mohapatra T (2009) Genic non-coding microsatellites in the rice genome: characterization, marker design and use in assessing genetic and evolutionary relationships among domesticated groups. BMC Genom 10:140
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428
Ray S et al (2016) Development and validation of cross-transferable and polymorphic DNA markers for detecting alien genome introgression in Oryza sativa from Oryza brachyantha. Mol Genet Genom 291:1783–1794. https://doi.org/10.1007/s00438-016-1214-z
Ray S, Vijayan J, Chakraborti M, Sarkar S, Bose LK, Singh ON (2018) Oryza officinalis complex. In: Mondal TK, Henry RJ (eds) The Wild Oryza genomes. Springer, Cham, pp 239–258
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sallaud C et al (2003) Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor Appl Genet 106:794–803
Sharma A, Chauhan R (2008) Identification of candidate gene-based markers (SNPs and SSRs) in the zinc and iron transporter sequences of maize (Zea mays L). Curr Sci 95(8):1051–1059
Singh H et al (2010) Highly variable SSR markers suitable for rice genotyping using agarose gels. Mol Breed 25:359–364
Sun R, Lin F, Huang P, Zheng Y (2016) Moderate genetic diversity and genetic differentiation in the relict tree Liquidambar formosana Hance revealed by genic simple sequence repeat markers. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01411
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thumilan BM, Sajeevan R, Biradar J, Madhuri T, Nataraja KN, Sreeman SM (2016) Development and characterization of genic SSR markers from Indian mulberry transcriptome and their transferability to related species of Moraceae. PLoS ONE 11:e0162909
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res 45(W1):W122–W129
van Berloo R (2008) GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered 99(2):232–236
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Yue X-y, Liu G-q, Zong Y, Teng Y-w, Cai D-y (2014) Development of genic SSR markers from transcriptome sequencing of pear buds. J Zhejiang Univ Sci B 15:303
Zhang S et al (2014) Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genom 15:78
Zheng X et al (2015) Development and characterization of genic-SSR markers from different Asia lotus (Nelumbo nucifera) types by RNA-seq. Genet Mol Res 14:11171–11184
Acknowledgements
Authors are thankful to the Director, ICAR-National Rice Research Institute for encouragement and providing all facilities to carry out research work. Thanks are due to the Genebank of ICAR-NRRI, Cuttack for providing the germplasm.
Funding
The work was funded by Indian Council of Agricultural Research (ICAR), New Delhi. The Funding body has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
KAM conceptualized, designed and planned the study. KAM, TPMA, SR, AS and SS performed the experiments. KAM, TPMA, SR, SS, MC, JV, ONS, MJB and AKM analysed the data. KAM wrote the manuscript. TPMA, MC, SR and SS edited the manuscript. JV, ONS, MJB and AKM critically revised the manuscript. KAM, TPMA, SR and AS are responsible for preparing different tables and figures. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article and in additional files (Additional file 1-6).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Molla, K.A., Azharudheen, T.P.M., Ray, S. et al. Novel biotic stress responsive candidate gene based SSR (cgSSR) markers from rice. Euphytica 215, 17 (2019). https://doi.org/10.1007/s10681-018-2329-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2329-6