Abstract
Wild Cicer species are known to be more responsive to vernalization (induce early flowering when exposed to low temperatures) than the cultivated chickpea. This study was aimed at molecular mapping of vernalization response quantitative trait loci (QTLs) in chickpea. An interspecific recombinant inbred line (RIL) population [ICC 4958 (Cicer arietinum) × PI 489777 (Cicer reticulatum)] was phenotyped for vernalization response for two consecutive years (2009–2010 and 2010–2011) under field conditions. A linkage map already available for this population was used for QTL analysis. A major QTL contributing 55 % of phenotypic variation for vernalization response trait was identified on LG 3 at LOD score of 27. The simple sequence repeat (SSR) markers TA64 and CaM1515 were flanking the QTL which spans a distance of 22 cM. Six Bacterial Artificial Chromosome (BAC)-end sequence derived SSRs (CaM0717, CaM2086, CaM1760, CaM1364, CaM1122, and CaM0886) are present in this QTL region and can be used for isolation of vernalization response genes. Among 84 flowering related genes present in this region, Ca_06280 related to MADS box genes, was reported to play important role in vernalization in cereals. Understanding the genetic control of vernalization response in chickpea will help in exploitation of wild Cicer species in chickpea improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chickpea (Cicer arietinum L.) is an important cool-season food legume crop mainly grown in the arid and the semi-arid regions of the world. It is a highly self-pollinated diploid (2n = 2x = 16) crop with genome size of 740 Mb (Arumuganathan and Earle 1991). It serves as an important source of protein, minerals and vitamins in human diet and provides a diverse array of nutritional and health benefits (Jukanti et al. 2012). Being a legume crop, it fixes atmospheric nitrogen through biological nitrogen fixation (BNF) and provides economic, environmental, and agronomic benefits when included in the cropping systems, particularly in rotation with cereals.
The genus Cicer includes 43 species, of which nine are annual, 33 are perennial and one with unspecified life cycle (van der Maesen and Pundir 1987). The species C. arietinum is the only cultivated species of this genus. Annual species of Cicer have been subjected to numerous taxonomic studies. Based on their morphological traits, life cycle and geographical distribution Cicer species were classified broadly into four sections (van der Maesen 1987). Eight annual species namely C. arietinum, C. reticulatum, C. echinospermum, C. pinnatifidum, C. bijugum, C. judaicum, C. yamashitae and C. cuneatum were categorized in section Monocicer, two perennial species C. chorassanicum and C. incisum in section Chamaecicer, 23 perennial species in section Polycicer and seven woody perennial species in section Acanthocicer. Based on crossability and seed protein banding patterns, C. reticulatum has been proposed as wild progenitor of the cultivated chickpea (Ladizinsky 1975).
The wild Cicer species are valuable gene pools, particularly for resistance to biotic (Nene and Haware 1980; Singh et al. 1981; Singh et al. 1998) and abiotic (Toker 2005; van der Maesen and Pundir 1984; Singh et al. 1990; Robertson et al. 1995) stresses. These have largely remained under-utilized due to crossability barriers, but there are some examples of successful introgression of their genes into the cultivated species. Two wild Cicer species, C. reticulatum and C. echinospermum, closely related to the cultivated chickpea, have been used for widening genetic base of the cultigen and introgressing genes for resistance/tolerance to phytophthora root rot, cyst nematode, root-lesion nematode, pod borer, ascochyta blight, botrytis grey mold and low temperatures (Gaur et al. 2010).
Chickpea is quantitatively long-day plant; longer periods of light alternating with shorter periods of dark each day promote early flowering in most genotypes (Sethi et al. 1981). Extended photoperiod is given to the crop before flowering for rapid generation turnover (Gaur et al. 2007). Variation exists among genotypes for responsiveness to photoperiods. Chickpea cultivars from higher latitudes often exhibit greater response to photoperiod and flower late under shorter day lengths relative to cultivars from lower latitudes (Berger et al. 2011). Such kind of photoperiod response was also observed in oat crop, which is also categorized as long-day plant (Sorrells and Simmons 1992).
Flowering time and crop duration are the most important traits for adaptation of chickpea to different agro-climatic conditions (Berger et al. 2004, 2006; Gaur et al. 2008, 2015). Early flowering is generally associated with early maturity in chickpea and is the key trait for adaptation of chickpea to short-season environments as it helps the crop in escaping terminal or end-of-season drought and heat stresses (Gaur et al. 2008, 2015). Several early and extra-early chickpea cultivars have been developed (Gaur et al. 2008) and four genes (efl-1, efl-2, efl-3, and efl-4) for flowering time have been identified (Gaur et al. 2015). Recent studies conducted at ICRISAT, Patancheru, India, identified the location of these four loci on chickpea linkage maps using four different F2 populations. QTL analysis showed the presence of major QTLs on linkage groups 4, 6 and 8, and a few minor QTLs on LG 3 (BP Mallikarjuna, personal communication). Moreover, flowering time loci were mapped on all the LGs (except LG 7) in chickpea of which LG 3 was found to harboring flowering time genes by many researchers (Cobos et al. 2009; Aryamanesh et al. 2010; Hossain et al. 2010; Rehman et al. 2011; Varshney et al. 2014).
Exposure to the prolonged cold during winter is an important environmental factor that favours flowering in many spring sown crop plants. Wild chickpea accessions show response to vernalization (Abbo et al. 2002), the process by which exposure to cold promotes early flowering. The flower initiation and reproductive development of these wild relatives depend on the extent of cold treatment imposed (Abbo et al. 2002). Thus, plants typically achieve a vernalized state when these are exposed to low temperatures for optimum duration and initiate flowering after winter has passed. The flowering in many vernalization-requiring plants is also induced by long days. This provides an extra level of insurance that flowering does not occur during fall when the day lengths become shorter.
Genes responsible for vernalization and photoperiod play a significant role in geographical adaptation of chickpea (Abbo et al. 2002) and other crops like wheat (Gororo et al. 2001) and Oat (Holland et al. 2002). Genes affecting photoperiod and vernalization responses have been mapped in barley and wheat (Laurie 1997). The major vernalization response genes Vrn-A1 and Vrn-D1 from bread wheat (Triticum aestivum L.), Vrn-H1 from barley (Hordeum vulgare), Vrn-R1 from rye (Secale cereale L.) and Vrn-Am1 from Triticum monococcum are orthologous (Dubcovsky et al. 1998). Similarly, the photoperiod response genes Ppd-H1 from barley and Ppd from wheat map to syntenous positions, suggesting that they are orthologous genes (Laurie 1997). These results suggest that vernalization response along with photoperiod response are adaptive mechanisms in winter grown crops and the genomic regions controlling these traits are conserved across crops that respond to cold treatment and day length.
In order to understand the evolutionary importance of cold treatment and to bring the novel genes and alleles from wild chickpea into the cultivated species, understanding the genetic control of vernalization response will be useful. Hence, the present study was conducted to map the genomic region(s) responsible for vernalization response in a recombinant inbred line (RIL) population developed from an inter-specific cross.
Materials and methods
An F10 population comprising of 131 recombinant inbred lines, derived from the inter-specific cross of ICC 4958 (Cicer arietinum) and PI 489777 (C. reticulatum), was used in this study. ICC 4958 is a desi type drought tolerant breeding line and has vigorous root system (Saxena et al. 1993). PI 498777 is an accession of wild species C. reticulatum with hard seed coat and high response to vernalization.
Phenotyping for vernalization response
Seeds of all RILs and the wild parent were scarified before sowing. After scarification, the seeds were treated with fungicide mixture (2 g thiram and 1 g carbendazim per kilogram of seeds) to avoid infestation by seed or soil-borne pathogens. Seeds were placed on filter paper soaked with distilled water in petri plates (9 cm diameter) and germinated by keeping petri plates at 21 °C for 96 h in dark. For vernalization treatment the 5-day old seedlings were transferred to new petri plates and sealed with parafilm; then these petri plates were kept in an incubator at 4 °C for 30 days in the dark. In non-vernalization treatment (control), after germination in petri-plates, the 5-day old seedlings of each entry were transplanted into the soil in field. After 30 days of vernalization treatment, the seedlings were first kept indoor at room temperature (21 °C) for 48 h and then outdoor in shade for 24 h for hardening and then transplanted into the soil in field. Ten seedlings of each RIL were transplanted on 2 m row ridges in the field. The experimental design used was alpha-lattice with two replications. RILs were randomized within each replication in both the treatments. The experiment was conducted at research farm of ICRISAT at Patancheru in vertisols during post-rainy season 2009-10 and 2010-11. Weather parameters like maximum and minimum temperatures (°C), bright sunshine hours, humidity (%) and rainfall (mm) were recorded during the experiment period in both the years (supplementary Table 1).
Observations were recorded on days to flower and days to maturity on plot basis, and seed yield and shoot biomass on 5 consecutive plants in both vernalized and control treatments. For each RIL, difference in flowering time between control and vernalization treatments was used as vernalization response, and these values recorded for two consecutive years were used in QTL mapping. Seed yield and shoot biomass traits were recorded in 2010–2011 crop season and were used to study the relationship of flowering time with other agronomic traits. Broad-sense heritability (H) values were calculated as a ratio of genotypic and phenotypic variances.
QTL mapping
The inter-specific genetic map based on ICC 4958 × PI 489777 developed by Thudi et al. (2011), comprising of 1291 loci and the phenotyping data on vernalization response generated for two years (2009–2010 and 2010–2011) was used for identification of QTL(s) for vernalization response. QTL Cartographer Ver. 2.5 was used to detect vernalization response QTL(s) employing composite interval mapping analysis (Wang et al. 2010). Model 6 was applied, and control marker number and window size were 5 and 10 cM, respectively. Walk speed was 1 cM and the forward regression method was used. LOD-score >3.0 considered the presence of QTLs. The coefficient of determination (R2) for the marker most tightly associated to a QTL was used to estimate the proportion of phenotypic variation explained by that QTL. Statistical significance of QTL was assessed by permuting each data set 1000 times, with a significance level of 0.05.
In order to identify candidate genes present in the major QTL region, the BES-SSR markers present in this QTL region were subjected to BLAST against chickpea genome assembly (Varshney et al. 2013) and the corresponding UniProt IDs were retrieved. For functional categorization of the genes, the UniProt IDs of the genes were mapped onto UniProt KB database (http://www.uniprot.org/).
Results and discussion
Vernalization response in parents and RIL population
Response to vernalization varied between cultivated and wild parents used in the present study. The cultivated parent (ICC 4958) did not show any difference (P = 0.315) in days to flowering under control and vernalization treatments, whereas wild parent (PI 489777) flowered 34 days earlier under vernalization treatment (P < 0.01) compared to control (Table 1). Similar kind of response was observed for days to maturity in parental lines. These results support the findings of Abbo et al. (2002), where cold treatment advanced the flowering time by 19 days in wild chickpea genotypes and 3 days in cultivated chickpea. Because of vernalization treatment the wild parent was able to flower early and produced 10.3 times higher seed yield than when grown under control. On the other hand, vernalization treatment reduced seed yield 3.1 times and shoot biomass 2.0 times in the cultivated parent (Table 1). Thus, cold treatment had a positive effect on flowering time (early initiation of flowering) and seed yield in the wild genotype, and negative effect on seed yield in the cultivated genotype.
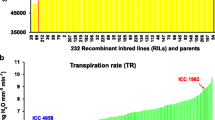
From the total 131 RILs evaluated, 124 RILs were used to study vernalization response where complete phenology data were available in both control and vernalization treatments. Vernalization response (difference in flowering time between control and vernalization treatments) of RILs was categorized into classes to evaluate the distribution pattern (Fig. 1). Interestingly the lines that responded (>16 days) and non-responded (<16 days) to vernalization treatment were clearly grouped separately from each other as shown in the Fig. 1. Twelve genotypes (10 %) completely failed to flower under control conditions even at 100 days after sowing, but produced flowers when vernalization treatment was given. Chi square analysis gave a good fit to 1:1 ratio for vernalization responsive (VR) and vernalization non-responsive (VNR) lines in this RIL population, indicating presence of a major gene for vernalization response in this population. However, Abbo et al. (2002) suggested that this trait was under the control of polygenes as the F2 populations studied by them showed continuous distribution for vernalization response.
Observations were recorded on phenology, seed yield and shoot biomass traits in both VR and VNR lines. Under vernalization treatment, the average flowering time of VR lines was similar to that of wild parent and the average flowering time of VNR lines was similar to that of cultivated parent (Table 1). The VR lines produced shoot biomass equivalent to VNR lines (P = 0.19), but lesser seed yield than VNR lines (P < 0.00). In control treatment, all observed traits showed a significant difference (P < 0.00) between VR and VNR lines in RIL population. VR lines took 41 days extra time to flower than VNR lines when grown without vernalization (control treatment). In vernalization treatment, the difference in average flowering time between VR and VNR lines reduced to 17 days. Moreover, as anticipated, the flowering time of VNR lines in RIL population was almost similar to flowering time of cultivated parent.
Relationship between flowering time and seed yield was evaluated under both control and vernalization conditions (Fig. 2a, b). Flowering time (R2 = 0.79) and seed yield (R2 = 0.30) showed a significant polynomial relationship under both growing conditions. VR lines produced less than 20 g seed yield under control conditions, and 4–70 g under vernalization treatment. Whereas VNR lines produced higher seed yield under control than vernalization treatment. Similarly, flowering time did not show any relationship with shoot biomass or seed yield in vernalization treatment and shoot biomass in control. A highly significant relationship (R2 = 0.74) was observed between vernalization response of RILs in both the years (Fig. 2c) which shows the consistency of phenotypic data. In summary, under control and vernalization treatment flowering time and seed yield are related, and the relationship was mainly influenced by extent of variation caused in VR lines (Fig. 3).
QTL mapping
The present ICC 4958 × PI 489777 population is a widely-used resource for genetic analysis in chickpea (Pfaff and Kahl 2003; Nayak et al. 2010; Hiremath et al. 2012; Roorkiwal et al. 2013). The interspecific genetic map developed by Thudi et al. (2011) comprises of 1291 loci (157 BAC end sequence derived (BES) - SSRs, 11 EST-SSRs, 18 CISR, 35 CAPS, 81 COS-SNPs, 621 DArT and 368 legecy markers). QTL analysis using QTL Cartographer identified a major QTL region spanning 22 cM on Linkage group (LG) 3. This LG consists of 193 markers spanning a distance of 99 cM with an average marker density of 0.51 markers/cM (Fig. 4). This QTL explained 47.9 % of phenotypic variation in 2009–2010 and 54.9 % in 2010–2011 for vernalization response trait in chickpea with a maximum LOD value of 27. RIL population was screened for 2 years and the major QTL identified was stable and consistent in both the years. A total of six Bacterial Artificial Chromosome (BAC)-end sequence derived SSRs (CaM0717, CaM2086, CaM1760, CaM1364, CaM1122, and CaM0886) were mapped in the QTL region. Integration of these BES-SSRs on to physical map may help in isolation of vernalization genes. Further, one expressed sequence tag (EST)—SSR marker (ICCeM028; Varshney et al. 2009) was also located in this region, which can be used as a functional marker. Flowering time of RILs under control and after vernalization treatment was subjected to QTL analysis. A minor QTL (LOD 3) was identified for flowering time after vernalization on LG 4 with phenotypic variation of 13 % (Supplemental Fig. 1). Similarly, a minor QTL with 8.7 % phenotypic variation was observed for flowering time on LG 8 in control treatment (Supplemental Fig. 2). These results show that the vernalization response trait was controlled by a QTL on LG 3 independent of the flowering time QTLs located on LG 4 and LG 8. Similarly, in another recent study (BP Mallikarjuna, personal communication) using four sources of early flowering genes, none of the flowering time QTLs was mapped on LG 3. However, there are reports which suggest that LG 3 harbors several loci controlling flowering time in chickpea (Hossain et al. 2010, Rehman et al. 2011, Varshney et al. 2014). Cobos et al. (2009) and Aryamanesh et al. (2010) identified flowering time QTLs on LG 3 using inter-specific mapping populations and the SSR marker TA 64 linked to these QTLs is also linked to vernalization response QTL identified in this study. Thus, it appears that LG 3 harbors several QTLs for flowering time and vernalization response in chickpea. Days to flowering is a trait with high heritability and in present study also it gave high heritability values both in control (H = 97.4 in 2010–2011) and vernalization treatment (H = 94.2 in 2010–2011). Thus, despite the small size of the mapping population (n = 124 lines), we can expect reliable results from QTL analysis.
A major QTL region explaining 47.9 and 54.9 % of phenotypic variation for vernalization response during 2009–2010 and 2010–2011, identified on LG 03 based on ICC 4958 × PI 489777 mapping population. The markers located in QTL region are shown in red colour while BAC end derived SSR markers in the QTL region are shown in blue colour
Mining candidate genes in vernalization response QTL region
An effort was made to mine the candidate genes in the vernalization QTL region by aligning the BES-SSRs on to reference chickpea genome. The flanking markers identified a 22.03 Mb region on Ca3 chromosome. A total of 1132 genes present in this region were functionally categorized based on Gene Ontology (GO) descriptions (UniProt database, http://www.uniprot.org/). Interestingly 84 flowering related genes were present in this region (Data not shown). Among these genes, Ca_06280 is related to MADS box genes, which were reported to vernalization induced flowering in cereals (Trevaskis et al. 2003).
In the few species that have been studied, the biennial or winter-annual habit is governed by a relatively small number of loci, either dominant or recessive depending on the species (Sung and Amasino 2005). The preliminary results based on significance of QTL and high heritability values in both environments, the vernalization response appears to be a qualitative trait controlled by one or two major genes. Similarly, the vernalization requirement of many cereals, including wheat and barley, is controlled by one dominant and one recessive locus (Dubcovsky et al. 1996; Laurie 1997; Tranquilli and Dubcovsky 2000).
Different crops are adapted to a variety of winter climates; therefore, it is not surprising that the range of effective cold temperatures varies among species. In chickpea, 4 °C was found promising in the present study and also reported by Abbo et al. (2002). Similarly, 2–4 °C is optimal in Arabidopsis, whereas slightly higher temperature in the range of 8–17 °C is optimal in onion (Allium cepa) (Lang 1965). However, it is important to note that in the laboratory, the duration of cold requirement is often measured as the time of continuous exposure to the optimum cold temperature, which does not mimic conditions in the field. However, the results from such experiments can be effectively utilized at places where the wild species are not seen or not generally cultivated, and the minimum temperatures of a location are not often coincide with the required optimum vernalization temperature.
The facultative nature of the promotion of flowering by vernalization in winter-annual species makes chickpea an attractive system to study vernalization because plants containing mutations in this process will eventually flower and thus homozygous mutants are amenable to genetic studies. The information obtained from this study on identification of a major QTL for vernalization response in wild species of chickpea and molecular mapping of this QTL will be useful in chickpea breeding programs for exploitation of wild species in chickpea improvement.
References
Abbo S, Lev-Yadun S, Galwey N (2002) Vernalization response of wild chickpea. New Phytol 154:695–701
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol 9:208–218
Aryamanesh N, Nelson MN, Yan G, Clarke HJ, Siddique KHM (2010) Mapping a major gene for growth habit and QTLs for ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum. Euphytica 173:307–319
Berger JD, Turner NC, Siddique KHM, Knights EJ, Brinsmead RB, Mock I, Edmondson C, Khan TN (2004) Genotype by environment studies across Australia reveal the importance of phenology for chickpea (Cicer arietinum L.) improvement. Aust J Agric Res 55:1–14
Berger JD, Ali M, Basu PS, Chaudhary BD, Chaturvedi SK et al (2006) Genotype by environment studies demonstrate the critical role of phenology in adaptation of chickpea (Cicer arietinum L.) to high and low yielding environments of India. Field Crops Res 98:230–244
Berger JD, Milroy SP, Turner NC, Siddique KHM, Imtiaz M, Malhotra R (2011) Chickpea evolution has selected for contrasting phenological mechanisms among different habitats. Euphytica 180:1–15
Cobos MJ, Winter P, Kharrat M, Cubero JI, Gil J, Milian T et al (2009) Genetic analysis of agronomic traits in a wide cross of chickpea. Field Crops Res 111:130–136
Dubcovsky J, Luo MC, Zhong GY, Bransteitter R, Desai A, Kilian A, Kleinhofs A, Dworak J (1996) Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143:983–989
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
Gaur PM, Samineni S, Gowda CLL, Rao BV (2007) Rapid generation advancement in chickpea. J SAT Agric Res 3(1). Available online at http://www.icrisat.org/journal/
Gaur PM, Kumar J, Gowda CLL, Pande S, Siddique KHM, Khan TN, Warkentin TD, Chaturvedi SK, Than AM, Ketema D (2008) Breeding chickpea for early phenology: perspectives, progress and prospects. In: Kharkwal MC (ed) Food legumes for nutritional security and sustainable agriculture, vol vol 2. Indian Society of genetics and Plant Breeding, New Delhi, pp 39–48
Gaur PM, Mallikarjuna N, Knights T, Beebe S, Debouck D, Mejía A, Malhotra RS, Imtiaz M, Sarker A, Tripathi S, Gowda CLL (2010) Gene introgression in grain legumes. In: Gupta S, Ali M, Singh BB (eds) Grain legumes: genetic improvement, management and trade. Indian Society of Pulses Research and Development. Indian Institute of Pulses Research, Kanpur, pp 1–17
Gaur PM, Samineni S, Kamatam S, Thudi M, Varshney RK, Gowda CLL (2015) Allelic relationships of flowering time genes in chickpea. Euphytica 203:295–308
Gororo NN, Flood RG, Eastwood RF, Eagles HA (2001) Photoperiod and vernalization responses in Triticum turgidum × T. tauschii synthetic hexaploid wheats. Ann Bot 88:947–952
Hiremath PJ, Kumar A, Penmetsa RV, Farmer A, Schlueter JA, Chamarthi SK, Whaley AM, Carrasquilla-Garcia N, Gaur PM, Upadhyaya HD et al (2012) Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol J 10:716–732
Holland B, Portyanko A, Hoffman L, Lee M (2002) Genomic regions controlling Vernalization ans photoperiod responses in oat. Theor Appl Genet 105:113–126
Hossain S, Ford R, McNeil D, Pittock C, Panozzo JF (2010) Development of a selection tool for seed shape and QTL analysis of seed shape with other morphological traits for selective breeding in chickpea (Cicer arietinum L.). Aust J Crop Sci 4:278–288
Jukanti AK, Gaur PM, Gowda CLL, Chibbar RN (2012) Chickpea: nutritional properties and its benefits. Br J Nutr 108:S11–S16
Ladizinsky G (1975) A new Cicer from Turkey. Notes Res Bot Gard 34:201–202
Lang A (1965) Physiology of flower initiation. In: Ruhland W (ed) Encyclopedia of plant physiology. Springer, Berlin, pp 1371–1536
Laurie DA (1997) Comparative genetics of flowering time. Plant Mol Biol 35:167–177
Nayak SN, Zhu H, Varghese N, Datta S, Choi H-K, Horres R, Jungling R, Singh J, Kishore PB, Sivaramakrishnan S et al (2010) Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor Appl Genet 120:1415–1441
Nene YL, Haware MP (1980) Screening chickpea for resistance to wilt. Plant Dis 64:379–380
Pfaff T, Kahl G (2003) Mapping of gene-specific markers on the genetic map of chickpea (Cicer arietinum L.). Mol Gen Genomics 269:243–251
Rehman AU, Malhotra RS, Bett K, Tar’an B, Bueckert R, Warkentin TD (2011) Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress. Crop Sci 51:450–463
Robertson LD, Singh KB, Ocampo B (1995) A catalog of annual wild Cicer species. ICARDA, Aleppo, Syria
Roorkiwal M, Sawargaonkar S.L, A. Chitikineni, M. Thudi, R.K. Saxena, H.D. Upadhyaya, M.I. Vales, O. Riera-Lizarazu, R.K. Varshney. (2013) Single nucleotide polymorphism genotyping for breeding and genetics applications in chickpea and pigeonpea using the beadxpress platform plant Gen. 6:2. doi:10.3835/plantgenome2013.05.0017
Saxena NP, Krishnamurthy L, Johansen C (1993) Registration of a drought-resistance chickpea germplasm. Crop Sci 33:1424
Sethi SC, Byth DE, Gowda CLL, Green JM (1981) Photoperiodic response and accelerated generation turnover in chickpea. Field Crop Res 4:215–225
Singh KB, Hawtin GC, Nene YL, Reddy MV (1981) Resistance in chickpeas to Ascochyta rabiei. Plant Dis 65:586–587
Singh KB, Malhotra RS, Saxena MC (1990) Source for tolerance to cold in Cicer species. Crop Sci 30:1136–1138
Singh KB, Ocampo B, Robertson LD (1998) Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Genet Resour Crop Evol 45:9–17
Sorrells ME, Simmons SR (1992) Influence of environment on the development and adaptation of oat. In: Marshall HG, Sorrells ME (eds) Oat science and technology. American Society of Agronomy, Madison, pp 115–163
Sung S, Amasino RM (2005) Remembering winter: toward a molecular understanding of vernalizations. Annu Rev Plant Biol 56:491–508
Thudi M, Bohra A, Nayak SN, Varghese N, Shah TM, Penmetsa RV, Thirunavukkarasu N, Gudipati S, Gaur PM, Kulwal PL et al (2011) Novel SSR markers from BAC-End sequences, DArT Arrays and a comprehensive genetic map with 1291 marker loci for chickpea (Cicer arietinum L.). PLoS One 6:e27275
Toker C (2005) Preliminary screening and selection for cold tolerance in annual wild Cicer species. Genet Resour Crop Evol 52:1–5
Tranquilli G, Dubcovsky J (2000) Epistatic interaction between vernalization genesVrn-Am 1 and Vrn-Am2 in diploid wheat. J Hered 91:304–306
Trevaskis B, Bagnall DJ, Ellis MH, James Peacock W, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. PNAS 100:13099–13104
van der Maesen LJG (1987) Origin, history and taxonomy of chickpea. In: Saxena MC, Singh KB (eds) The Chickpea. CAB International, Wallingford, pp 11–34
van der Maesen LJG, Pundir RPS (1984) Availability and use of wild Cicer germplasm. Plant Genet Resour Newslett 57:282–285
van der Maesen LJG, Pundir RPS (1987) Availability and use of wild Cicer germplasm. Plant Genet Resour Newslett 57:19–24
Varshney RK, Hiremath PJ, Lekha P, Kashiwagi J, Balaji J, Deokar AA, Vadez V, Xiao Y, Srinivasan R, Gaur PM, Siddique KHM, Town CD, Hoisington DA (2009) A comprehensive resource of drought-and salinity responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genom 10:523
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, Millan T et al (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246
Varshney R, Thudi M, Nayak S, Gaur P, Kashiwagi J, Krishnamurthy L et al (2014) Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor Appl Genet 127:445–462
Wang S, Basten CJ, Zeng ZB (2010) Windows QTL Cartographer 2.5, Department of Statistics, North Carolina State University, Raleigh, NC, USA (Available from http://statgen.ncsu.edu/qticart/WQTLCart.htm)
Acknowledgments
Thanks are due to Dadakhalandar Doddamani for useful discussions on Gene Ontology. This work was supported by International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) and undertaken as part of the CGIAR Research Program on Grain Legumes.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10681_2015_1571_MOESM1_ESM.pptx
Supplemental Fig. 1: A QTL for flowering time identified on LG 4 under vernalization treatment during 2009–2010. Supplementary material 1 (PPTX 94 kb)
10681_2015_1571_MOESM2_ESM.pptx
Supplemental Fig. 2: A QTL for flowering time identified on LG 8 under control treatment during 2009–2010. Supplementary material 2 (PPTX 87 kb)
Rights and permissions
About this article
Cite this article
Samineni, S., Kamatam, S., Thudi, M. et al. Vernalization response in chickpea is controlled by a major QTL. Euphytica 207, 453–461 (2016). https://doi.org/10.1007/s10681-015-1571-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1571-4