Abstract
This study sought to identify new sources of resistance to cowpea aphids (CPA) using molecular and phenotypic approaches and the inheritance pattern. Sixty cowpea genotypes were phenotyped for resistance to CPA using insect proof cages and further confirmed using markers linked to aphid resistance. Result revealed that among the cowpea genotypes, TVu 2897 and TVNu 1158 supported lowest number of aphids and plant damage scores. The seedlings of these genotypes also had high level of survival rates and were completely healthy with normal growth. This indicates that these genotypes are resistant to aphid attacks. However, the resistance in TVNu 1158 did not seem strong compared to the genotype TVU 2897 that was confirmed to be resistant to multiple aphid biotypes. The mechanism of resistance in TVu 2897 and TVNu 1158 were expressed as a hypersensitive response at the site of infestation on the leaves. The other genotypes especially Aloka local and keffi local supported the highest number of aphids, damage score and low level of survival rate, suggesting that they are susceptible to aphid attack. The cowpea genotype IT84S-224-6 previously reported to be resistant to aphids supported high number of aphids and was marked by stunted growth and high mortality rate. Molecular and phenotypic screening revealed that TVu-2876 has a strong resistance to cowpea aphid and should be a good source of resistance gene that can be used in breeding to develop new aphid resistant cowpea cultivars. Although, the results of phenotypic tests and molecular marker detection agreed in most cases, molecular markers detection was found more reliable in identifying genotypes for resistance to CPA. The segregation in F2 and BC1 populations derived from the cross between TVNu 2876 and Keffi local indicated that resistance to cowpea aphids in TVu-2876 is controlled by a single dominant gene. Allelism test revealed that resistance gene in TVNu 2876 is non-allelic with the gene that confers resistance in SARC 1-57-2 and TVNu 1158.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea [Vigna unguiculata (L.) Walp.] is one of the most important food and forage legume crops in subtropical and tropical regions of the world, primarily in sub-Saharan Africa. It is a versatile crop cultivated between Latitude 35°N–30°S of the equator, covering Asia, Africa, southern Europe and some parts of southern America (Bata et al.1987). Cowpea is grown on 11.32 million ha worldwide, with an annual grain production of about 5.72 million tons (FAO 2014). Of this amount, Africa accounts for 94% of grain production. Nigeria is the largest cowpea producer in the world and accounts for over 2.5 million tons grain production from an estimated 4.9 million ha (FAO 2014). It is mainly cultivated for the seeds; however, the sale of the fodder as animal feed during the dry season also provides vital income for farmers (IITA 2009). More importantly, the unique ability of cowpea to fix nitrogen even in poor soils makes it compatible as an intercrop with cereals and root crops. Despite the importance of the crop, the productivity of cowpea in sub-Saharan Africa is low, less than 500 kg/ha due to a number of factors such as old varieties and poor agronomic practices, insect pests, diseases and parasitic weeds. Insect pests have been shown to constitute a serious setback in the cultivation of cowpea especially in the drier regions of the tropics. Every stage in the life cycle of cowpea has at least one major insect pest that can cause serious damage and impact yield negatively (Fatokun 2002).
The cowpea aphid (Aphis Craccivora Koch.) has been described as one of the most important pest of cowpea [Vigna unguiculata (L.) Walp.], causing significant yield losses when either young seedlings or the pods of adult plants are attacked (Annan et al. 1996). Cowpea aphid is a sap-sucking insect pest of cowpea (Gunilla 1985; Singh et al. 1990). It causes damage in susceptible cultivars, directly by modifying plant metabolism and ingesting plant nutrients and in many cases indirectly, through the transmission of plant-pathogenic viruses (Blackman and Eastop 2000). In addition to sucking sap from the young leaves stem tissues and pods, Aphids also act as vectors in the transmission of cowpea aphids-borne mosaic virus even at a low population density (Atiri et al. 1986; Kitch et al. 1999). At high infestation levels, honeydew released by CPA can block plant respiration and stimulate development of black mold, thereby reducing photosynthesis. Cowpea aphids are well distributed throughout the tropics, colony expand very quickly in hot and dry weather and have numerous hosts but primarily on legumes. Cowpea aphids produce eggs that develop within the adult aphid which gives birth to nymphs alive. Nymphs mature into reproductive adults between 2 and 3 days causing a high severity of infestation as their population density increase rapidly (Schreiner 2000). Aphids primarily attack young cowpea seedling, however large populations also infest flower buds, flowers and pods, and cause direct damage to the plants by sucking its sap (Singh et al. 1996). Little damage may be seen on plant with small population of the insect. However, large population which results in heavy feeding kills young plant through the distortion of leaves, stunting of plant, thus affecting the overall plant vigour. There is also the delay in flower initiation, significant reduction in the nodulation of the root system as well as reduced pod set in plants which survive attack (Singh et al. 1996).
A number of aphid control measures (cultural, chemical and bio-control measures) have been suggested for the control of A. craccivora to prevent its impact on cowpea yield and spread of different viruses but are of limited value to subsistence farmers. Among the control options, the use of resistant varieties appears to be more viable and economical for resource poor farmers. Biological control alone is not adequate because natural enemies often appear when CPA infestation is already high and causing serious damage. Applying pesticides early in the season prevents CPA infestation and colonization but beneficial insects can be destroyed, leading to outbreaks of other insect pests. In fact, pesticide application is not a common practice in low-input farming systems in Africa (Souleymane et al. 2013) because of high cost and unavailability. Improving cultivars by adding in resistance genes through breeding promises a sustainable strategy for aphid control not only in cowpea but also in many other crop species (Huynh et al. 2013; Smith and Chuang 2014). The cumulative effect of several different mechanisms often provides effective resistance to insect by deterring, delaying or tolerating attack, feeding and reproduction. Even if the cumulative phenomena of resistance are not inherited together, individual mechanisms are often simply inherited and can be transferred in a stepwise manner into susceptible varieties (Bidinger 1992).
Many cowpea cultivars grown in the West African sub-region are susceptible to CPA and require pesticide treatments during early vegetative and flowering stages. Breeding resistant cowpea cultivars must rely on African cowpea genotypes that can act as aphid resistant donors (Hall et al. 2003). To successfully breed for resistance to cowpea aphids; suitable sources of resistance are a basic requirement. Resistance sources can be obtained from cultivated varieties, germplasm collection, wild species as well as by induced mutation. Sources of resistance to the cowpea aphids have been identified in some varieties of cowpea. However, the problem of biotype occurrence still persists as most resistance has broken-down. This is because high level resistance found in some wide relative and cultivated genotypes are conditioned by single dominant gene, which is not durable. For example, in Nigeria, there have been reports of breakdown of aphid resistance in previously resistant cowpea cultivars (Singh personal communication). This, therefore, call for the need to conduct screening to identify new sources of resistance to the cowpea-aphids.
Previous cowpea screening for aphid resistance was done by visual rating and counts which is difficult, complex, and often unreliable. There has been instance in which some lines that were earlier found to be resistant based on phenotyping in one location turned out to be susceptible in another location. The recent availability of molecular markers that are linked to aphid resistance can improve efficiency and precision in selecting cowpea genotypes for aphid resistance (Mayers et al. 1996). In the past, molecular markers have been used in cowpea to identify traits such as Striga resistance (Omoigui et al. 2012, 2016). However, progress in utilizing molecular markers associated with aphid has been limited due to the lack of reliable marker for the trait. Fortunately, recent advances in crop genomics have facilitated the identification of molecular markers associated with aphid resistance (Kusi et al. 2010). Integration of molecular markers in aphid resistance screening will greatly enhance the selection efficiency since field screening is difficult, complex, and often unreliable.
Aliyu and Ishiyaku (2013) reported differential reaction among seven (7) cowpea genotypes screened for resistance to cowpea aphids using conventional method. The differential reaction was observed in fecundity, larval development, adult longevity as well as the intrinsic rate of increase and multiplication of aphid. In a similar study, Obopile and Ositile (2010), reported differences in aphid fecundity among resistant and susceptible cowpea varieties. Laamari et al. (2008) reported antibiosis-mediated resistance to aphids on some landraces of broad bean, which was evidently observed in reduced fertility, multiplication rate and duration of reproductive life as well as in the observed longer duration of larval development. Kamphuis et al. (2012) reported the involvement of antibiosis, antixenosis and tolerance to CPA resistance in accessions of Barrel clover Medicago truncatula. In testing cowpea for resistance to aphid, Souleymane et al. (2013) screened some cowpea lines for aphid resistance and reported resistance by tolerance in TVNu-1158 (a wild cowpea line) compared to the cultivated lines. High flavonoid content has also been reported to be responsible for aphid resistance in cowpea (Cardinali et al. 1995) which is mediated by gene. This therefore, suggests the possibility of breeding resistant cowpea varieties against aphid attack. The different methodologies developed in screening cowpea germplasm and breeding lines, have provided the building blocks for the development of resistant varieties to insect pests (Singh et al. 1996). Some of the known resistant cowpea lines and germplasm have been found to be susceptible (Timko personal communication) either due to different biotypes or breakdown in aphid resistance or due to the approach used for screening for resistance to aphid attack. Most researchers (references) have used the conventional phenotyping approach to screen cowpea genotypes for resistance to aphis attack which may not be efficient. Another possible source for the different biotype could be the unrestricted movement of research materials from one part of the country to another and beyond.
There is the need to screen the previously reported resistant germplasm lines alongside new collections to revalidate resistant status and identify strong sources of resistance to aphids that will serve as resistant donors in breeding programs. Although phenotypic screening has been used in the past in selecting cowpea for aphid resistance (Souleymane et al. 2013; Ofuya 1993), it is a difficult process and often unreliable because of the strong influence of the environment on its expression. However, selection efficiency can be greatly improved by integrating molecular markers with the phenotyping screening for precision in selecting for aphid resistance.
Over the last 8 years many efficient markers for aphid resistance genes have been described. Kusi et al. (2010) reported CP171/172 a PCR based marker linked to aphid resistance while Huynh et al. (2015) reported a SNP marker for aphid resistance in cowpea. Some of these markers are being used in marker-assisted selection (MAS) and to identify resistance genes in varieties and lines where the genetic background is unknown.
In this study, we screened cowpea germplasm collections to identify new sources of resistance to aphid infestation in cowpea using phenotypic data collected from germplasm materials grown under aphid kept in insect-proof cages with fine saran mesh and molecular markers validation. Gene-associated PCR markers (Kusi et al. 2010) and newly identified marker (unpublished data) were used to distinguish between resistant and susceptible genotypes.
Materials and methods
The experiment was conducted at the University of Agriculture, Makurdi, Benue State, Nigeria. The cowpea genotypes used for this study consisted of 60 cowpea genotypes obtained from the Genetic Resources Unit (GRU) of the International Institute of Tropical Agriculture (IITA) and the Molecular Biology Laboratory of the University of Agriculture Makurdi (UAM) (Table 1). The experiment was carried out during the months of April and June 2014 and 2015. Temperature profile at this time ranges between 23 and 30 °C with daily relative humidity ranging between 65 and 70%. The climatic profile was collected from our nearby weather station. The cowpea aphids used in this study were collected from Ukange Local Government area of Benue State. The aphid culture was maintained on a highly susceptible cowpea cultivar, TVx-3236 planted in an insect proof cage as previously described (Souleymane et al. 2013).
Experimental design
The experiment was laid out in completely randomized design (CRD) with three replications. Cowpea seeds were planted in wooden trays measuring 57.5 cm × 37.5 cm × 14.5 cm filled with top soil up to 10 cm depth and kept in insect-proof cages with fine saran mesh, small enough to allow passage of air but not insects. The seeds of each genotype were planted in single row, comprising five rows per tray, spaced 10 cm apart, while the distance between plant stands was kept at 5 cm. The plant growth conditions were as described previously by Bata et al. (1987), Githiri et al. (1996) and Kusi et al. (2010). Five 4th-instar nymphs (apterous adult) aphids were transferred on each plant at seven (7) days after planting using a camel’s hair brush to reduce mechanical injury on the insect. Five plants each, of the genotypes were maintained per row. The plants were irrigated as at when necessary. The trays remained in the insect-proof cages for 21 days after which the plants were assessed for damage by aphids. Dead plants were regarded as susceptible while those still alive, with first trifoliate leaves developing, as resistant.
Counting of aphid and scoring for the aphid population per plant were used in screening of cowpea genotypes during the 21-day screening. Aphids per plant were counted at 5, 9, 13, 17 and 21 days after infestation. The aphid population build-up on each plant and the survival rate of CPA was measured at 21 days after infestation using a 1–5 rating scale based on visual and inspection counts and its symptoms given by Souleymane et al. (2013) and El-Defrawi and Bishara (1992) (Table 2).
Data collection
The following data were collected to assess the cowpea genotypes for resistance and susceptibility:
-
number of aphids on individual plants was counted at 5, 9, 13, 17, and 21 days after infesting seedlings with aphids.
-
visual score damages on each plant were recorded on 7 and 14 days after infestation.
-
the survival rate of CPA was measured at 21 days after infestation.
-
aphid population pressure on each plant was weighed using a 1–5 rating scale (1 = a few individual aphids, 2 = few small individual colonies, 3 = several small colonies, 4 = large individual colonies, 5 = large continuous colonies) taken at interval of 3, 6, 9 and 12 days after infestation.
-
Total plant damage rating on scales 1–5 was measured at 16 days after aphid infestation, when aphid damage caused distinct phenotypic variation among cowpea plants based on crown damage and the extent of aphid occurrence applied to more than 60% of plants in each row number of dead plants was counted at 21 days after infestation. That is, seedlings killed or severely damaged by aphid infestation were regarded as susceptible while those still alive, with first trifoliate leaves developing, as resistant (Souleymane et al. 2013; Huynh et al. 2015).
Molecular marker analysis
Following insect cage proof phenotyping, DNA analysis was carried out to validate phenotypic data. For PCR assays, two SSR markers (CP171/172 and KAD61) previously reported to be linked to aphid resistance were used (Kusi et al. 2010; unpublished data). KAD16 has previously been validated in F2 segregating populations derived from the cross between TVNu 2876 × Keffi local. KAD61 mapped 7.0 cm from the aphid resistance gene. Genomic DNA extracted from young leaves of parental plants was made ready for PCR as described previously (Omoigui et al. (2012). The sequences of the used primers and size fragment are present in Table 3.
DNA extraction
DNA was extracted from young leaf tissues of 14-day old plantlets from 60 cowpea germplasm and stored at −20 °C till DNA extraction. The genomic DNA was extracted using the CTAB extraction protocol (Doyle and Doyle 1987). Leaf samples collected in liquid Nitrogen were ground using the tissue-lyser until a fine powder was obtained. Pre-warmed 950 µl of C-TAB buffer and 2 µl of 2-mercaptoethanol was added into each sample before incubating at 60 °C for 30 min. The solution was mixed by inverting tubes gently at intervals of 10 min. 700 µl of 24:1 chloroform: isoamyl alcohol was added into the solution and incubated for 5 min at room temperature. The mixture was then centrifuged for 10 min at 7500 rpm, after which the supernatant was carefully transferred into newly labeled 1.5 ml tubes. 500 µl of ice cold isopropanol was added to the transferred supernatant to obtain a white precipitate and then stored at 20 °C for 30 min. The precipitate was then centrifuge for 20 min at 10,000 rpm to pelletize the DNA. To dissolve the DNA, 1× TE buffer was added to the pellet and stored at 4 °C. After dissolution, the extracted DNA was treated with RNase by incubating at 37 °C for 45 min. The concentrate DNA extract was stored at −20 °C until required.
Polymerase chain reaction (PCR) was performed using the Eppendorf master cycler gradient thermocycler in a total volume of 25 µl containing 2.5 µl 10× PCR buffer, 1.5 mM MgCl2−, 0.5 µl dNTP’s, 0.2 µl Taq polymerase, 18 µl of distilled water, 1 µl of each primer and about 50 ng of template DNA. Amplifications were performed at 94 °C for 10 min, followed by 40 cycles of 94 °C for 25 s, 55 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR products were separated in 2% agarose gel pre-stained with ethidium bromide and visualized using UV transilluminator. The image was photographed and gel images were used for scoring.
Data analysis
Data collected were analysed using descriptive statistics. Average number of aphids per genotype was calculated and means level of infestation scores of each genotype was determined. The count data collected was transformed using logarithm transformation. Data was subjected to analysis of variance (ANOVA) using the general linear model statistical procedures with the SAS system for Windows (SAS Institute 2014). Treatment means separation was done using the Student–Newman–Keuls (SNK) method.
Based on the result of screening cowpea germplasm for resistance/susceptibility, the mode of inheritance of the gene in TVu-2876 was determined using two genetically divergent parents that show clear differences for aphid resistance and susceptibility. Resistant germplasm (TVNu 2876) was crossed with the susceptible germplasm (Keffi local) to generate F1. The seeds of the F1 were selfed in the screenhouse to generate F2 segregating population, while some were used in a backcross scheme to raise BC1F1 population for the inheritance study.
To test the allelic relationship among TVNu 2876 an aphid resistant germplasm identified in the study location, TVNu 1158 and F-Ghana (SARC 1-57-2) earlier reported to be resistant to aphid in Ghana, segregation ratios for each resistant × resistant (TVNu 2876 by F-Ghana (SARC 1-57-2) and TVNu 1158) progenies were computed. Genetic hypotheses were tested for significance using the Chi squared goodness-of-fit test to determine the deviation of observed frequencies from the hypothesized ratios.
Results and discussion
There were significant (P < 0. 01) differences among the cowpea genotypes for insect population pressure (PPI), plant damage (PTDMG), and number of dead plant (NDP), (Table 4). There was no genotype × year interaction effect for all the parameters measured, indicating that all the genotypes behaved the same in the different years. The rate of aphid population build-up on seedlings was very rapid and the plants were fully colonized within 6 and 9 days after infestation. Most of the genotypes showed severe stunting and wilting, and damage symptoms appearing as yellow patches or leaf chlorosis surrounding the aphid infestation sites within 15 days after infestation, with the exception of TVNu 2876. The cowpea genotypes, Aloka local and Keffi local had the highest number of aphids at 3, 6, 9 and 12 days after infestation based on the insect population pressure scored at these time intervals with mean scores of 4.5 and 4.6, respectively. The cowpea genotypes, TVNu 2876, TVNu 1158 and Sierra-Leone local had aphid population pressure of 2.2, 1.5 and 2.7, respectively that were significantly lower than other genotypes (Table 5). There were no significant differences among the genotypes for PP3DAI, PP6DA, and PP9DAI. However, TVNu 2876 had a strong resistance compared to the other two genotypes.
The strong differences in population pressure observed between susceptible and resistant cowpea genotypes tested corroborated the findings of Souleymane et al. (2013) who reported a rapid multiplication and full colonization on susceptible plants within 7 to 10 days after infestation. The aphid build up observed between 3 and 6 days after infestation for all the genotypes evaluated suggests that young plants are unable to elicit their resistance to prevent reproduction of aphid at the early stage of plant growth. However, the aphids’ populations die out on the resistant plants as they grew older suggesting antibiosis resistance mechanism. This observation supports the findings of Alabi et al. (2012) who reported that all resistant and susceptible cowpea genotypes cluster in the same group with respect to the overall aphid build up in a free-choice and no-choice tests at seedling stage. Similarly, antibiosis resistance has been reported to result in increased mortality or reduced longevity and reproduction of the insect (Teetes 2007). However, significant difference in population pressure was observed at twelve 12 days after infestation. This difference in time interval may be attributed to the innate ability of the plant to illicit the production of plant resistant chemicals (antibiosis) in response to aphid attack, a condition commonly referred to as hypersensitive reaction.
Although, aphid multiplication rate did not differ significantly in the evaluated genotypes at the early stage after infestation (within 3–6 days after infestation), the differential response of genotypes indicated by aphid population pressure at 12 days after infestation suggests an increase in fecundity. Fecundity increase was highest on TVu-801 with a mean score of 4.75 and lowest on TVNu-2876 with a mean score of 1.5 (Table 5).
Significant difference was also observed for a number of dead plants recorded at 16 days after infestation (Table 3). Keffi local had the highest mean number of dead plants followed by TVu 134 in comparison with TVNu 1158, TVNu 2876 and Sierra-Leone local that recorded high percentage survival rate (Table 6).
Ofuya (1993), also reported significant differences in number of aphids on susceptible and resistant varieties. The leaves of susceptible genotypes in the present study turned yellow and the plant became stunted and died between 15 and 21 days after infestation. The present study revealed that IT97K-556-6, a cultivated cowpea was tolerant to aphid. Despite the high number of aphids supported by this genotype, the genotype had high survival rate indicating that the genotype is tolerant to aphid. Resistance in TVNu 2786 and TVNu 1156 were expressed as hypersensitive reaction through the elicitation of antibiosis against the insect pest. Past studies have also showed that cowpea resistance against aphids was expressed through antibiosis and antixenosis in some cultivars (Arturo et al. 1988).
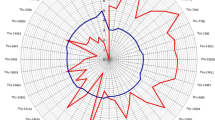
The mean plant damage score of cowpea genotypes is presented in Fig. 1. Among the 60 cowpea genotypes screened for resistance to cowpea aphids, two genotypes TVNu-1158 and TVu-2876, consistently expressed high resistance to cowpea aphid by recording less damage scores ranging from 1.4 to 2.8. The most devastating effect caused by the cowpea aphid was observed on Keffi local, with a mean damage score of 5.0, most of the plants died before reaching 21 days after infestation (Fig. 1). This genotype is thus classified as highly susceptible to cowpea aphid. The other cowpea genotypes that suffered similar damage effects were TVu 134 (4.9), TVu 4539, TVu 36, IT98K-573-2-1, and IT90 K-277-2 had damage score of 4.8 each.
The result of the phenotyping screening identified TVu 2786 and TVNu-1158 as having strong sources of resistance to cowpea aphids. The genotype recorded lower population pressure, lower plant damage score, and lower number of dead plants than the other genotypes at 21 days after infestation. The ability of these parameters to distinctly discriminate between resistant and susceptible genotypes shows their potential for use in identifying differences in cowpea reaction to aphids.
High plant damage, increased CPA population pressure and high number of dead plants resulting from the direct feeding activities of cowpea aphids characterized the reactions of susceptible cowpea genotypes. Evidence of susceptibility on cowpea genotypes was prevalent on Keffi local, and Aloka local which were distinctly marked by stunted growth, increased secretion of honey dew, resulting in an early yellowing of older leaves and high plant mortality.
The cowpea genotype, TVNu-1158, a wild relative, also recorded the lowest mean plant damage score and mean population pressure score of 2.2 and 2.8 respectively. In addition to its high survival percentage (85%), TVNu-1158 also possesses resistant characteristics attributable to the plants innate ability not to support build-up of the insect population up to damaging levels. This present study agrees with the findings of Souleymane et al. (2013) who reported a slower aphid population build-up on the wild cowpea genotype and suggested antibiosis as the mechanism of resistance. Similarly, the current study reveals a survival rate of 85% which is close to 80% survival rate earlier reported for this genotype by Souleymane et al. (2013). One other resistant variety identified in this study that showed high percentage survival rate and low plant damage score include Sierra-Leone Local.
The F2 segregating population derived from the cross between TVu-2876 (resistant parent) and Keffi local (susceptible parent) was used to determine the inheritance pattern. Segregation in the F2 population segregated 168 resistant to 59 susceptible individuals (Table 7). The χ2 analysis for goodness-of-fit revealed that the segregation pattern fits the 3:1 genetic ratio, indicating that resistance to cowpea aphids in TVNu-2876 is controlled by a single dominant gene. The backcross populations involving the F1 plants and susceptible parents segregated into 1:1 ratio, which further confirmed that a single dominant gene confers resistance in TVNu 2876.
Marker assay
The thirty cowpea genotypes that were found to show some level of resistance or tolerance using phenotypic data were further screened for presence of markers conferring resistance to cowpea aphids with two different molecular markers linked to aphid resistance in cowpea. KAD16 has previously been validated in F2 segregating populations derived from the cross between TVNu 2876 × Keffi local. KAD61 mapped 7.0 cm from the aphid resistance gene.
In contrast to the plant phenotype, there were some distinct differences in the genotypes based on the marker score. Some genotypes that were found to be resistant based on phenotypic data were found to be susceptible with the marker score. However, the two markers (CP171/172 and KAD61) gave reproducible and score-able bands with known resistant and susceptible genotypes (Figs. 1, 2). The two markers amplified the cowpea genotype, TVNu 2876, whereas TVNu 1158 was only amplified by KAD61. CP171/172 is a co-dominant marker developed for aphid resistance biotype in Tamale, Ghana while KAD61 a dominant marker was linked with aphid biotypes from Nigeria (Table 8). Since CP171/172 marker is linked with Ghana aphid biotype it is not surprising that this marker did not amplify TVNu 1158, indicating that TVNu 2876 could be resistant to different aphid biotypes in the study location. Also, cowpea genotype TVNu 2876 was highly diverse from TVNu1158 on the basis of marker assay and band size difference. The molecular weight amplified in TVNu 2876 was unique with a 100 bp while that in TVNu 1158 was 120 bp suggesting a novel gene in TVNu 2876. The result of this present study is interesting because most aphid resistant cowpea genotypes developed at IITA have been reported to be susceptible in other countries (Ofuya 1997). Kusi et al. (2010) recently reported high susceptibility of IITA lines and suggested the existence of cowpea aphid biotype in northern Ghana which is more virulent than the Nigerian biotypes. Identification of new genetic sources of resistance to aphid by TVNu 2876 will ensure gene pyramiding to guide against aphid biotypes. The marker score highly correlated with the phenotypic data for the susceptible genotypes as the marker presence were completely absent in those genotypes. The discrepancies that were observed were in relation to the moderately resistant genotypes. The presence of polymorphic markers between resistant and susceptible parents (Fig. 3) as demonstrated in this study reveals the possibility in tracking the gene for resistance to A. craccivora in cowpea.
Many of the cowpea genotypes that were identified as resistant in insect proof screening was confirmed susceptible by the markers applied. This result stresses the effectiveness of DNA marker characterizing genotypes for resistance and susceptibility to aphids rather than selecting cowpea for aphid resistance on the basis of phenotypic screening alone which may be influenced by the environment. Also, some of the genotypes that were earlier classified to be moderately resistant under phenotypic score were classified as susceptible when markers were applied (Table 8). Similar finding had been reported by Omoigui et al. (2016) who found SSR and SCAR markers to be effective in discriminating between Striga resistance and Striga susceptible cowpea genotypes. In many cases resistant genes can only be identified using molecular markers (Melchinger 1990). The utility of such findings is further authenticated by other studies, where the presence of rust resistance genes was confirmed with molecular marker (Li et al. 2017; Imbaby et al. 2014), Aphid resistance genes (Huynh et al. 2015; Kusi et al. 2010). Marker-assisted selection offers the opportunity to select desirable lines on the basis of genotype rather than phenotype. In the current studies, the discrepancies observed in the reaction of the genotypes may suggest a gap in our present knowledge of genetics of aphid resistance in cowpea using phenotypic data alone.
KAD61 is a dominant marker, it amplifies resistant lines with a single band while susceptible lines had no band. Whereas, cp117/16 is a co-dominant marker and showed different bands size in resistant and susceptible parents. The limitation of a dominant marker is that it can only classify segregating population into two categories (resistant: susceptible), it cannot differentiate alleles of the same genotype. Unlike a co-dominant marker that can different alleles of the same genotype. A co-dominant marker will amplify both resistant and susceptible parents. When used in a segregating population, three possible genotypes are seen (AA, Aa, and aa). The advantage of this is that homozygous dominant (AA) can be distinguish from heterozygous dominant (Aa). Consequently, categorize the segregating population into three classes (homozygous resistant with single band: heterozygous resistant with double band: homozygous recessive susceptible with a single band), which is very critical for selection in F3 population. This limitation, however, does not mean that dominant marker cannot be used in marker-assisted backcrossing program. The marker can be employed in F2 segregating population to quickly eliminate the susceptible lines, so that only resistant lines (homozygous resistant and heterozygous resistant lines) are carried forward to F3 for further screening and selection. Dominant markers can also be employed in successive backcrossing program to check each backcross if the resistant lines are carried forward. For each backcross, the successful backcross progenies being carried forward are heterozygous. The utility of dominant markers (C42B) in discriminating Striga resistant and susceptible lines had been successfully demonstrated in cowpea (Omoigui et al. 2012, 2016).
After the identification of TVNu 2876 as good source of resistant to aphid, allelic relationship was conducted to confirm if the gene that confer resistant in TVNu 2876 and F-Ghana (SARC 1-57-2) are the same. TVNu 2876 was crossed with SARC 1-57-2 and TVNu 1158 to obtain F1. The F1 plants were selfed to obtain F2 populations. One hundred and twenty individual plants of the F2 populations derived from the cross between TVNu 2876 and SARC 1-57-2 were planted and screened for aphids under artificially infested plants with aphids. Twenty-Five F1 seeds from both the straight and reciprocal cross, twenty seeds of parents: TNVu 2876, and SARC 1-57-2 included as checks were assessed for aphid resistance.
In the F2 cross, the segregation for aphid resistance in the allelism test showed that 115 plants were resistant and 10 plants were susceptible, which exhibited the action of dominant genes conferring resistance to TVNu 2876 and SARC 1-57-2 (Table 9). The Chi square values showed a good fit for a segregation ratio of 15 resistant to 1 susceptible, which demonstrates the presence of two independent dominant genes. This result supported the hypothesis that the gene conferring resistance to aphids in TNVu 2876 is independent, harboured in SARC 1-57-2. Similarly, in the cross between TNVu 2876 and TVNu 1158, segregation was observed in the F2 population (110 resistant plants and 10 susceptible plants), which suggests that resistance gene in TVNu 2876 and TVNu 1158 are not the same. This finding also corroborates the marker data, which showed different banding pattern in the two cowpea genotypes.
Conclusions
This study identified new cowpea genotype TVNu 2876 as a strong source of resistance to CPA. The application of molecular markers in this study has revealed that phenotypic data alone is not always comprehensible in identifying genotypes for resistance to CPA. This inconsistency shows that markers should be used in conjunction with phenotypic data for selection of genotypes for resistance to aphids. The mechanisms of resistance in cowpea genotype, TVNu 2876 and TVNu 1158 suggest that of hypersensitive response. The susceptibility of IT84S-2246-4 indicates the possibility that there is a new variant of cowpea aphid that is capable of infesting known resistant sources or that the resistance gene in IT84S-2246-4 has broken down. With the frequent breakdown of single dominant gene in our extensive monoculture agricultural systems, it is important to pyramid multiple resistant genes into crops with major R-genes to help inhibit the occurrence of new virulent biotypes or against gene breakdown. The segregation in F2 and BC1 populations derived from the cross between TVNu 2876 and Keffi local indicated that resistance to cowpea aphids in TVu-2876 is controlled by a single dominant gene. Allelism test revealed that resistance gene in TVNu 2876 is non-allelic with the gene that confers resistance in SARC 1-57-2 and TVNu 1158. The cowpea genotype, TVNu 2876 identified in this study has a great potential as source of additional resistant gene or for gene pyramiding against the cowpea aphid in Makurdi and its environs.
References
Alabi OY, Aziza E, Omoloye AA (2012) Preliminary evaluation of selected cowpea varieties for resistance to cowpea aphid, Aphis craccivora. Niger J Ecol 12:45–55
Aliyu H, Ishiyaku MF (2013) Identification of Novel Resistance Gene Sources to Cowpea aphids (Aphid craccivara Koch) in cowpea [Vigna unguiculata (L.) Walp.] at Zaria. PJBS 16(15):743–746
Annan IB, Schafers GA, Tingey WM (1996) Impact of density of Aphids craccivora (Aphididae) on growth and yield of susceptible and resistant cowpea cultivars. Ann Appl.Biol 128:183–193
Arturo G, Jens W, Jan P (1988) Cowpea aphid performance and behaviour on two resistant cowpea lines. Entomol Experi Appl 49:259–264
Atiri GI, Enobakhare DA, Thottappilly G (1986) The importance of colonizing and non-colonizing aphid vectors in the spread of cowpea aphid-borne mosaic virus in cowpea. Crop Prot 5:406–410
Bata HD, Singh BB, Singh SR, Ladeinde TAO (1987) Inheritance of resistance to aphid in cowpea. Crop Sci 27:892–894
Bidinger FR (1992) Utilization of plant genetic resources in crop breeding programmes. Dinteria 23(1992):70–81
Blackman RL, Eastop VF (2000) Aphids on the world’s crops: an identification and information guide. Wiley, Chichister
Cardinali A, Linsalata V, Perriono P, LattanzioV, Brouillard R, Jay M, Scalbert A (1995) Chemotaxonomy of wild Vigna species as potential sources of resistance to insects. Polyphenols 94: 17th International Conference, Palma de Mallorca, Spain. 375–376
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
El-Defrawi GM, Bishara SI (1992) Resistance to Aphis Craccivora Koch in faba bean. Zagazig J Agric Res 19(6):2647–2655
FAO (Food and Agricultural Organization of the United Nations) (2014) FAOSTAT Data
Fatokun CA (2002) Breeding cowpea for resistance to insect pests: attempted crosses between cowpea and Vigna vexillata. Challenges and opportunities for enhancing sustainable cowpea production. IITA, Ibadan, pp 52–61
Githiri SM, Ampong-Nyarko K, Osir EO, Kimani PM (1996) Genetics of resistance to Aphis craccivora in cowpea. Euphytica 89:371–376
Gunilla E (1985) Studies on Aphis craccivora (Koch); preference test using different cowpea extracts and rearing experiments on artificial diets
Hall AE, Cisse N, Thiaw S, Elawad HOA, Ehlers JD, Ismail AM, Fery RL, Roberts PA, Kitch LW, Murdock LL, Boukar O, Phillips RD, McWatters KH (2003) Development of cowpea cultivars and germplasm by the bean/cowpea CRSP. Field Crops Res 82:103–134
Huynh B-L, Ehlers JD, Close TJ, Cisse´ N, Drabo I, Boukar O, Lucas, Wanamaker S, Pottorff M, Roberts PA (2013) Enabling tools for modern breeding of cowpea for biotic stress resistance. Translational genomics for crop breeding. Wiley, London, pp 183–199
Huynh B-L, Ehlers JD, Ndeye A, Wanamaker S, Lucas, Close TM, Roberts PA (2015) Genetic mapping and legume synteny of aphid resistance in African cowpea (Vigna unguiculata L. Walp.) grown in California. Mol Breeding. doi:10.1007/s11032-015-0254-0
Imbaby IA, Mahmoud MA, Hassan MEM, Abd-El-Aziz ARM (2014) Identification of leaf rust resistance genes in selected egyptian wheat cultivars by molecular markers. Scientific World Journal. doi:10.1155/2014/574285
International Institute of Tropical Agriculture (2009) IITA research to nourish Africa-Cowpea. IITA, Ibadan
Kamphuis LG, Gao L, Singh KB (2012) Identification and characterization of resistance to cowpea aphid (Aphis craccivora Koch) in Medicago truncatula. BMC Plant Biology 12:101. http://www.biomedcentral.com/1471-2229/12/101
Kitch LW, Battenberg H, Wolfson JL (1999) Indigenous knowledge and cowpea pest management in sub-Saharan Africa: entomology department. Purdus University, west Lafayette
Kusi F, Obeng-Ofori D, Asante SK, Padi FK (2010) New sources of resistance in cowpea to the cowpea aphid (Aphis craccivora Koch) (Homoptera: Aphididae). J Ghana Sci Assoc 12(2):95–104
Laamari M, Khelfa L, Coeur d’Acier A (2008) Resistance source to cowpea aphid (Aphis craccivora Koch) in broad bean (Vicia faba L.) Algerian landrace collection. Afr J Biotech 7:2486–2490
Li G, Xu X, Bai G, Carver BF, Hunger R, Bonman JM (2017) Novel sources of leaf rust resistance in winter wheat. Crop Sci 57:1–12. doi:10.2135/cropsci2016.08.0725
Mayers GO, Fatokun CA, Young ND (1996) RFLP mapping of an aphid resistance gene in cowpea (Vigna unguiculata L. Walp.). Euphytica 91:181–187
Melchinger AE (1990) Use of molecular markers in breeding for oligogenic disease resistance. Plant Breed 104:1–19. doi:10.1111/j.1439-0523.1990.tb00396.x
Obopile M, Ositile B (2010) Life table and population parameters of cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae) on five cowpea, Vigna unguiculata (L. Walp) varieties. J Pest Sci 83:9–14
Ofuya TI (1993) Evaluation of selected cowpea varities for resistance to Aphis craccivora Koch. (Homoptera: Aphididae) at seedling and podding phase. Ann Appl Biol 123:19–23
Ofuya TI (1997) Control of the cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae), in cowpea, Vigna unguiculata (L.) Walp. Integr Pest Manag Rev 2(4):199–207
Omoigui LO, Yeye M, Ousmane B, Gowda BS, Timko MP (2012) Molecular characterization of cowpea breeding lines for Striga resistance using SCAR markers. J Agric Sci Tech B 2:362–376
Omoigui LO, Kamara AY, Alunyo GI, Bello LL, Oluchi M, Timko MP, Boukar O (2016) Identification of new sources of resistance to cowpea Vigna unguiculata accessions. Gene Resour Crop Evol. doi:10.1007/s10722-016-0410-3
SAS Institute (2014) The SAS system for Windows v. 9.3. SAS Inst., Cary
Schreiner I (2000) Cowpea Aphids, Agricultural pests of the pacific
Singh SR, Jakai LEN, Santos JHR, Adalla CB (1990) Insect pests of cowpea. In: Singh SR (ed) Insect pest of tropical food legumes. Wiley, Chichster, pp 43–89
Singh BB, Chambalis OL, Sharma B (1996) Recent advance in cowpea breeding. Advance in cowpea research. IITA, Ibadan, pp 182–189
Smith CM, Chuang WP (2014) Plant resistance to aphid feeding: behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest Manag Sci 70:528–540
Souleymane A, Aken’Ova ME, Fatokun CA, Alabi OY (2013) Screening for resistance to cowpea aphid (Aphis craccivora Koch) in wild and cultivated cowpea (Vigna unguiculata L. WALP.) accessions. Int J Sci Environ Technol 2(4):611–621
Teetes GL (2007) Plant resistance to insects: a fundamental component of IPM. In: Radcliffe EB, Hutchison WD, Cancelado RE (eds) Radcliffe’s IPM world textbook. University of Minnesota, St. Paul
Acknowledgements
This work was supported by research grants provided by the Kirkhouse Trust, UK to the University of Agriculture Makurdi. The authors wish to thank the Genetic Resource unit of the International Institute of Tropical Agriculture (IITA) for providing the germplasm materials.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Omoigui, L.O., Ekeuro, G.C., Kamara, A.Y. et al. New sources of aphids [Aphis craccivora (Koch)] resistance in cowpea germplasm using phenotypic and molecular marker approaches. Euphytica 213, 178 (2017). https://doi.org/10.1007/s10681-017-1962-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1962-9