Abstract
Potassium (K) deficiency and salinity are major environmental stresses affecting the growth and agricultural production of cotton (Gossypium hirsutum) plants worldwide. In an effort to improve potassium uptake and tolerance to salt stress through genetic engineering, we inserted AlHAK1 into cotton via Agrobacterium-mediated transformation, and then transgenic lines were obtained. To test their response to K-starvation and salt stress, we cultured seedlings from the wild type (WT) and three homozygous overexpression lines (T3 generation) in plastic pots and treated them with a modified half-strength Hoagland’s solution supplemented with different concentrations of potassium or sodium: NT (normal treatment with standard amounts of K and Na), KT (additional 0.05 mM KCl), ST (additional 150 mM NaCl), or KST (0.05 mM KCl plus 150 mM NaCl). After 15 days, all transgenic lines exhibited significantly larger values for shoot and root lengths and biomass (shoot dry weight or root dry weight) when compared with WT plants. Most root morphological parameters for the transgenics were also increased, e.g., total lengths, specific root lengths and surface areas. However, average root diameters were significantly lower than that of the WT (P < 0.05 or P < 0.01). Under salt-stress conditions, the ratios for K+/Na+ were higher in the leaves and roots of transgenic plants, and they also had less malondialdehyde and hydrogen peroxide (H2O2) than the WT tissues. Those responses paralleled greater activities by the antioxidant enzymes superoxide dismutase and peroxidase. We clearly demonstrated that cotton plants transformed with a high-affinity K+ transporter gene have enhanced K+ uptake and salt tolerance. These findings could serve as a promising step toward the development of new cotton cultivars with improved potassium uptake and tolerance to salt stress, and they have significant implications for increasing crop yields on high-salinity soils where potassium levels are low.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potassium (K) is an essential nutrient that is the most abundant cation in plants, where it constitutes up to 10 % of the total dry matter (Leigh and Wyn Jones 1984). A deficiency in K+ causes regional withering of old leaves and death of meristems, eventually leading to serious reductions in shoot growth and crop yields (Gierth and Mäser 2007). In addition to its importance in developmental and reproduction processes, K+ acquisition is essential to defense mechanisms against salt stress that arises mainly because of excessive amounts of Na+ (Wu et al. 1996). The high demand for K can be met through effective uptake from the soil solution by roots, followed by transport into the aerial parts of the plant. Membrane transport of K can be mediated either by channels that utilize membrane potential to facilitate K transport down along its electrochemical gradient or else by secondary transporters (Gierth and Mäser 2007). Many genes encoding K+ channels and transporters have been identified (Horie et al. 2011; Xu et al. 2014; Nieves-Cordones et al. 2014; Hammou et al. 2014). These candidate genes have been confirmed in plant genomes as members of three major transporter families: the KT/HAK/KUP, TRK/HKT transporters; the cation proton antiporter family and K+ channels.
Numerous genes encode KT/HAK/KUP transporters in evolutionarily diverse organisms ranging from green algae to angiosperms (Cabrera et al. 2012). Their ubiquity implies that these genes play important roles in nutrient acquisition and survival in K+-poor environments (Grabov 2007). For example, Arabidopsis thaliana has 13 KT/HAK/KUP K+ transporters (Rubio et al. 2000) while Oryza sativa has at least 17 transporters within that family (Bañuelos et al. 2002). The HAK5 in Arabidopsis belongs to a HAK-1 type transporter that works as a high-affinity K+ transporter (Nieves-Cordones et al. 2010). Overexpression of those genes enhances potassium uptake and salt tolerance in transgenic plants (Horie et al. 2011; Xu et al. 2014; Mansour 2014). These results demonstrate that HAK genes are high-affinity K+ uptake components that function predominantly under K+-starvation.
Cotton (Gossypium hirsutum L.) is one of the most important cash crops in the world. Optimum yields depend upon the availability of potassium throughout the growing season (Pettigrew 2003). Compared with other field crops such as corn or soybean, cotton appears to be more dependent upon K-availability and shows symptoms of deficiency much earlier under limited soil-K (Cope 1981). The incidence of K-deficiencies in cotton cultivation is now increasing in many countries for three reasons: (1) a negative K-balance in the soil, (2) adoption of modern cultivars that feature accelerated fruit set and greater boll loads (Tian et al. 2008; Rengel and Damon 2008) and (3) the popularization of transgenic Bt (Bacillus thuringiensis Berliner)-cotton, which is more susceptible to K-deficiencies and salt stress (Zhang et al. 2007) Such deficiencies negatively influence photosynthesis (Wang et al. 2012), root development (Zhang et al. 2009a), overall plant growth, and premature senescence (Pettigrew 2003; Xia et al. 2011). These responses ultimately result in lower lint yields and poor fiber quality. Although these physiological phenomena have been studied extensively, no reports have been made about efforts to improve plant growth by expressing a high-affinity K+ transporter gene, such as AlHAK1 from Aeluropus littoralis Parl., to enhance potassium uptake.
Our study objective was to obtain a new cotton germplasm with better potassium uptake and salt tolerance. The goal was to increase crop productivity on potassium-deficient and high-salinity soils. To achieve this, we inserted AlHAK1 into the cotton genome. Overexpression lines not only had greater potassium uptake when compared with the wild type (WT), but also showed significantly stronger salt tolerance and activities by antioxidant enzymes. We believe that this is the first report of work done to improve those traits in a crop plant through constitutive expression of a high-affinity K+ transporter gene.
Materials and methods
Plant materials
Lumianyan28 is transgenic Bt (Bacillus thuringiensis Berliner)-cotton variety, and transgenic Bt (Bacillus thuringiensis Berliner)-cotton, which is more susceptible to K-deficiencies and salt stress, so we choose this varity as our transformation material. The procedures for seed germination, seedling growth, and isolation of shoot apices have been described previously (Gould and Magallanes-Cedeno 1998; Liu et al. 2011).
Isolation of AlHAK1 cDNA
Total RNA was isolated from K+-starved, 4-day-old A. littoralis plants by the acid guanidinium phenol chloroform method. As our PCR template, first-strand cDNA was synthesized from 1 μg of total RNA with a first-strand cDNA synthesis kit (Perkin Elmer, Branchberg, NJ, USA). The AlHAK1 cDNA was amplified by PCR using a sense primer (5′-GCAAGCGGCCTCCGTATCACG-3′) and an antisense primer (Oligo dT Primer) for AlHAK1. The PCR products were cloned into the pUC118 vector with a TaKaRa BKL kit (Takara Bio Inc., Tokyo, Japan) and then sequenced. The putative ORF sequence of AlHAK1 was identified in GenBank (Accession No. DQ645465). For amplifying the cDNA, a PCR primer (AF: 5′-CATCTAGAATGTCGCTCGAGGTCGAG-3′) and an antisense primer (AR: 5′-GCGAGCTCCTATATTTCATATGTGATCCC-3′) were designed based on the ORF sequence information.
Bioinformatics analysis
The cDNA sequences were analyzed with DNAstar software and the BLAST program (http://ncbi.nlm.nih.gov). Open reading frames (ORFs) were identified with an ORF finder (NCBI), and the protein sequences were deduced. Preliminary properties of the encoded protein were predicted by ProtParam (Appel et al. 1994) while membrane-spanning structures were estimated by TMHMM (Sonnhammer et al.1998). Sub-cellular localization was predicted by Psort (Horton and Nakai 1997).
Cotton transformation using shoot apices
The ORF of AlHAK1 was placed under the control of the CaMV 35S promoter in the pBI121 vector (Fig. 1). The resulting plasmid with an AlHAK1 expression cassette was used to transform the apices from cotton shoots according to the method of Gould and Magallanes-Cedeno (1998) and Liu et al. (2011). This transformation required four major steps: (1) obtaining shoot apex explants and pre-culturing them for 2 d in an MS medium; (2) co-cultivation with an Agrobacterium suspension containing the pBI121-AlHAK1 vector; (3) transferring the suspension to a co-cultivation medium, where it was held under darkness for 2 days; and (4) transfer to a shoot induction medium. The regenerated primary plants were designated as the T0 generation. These were self-pollinated and, ultimately, T3 transgenic lines were obtained for further analysis. Those seeds were germinated in a greenhouse at 28/20 °C (day/night) under a 16-h photoperiod.
Schematic representation of pBI121-AlHAK1 plasmid construction. pBI121-AlHAK1, carrying AlHAK1 and NPT II, was driven by 35S and NOS promoters, respectively. RB right border, AlHAK1 high-affinity K+ transporter gene, NPT II neomycin phosphotransferase. Locations for restriction maps and all primers are shown on construct
Molecular characterization of putative transgenic plants
Southern blotting was performed according to the procedure described by Sambrook et al. (1989). Genomic DNA samples (70 μg) were digested with EcoR I enzymes, then separated in 1 % (w/v) agarose gels and transferred to membranes (Hybond-N+ positively CHGD Nylon Transfer Membrane; Amersham Biosciences). For our probe a 500-bp fragment of the AlHAK1 amplified from pBI121 with primers PF (5′-TGCTCTTCTCAGTCCAGCGTTTCG-3′, forward) and PR (5′-ACCTCTCCGCAGGAATCACA-3′, reverse) was labeled with DIG (DIG High Prime DNA Labeling and Detection Starter Kit I; Roche) according to the manufacturer’s instructions.
Western blots were used to verify protein expression in leaf extracts. Anti-AlHAK1 antibody obtaining by the exogenous expression of whole protein was used as antigen, and this antigen could recognize the exogenous expression of AIHAK protein, and only a single band was observed in the vicinity of the 87Kd. Briefly, those proteins were resolved via SDS-PAGE and electrophoretically transferred to a PVDF membrane (Bio-Rad, USA). Each blot was first incubated with a rabbit anti-AlHAK1 antibody (1:1,000 dilution) and then with an alkaline phosphatase-tagged secondary antibody.
Plant growth conditions
Seeds of WT and transgenic cotton plants were surface-sterilized with 10 % H2O2 for 30 min and then germinated in a sand culture at 26 °C. At the trefoil stage (15 days after germination), uniformly sized seedlings were transferred for hydroponic culturing in 16- × 13- × 16-cm plastic pots filled with modified half-strength Hoagland’s solution. The following supplements were added to the solutions: NT (normal treatment with standard amounts of K and Na), KT (additional 0.05 mM KCl), ST (additional 150 mM NaCl), or KST (0.05 mM KCl plus 150 mM NaCl). For all treatment types, the Hoagland’s solution also contained 2.5 mM Ca (NO3)2, 1 mM MgSO4, 0.5 mM (NH4)H2PO4, 0.2 µM CuSO4, 1 µM ZnSO4, 0.1 mM EDTA-Fe–Na, 20 µM H3BO3, 5 ρM (NH4)6Mo7O24, and 1 µM MnSO4. Three seedlings were cultured in each box under a day/night temperature regime of 28/16° and a 12-h photoperiod provided by fluorescent light (240 μEm−2 s−1). Deionized water was added daily to replace the water lost through evapotranspiration. The pH was maintained at 6.5 by adding concentrated solutions of NaOH, and air was bubbled into the nutrient solution to provide O2 and achieve nutrient homogeneity.
Sampling method and assessment of physiological parameters
At 15 days after the stress experiments began, i.e., 25 days after germination, every fifth seedling was selected from each treatment. Entire root systems were scanned with an EPSON Transparency unit (Seiko Epson Corp., Tokyo, Japan) and then analyzed with WinRHIZO version 4.0b (Regent Instruments Inc., Quebec, Canada). The specific root length was calculated as total root length divided by root dry weight. The sampled plants were separated into their root and shoot portions. Tissues were oven-dried at 80 °C for 48 h and weighed on a digital balance.
Measurements of potassium and sodium levels
Leaf and root tissues were collected from each treatment type, then rinsed with distilled water, and oven-dried at 80 °C for 20 h. The dried samples (approximately 1.0 g each) were boiled in a mixture of nitric acid and perchloric acid (4:1, HNO3:HClO4) in the presence of glass beads for 5 h. The digested plant materials were filtered, diluted with distilled water, and analyzed for Na+ and K+ contents with an atomic flame photometer (FP6410).
Determination of hydrogen peroxide (H2O2) and malondialdehyde (MDA) contents, and assays for enzyme activity
Three transgenic lines (T3) and wild-type plants (9 plant/each line) were cultured in plastic pots filld with MS solution. Each plastic pot contained three seedlings, held in place with polystyrene foam, and three replications were across plastic pots. 10-day-old WT plants and T3 seedlings from AlHAK1-transgenic Lines OE2, OE5, and OE7 containing one insert locus were exposed to 150 mM NaCl for 72 h. Their leaves were harvested for biochemical analysis. Hydrogen peroxide was measured spectrophotometrically after reaction with KI (Alexieva et al. 2001). The reaction mixture consisted of 0.5 mL 0.1 % trichloroacetic acid leaf extract supernatant, 0.5 mL of 100 mM K-phosphate buffer and 2 mL reagent (1 M KI w/v in fresh double-distilled water H2O). The reaction was developed for 1 h in darkness and absorbance measured at 390 nm. Then the amount of hydrogen peroxide was calculated using a standard curve prepared with known concentrations of H2O2.
Leaf of transgenic and wild plants were initially incubated with 1 % potassium iodide and 0.1 % butylated hydroxytoluene at 50 °C for 20 min, and then with 0.4 % TBA at 60 °C for 60 min. The MDA-TBA complex formed was extracted with isobutyl alcohol and measured by high-performance liquid chromatography with fluorescence detection Zhang et al. (2009b).
Leaves were homogenized at 4 °C in 100 mM K-phosphate buffer (pH 7.8), 10 mM MgCl2, 0.2 mM EDTA. The homogenate was centrifuged at 17,000×g for 30 min to yield a crude enzyme extract. Peroxidase activity and superoxide dismutase activity (SOD) were monitored as described previously (Ranieri et al. 2000).
Statistical analysis
Data were examined by Origin software (Version 7.5) and one-way ANOVAs. Values were considered significantly different at P < 0.05 or P < 0.01 for all experiments. Treatment results were compared by tests for least significant differences.
Results
AlHAK1 cloning and bioinformatics analysis
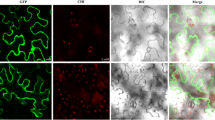
The cDNA of a high-affinity K+ transporter (AlHAK1) gene in an A. littoralis Parl was obtained according to the sequence of the GenBank uncleotide sequence database with the Accession Number DQ645465. By nucleic acid sequence analysis, AlHAK1 sequence and public sequence of DQ645465 contain the same nucleic acid sequence identity. The full-length AlHAK1 cDNA sequence comprises 2,844 bp, and its ORF encodes a protein of 777 amino acids with a calculated molecular mass of 87 kDa. The results of TMHMM predicted that the protein AlHAK1 is located mainly on plasma membrane with 12 transmembrane (Fig. 2a). Subsequently, the subcellular localization experiment showed that the transiently expressed AlHAK1–GFP fusion protein was clearly localized to the cytomembrane, whereas GFP occurred in both the nucleus and cytoplasm (Fig. 2b). Therefore, these results indicated that the AlHAK1 protein is indeed localized to the cytomembrane.
Generation of transgenic cotton
A total of 919 shoot apex explants, each five mm long taken from 5-day-old seedlings were cultured; of which, a total of 45 (5.0 %) plantlets survived on 75 mg/L kanamycin-containing medium. Plants expressing the nptII gene showed no symptoms, while the leaves of wild-type plants turned yellow and then mottled with brown spots (Fig. 3a). Subsequently, the kanamycin-resistant transformants were further confirmed by Southern blot analysis of EcoRI -digested genomic DNA. Examining their hybridization signals showed that OE2, OE5 and OE7 transformants each harbored one insert locus in the genome while OE6 had two. However, DNA isolated from the WT did not hybridize with the probe (Fig. 3b). The presence of AlHAK1 in these four lines was also proven through western blot analysis. Immunoreactive bands were observed in all proteins extracted from leaves of the transgenics but none was detected in the WT samples (Fig. 3c). Thus, the results verified that AlHAK1 was integrated into the transformants and expressed in these plants.
Detection of AlHAK1 in T0 transgenic and WT cotton. a Kanamycin leaf-painting assay. The arrow indicates the specific sites of kanamycin application. b Southern blot analysis. Genomic DNA was digested with EcoR I and probed with 500 bp of AlHAK1. c Detection of AlHAK1 protein in leaf extracts by Western blot analysis. M DNA Molecular Weight Marker II (Roche), P pBI121-AlHAK1 plasmid, WT wild-type plants, OE2, OE5, OE6, and OE7, overexpression lines. The arrow indicates the specific sites of kanamycin application
Overexpression AlHAK1 gene promoted growth of transgenic plants
The different phenotypes of transgenic cotton and WT plants were also observed under different treatments. Under NT conditions, morphological or developmental phenotypes were similar between the WT and all OE plants (Fig. 4a). However, under stress conditions, the transgenics displayed better performance whereas growth of WT plants was significantly reduced (Fig. 4a). For KT, only the lower leaves from OE lines exhibited minor deficiency symptoms compared with almost all of the WT leaves, which showed typical symptoms of K-deficiency, including brown scorching and chlorosis between the veins. When plants were subjected to ST conditions, leaf tips from OE lines were somewhat etiolated while almost all WT leaves were discolored and had dropped. Transgenic plants also had better performance than the WT under KST conditions (Fig. 4a). For root phenotypes of transgenic cotton and WT plants, root phenotypes were similar between the WT and OE plants (Fig. 4b) under NT conditions, However, under stress conditions, the transgenics displayed robust performance whereas growth by WT plants was significantly reduced (Fig. 4b). Therefore, all of these results demonstrated that overexpression of AlHAK1 enhanced the capacity for K+ uptake and conferred salt tolerance in transformed cotton seedlings.
Overexpression AlHAK1 gene in transgenic and promoted growth performance of transgenic plants. a Growth performance of T3 transgenic and WT plants after 15 days of treatment. b Root morphology of transgenic lines and WT plants. WT wild-type plants, OE overexpression lines, NT normal treatment, KT 0.05 mM KCl, ST 150 mM NaCl, KST 0.05 mM KCl plus 150 mM NaCl
Effects of AlHAK1 overexpression on biomass production and shoot and root morphology parameters
15 days later, transgenic and WT seedlings performance significantly different in their amounts of biomass. For example, in response to KT, shoot dry weights were 35.6, 22.0 and 32.2 % higher in the three transgenic lines while their root dry weights were 32.1, 25 and 21.4 % greater than in the WT (Table 1). Similar trends were noted under ST and KST conditions. Shoot and root lengths also differed significantly between OE and WT plants (Table 2). Under KT, shoots were 19.8, 16.9 and 19.9 % longer in the transgenics while roots were 24.7, 16.7 and 23.3 % longer than in the WT.
Further, those parameters in the OE lines also improved with treatments of KT, ST and KST. Total root lengths were 32.6–80.5 % greater in the transgenics under KT conditions and were also significantly increased under ST and KST conditions (Fig. 5a). Values for specific root length and root surface area were also higher in OE lines under all stress treatments, but their average root diameters were smaller under such conditions (Fig. 5b–d). Again, those findings showed that overexpression of AlHAK1 strengthened growth performance, enhanced the capacity to take up K+ and other nutrients, and conferred salt tolerance.
Effects of treatments on root morphology parameters of transgenic lines and WT plants. a Total root lengths. b Specific root lengths. c Root surface areas and d Average root diameters. WT wild-type plants, OE overexpression lines, NT normal treatment, KT 0.05 mM KCl, ST 150 mM NaCl, KST 0.05 mM KCl plus 150 mM NaCl. Data are presented as mean ± SD (n = 5). (*P < 0.05; **P < 0.01)
Na+ and K+ status
The contents of Na+ and K+ in leaves and roots of transgenic OE lines and WT plants were measured after subjecting to non-salt and salt stresses (150 mM NaCl). Under non-salt stress, both transgenic cotton plants and WT plants showed similar levels of Na+ in leaves and roots. Under salt stress, the Na+ levels in leaves and roots increased in both WT and transgenic plants. However, in the leaves and roots, the increase in transgenic lines was higher than that of WT plants (Fig. 6a). K+ levels in the leaves were more than twice the levels in roots under non-salt stress, and all plants showed similar levels under non-salt stress. Under salt stress, the K+ level in WT decreased to about 50 % in both leaves and roots, while transgenic plants appeared to maintain a level similar to those under non-salt stress for both leaves and roots (Fig. 6b). We further found that K+/Na+ ratio was similar between leaves and roots in all genotypes under non-salt stress. However, in the presence of 150 mM NaCl, that ratio was up to 3.22-fold and 3.35-fold higher in both the leaves and roots of transgenic plants when compared with WT ratios (Fig. 6c). This suggested that the OE plants had possibly invoked a mechanism for salt tolerance by which they could maintain a high K+/Na+ ionic balance in a saline environment by regulating Na+ efflux and K+ influx.
Comparison of Na+, K+ contents and K+/Na+ ratio between OE transgenic lines and WT plants after 15 days of 150 mM NaCl treatment. a Content of Na+ in leaves and roots of plants. b Content of K+ in leaves and roots of plants. c K+/Na+ ratios in leaves and roots of plants. Data are mean ± SD of 3 independent measurements (*P < 0.05; **P < 0.01)
Accumulations of H2O2 and MDA and activity by antioxidant enzymes under salt stress
To test whether contrasts in salt tolerance between OE and WT plants are related to oxidative stress, we measured their levels of H2O2 and MDA. Under non-salt stress, no differences were found among the genotypes (Fig. 7a). However, in response to excess NaCl, more H2O2 and MDA was accumulated in the WT. For example, OE plants had up to 34.6 % less in H2O2 and 39.2 % in MDA when compared with the WT (Fig. 7a). This indicated that overexpression of AlHAK1 conferred greater tolerance to oxidative stress that arose from excess salt.
To investigate the possible underlying cause for lower H2O2 accumulations in the transgenic lines, we monitored the activity of major antioxidant enzymes. Under non-salt stress, SOD and POD activities did not differ significantly among genotypes (Fig. 7b), but they were much higher (up to 59.0 % increases for both) in the OE plants than in the WT when excess NaCl was added to the Hoagland’s solution (Fig. 7b). These data indicated that overexpression of AlHAK1 was associated with a more efficient antioxidant system that counteracted the negative effects of oxidative and salt stresses.
Discussion
The HAK gene family plays critical roles in responses to potassium-starvation and salt stress (Gierth and Mäser 2007; Yang et al. 2009; Tomoaki et al. 2011). Moreover, the cotton remains difficult to genetic engineer primarily because of its resistance to Agrobacterium infection and its recalcitrance to in vitro regeneration. Therefore, an alternative approach using shoot apices as explants was developed for free of genotype-dependence, with time-saving in regeneration of cotton plantlet in vitro. So, in this study, this method was used to transfer the high-affinity K+ transporter gene (AlHAK1) into cotton to develop potassium efficient-uptake and salt-tolerant cotton cultivar.
K+ uptake in the high-affinity range of concentrations and its components have been widely studied. In previous study, In common ice plant, transcript levels of McHAK1 and McHAK4 is increased by K+ starvation and salt stress of 400 mM NaCl in leaves and roots (Su et al. 2002). Also, accumulation of barley HvHAK1 transcripts is enhanced by K+ deprivation and exposure to high salt concentrations (Fulgenzi et al. 2008). In addition, AtHAK5 gene of Arabidposis thaliana was observed that it is required for K+ absorption necessary to sustain plant growth at low K+ in the absence or in the presence of salinity (Nieves-Cordones et al. 2010). In this study, the AlHAK1 gene was first cloned and functionally expressed in Saccharomyces cerevisiae, it is believed that this gene enables plants to maintain a high cytosolic K+/Na+ ratio under increased salinity (Su et al. 2007). Therefore, we also study the role of this gene in cotton under two important stress conditions: low K+ supply or the presence of salinity. Notability, the growth was significantly improved in transgenic plants along with their uptake of ions and activities of antioxidant enzymes.
Plant members of the KT-HAK-KUP family of transport proteins were first identified in Arabidopsis and barley (Hordeum vulgare), and were classified into four major clusters (Santa-María et al. 1997). Transporters within Cluster I have key roles in K+ capture from diluted K+ solutions (Gierth et al. 2005). They are also likely involved in K+ homeostasis during acclimation to NaCl stress. One gene in that cluster is AlHAK1, which was first cloned and functionally expressed in S. cerevisiae (Su et al. 2007). Therefore, it is believed that this gene enables plants to maintain a high cytosolic K+/Na+ ratio under increased salinity. We found that AlHAK1 was successfully incorporated into the cotton genome and over-expressed in transgenic lines. Those transformed plants exhibited much greater tolerance to K-deficiency and salt stress when compared with the WT. Our findings support previous reports that expression of K+-transporters by other plant, such as OsHAK5 and OSHKT2 in rice, improves K+ uptake and confers salt tolerance (Yao et al. 2010; Tomoaki et al. 2011). Taken together, these observations unambiguously demonstrate that engineering of the HAK transporter is a useful tool for increasing K+ uptake and salt tolerance, thereby enhancing overall plant performance in K-starvation and saline environments.
Cotton requires high levels of potassium but has limited capacity for its utilization (Dong et al. 2010; Jiang et al. 2011). However, K-resources are very scarce in China and have long been supplemented through the application of fertilizers that are lower in K than in either N or P (Cassman et al. 1989). Therefore, introduction of a transporter gene into cotton would be an efficient measure for increasing the level of potassium nutrition in that crop. Indeed, technologies involving genetic modifications have already greatly contributed to better plant growth, such as seen with Bt-cotton, which is resistance to insect attack. However, those transgenics have also proven to be more sensitive to K-deficiencies and high salt than their untransformed parents (Zhang et al. 2007; Yang et al. 2011). Those deficiencies can inhibit root elongation and lateral root formation, thereby influencing the uptake of nutrients from the soil because of significant reductions in total lengths, surface areas, and volumes (Shin and Schachtman 2004; Armengaud et al. 2004). Brouder and Cassman (1994) have shown that a larger or more efficient root system plays a decisive role in determining cotton tolerance to K+-deficiency. In this study, we found that, under high salinity, OE lines had well-developed root systems and more fine roots, which meant that the interface between root and solution was larger than that for WT plants. We are currently analyzing the molecular mechanism by which overexpression of AlHAK1 promotes root development.
Plants can generally survive better under saline conditions if they remain able to regulate their ionic homeostasis. Maintaining a high K+/Na+ ratio in the cells is a crucial aspect of salt adaptation. The extent to which a plant is salt-tolerant depends upon its ability to export Na+ from the shoots and maintain an elevated K+/Na+ ratio in the cells (Martinez-Atienza et al. 2007). In an environment with excess NaCl, ion transporters such as members of the KT-HAK-KUP family, the low-affinity cation transporter LCT1, and the Na+/H+ antiporter NHA must be activated (Zhu 2001). Among these, the HAK1 transporters demonstrate a stronger tendency to move K+ over Na+ (Santa-María et al. 1997). We noted here that transgenic plants had higher K+/Na+ ratios in both leaves and roots, which implied that overexpression of the transporter gene changed the metabolism of potassium so that the cells could maintain a higher K+ content. Saline conditions often lead to the accumulation of ROS, which can damage cellular membranes and mediate lipid peroxidation, causing oxidative stress (Xiong et al. 2002). Malondialdehyde content is widely recognized as an index of this peroxidation (Mittler 2002). We found that, under salt stress, levels of both H2O2 and MDA were markedly lower in OE plants than in the WT. This response may have been correlated with increased activities by ROS-scavenging SOD and POD. In addition, the greater amounts of AlHAK1 transcript in OE plants induced a rise in enzyme activities, thereby conferring greater salt tolerance and enhancing the stability of proteins and cell membranes within our experimental saline environment (150 mM NaCl). These improvements in the capacity for K+ uptake and salt tolerance by cotton are a promising step toward the development of new cultivars for agricultural production in soils where potassium supplies are deficient.
References
Alexieva V, Sergiev I, Mapelli S, Karanov E (2001) The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ 24:1337–1344
Appel RD, Bairoch A, Hochstrasser DF (1994) A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci 19:258–260
Armengaud P, Breitling R, Amtmann A (2004) The potassium dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136:2556–2576
Bañuelos MA, Garcia de Blas B, Cubero B, Rodriguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130:784–795
Brouder SM, Cassman KG (1994) Evaluation of a mechanistic model of potassium uptake by cotton in vermiculitic soil. Soil Sci Soc Am J 58:1174–1183
Cabrera E, Álvarez MC, Martín Y, Siverio JM, Ramos J (2012) K+ uptake systems in the yeast Hansenula polymorpha. Transcriptional and post-translational mechanisms involved in high-affinity K+ transporter regulation. Fungal Genet Biol 49:755–763
Cassman KG, Roberts BA, Kerby TK, Bryant DC, Higashi SL (1989) Soil potassium balance and cumulative cotton response to annual potassium additions on a vermiculitic soil. Soil Sci Soc Am J 53:805–812
Cope JT (1981) Effects of 50 years of fertilization with phosphorus and potassium on soil test levels and yields at six locations. Soil Sci Soc of Am J 45:342–347
Dong HZ, Kong XQ, Li WJ, Tang W, Zhang DM (2010) Effects of plant density and nitrogen and potassium fertilization on cotton yield and uptake of major nutrients in two fields with varying fertility. Field Crops Res 119:106–113
Fulgenzi FR, Peralta ML, Mangano S, Danna CH, Vallejo AJ, Puigdomenech P, Santa-Maria GE (2008) The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition. Plant Physiol 147:252–262
Gierth M, Mäser P (2007) Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581:2348–2356
Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high affinity K+ uptake and AKT1 channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137:1105–1114
Gould JH, Magallanes-Cedeno M (1998) Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Mol Biol Rep 16:1–10
Grabov A (2007) Plant KT/KUP/HAK potassium transporters: single family–multiple functions. Ann Bot 99:1035–1041
Hammou KA, Rubio L, Fernández JA, García-Sánchez MJ (2014) Potassium uptake in the halophyte Halimione portulacoides L. Aellen. Environ Exp Bot 107:15–24
Horie T, Sugawara M, Okada T, Taira K, Kaothien-Nakayama P, Katsuhara M, Shinmyo A, Nakayama H (2011) Sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioengr 111:346–356
Horton P, Nakai K (1997) Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol 5:147–152
Jiang CC, Xia Y, Chen F, Lu JW, Wang YH (2011) Plant growth, yield components, economic responses, and soil indigenous K uptake of two cotton genotypes with different K-efficiencies. Agric Sci China 10:705–713
Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and function of this ion in the plant cell. New Phytol 97:1–13
Liu JF, Zhao CY, Ma J, Zhang Y, Li MG, Yan GJ, Wang XF, Ma ZY (2011) Agrobacterium-mediated transformation of cotton (Gossypium hirsutum L.) with a fungal phytase gene improves phosphorus acquisition. Euphytica 181:31–40
Mansour MM (2014) The plasma membrane transport systems and adaptation to salinity. J Plant Physiol 171:1787–1800
Martinez-Atienza J, Jiang X, Garcia de Blas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Nieves-Cordones M, Alemán F, Martinez V, Rubio F (2010) The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Plant Mol 3:326–333
Nieves-Cordones M, Alemán F, Martínez V, Rubio F (2014) K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J Plant Physiol 171:688–695
Pettigrew WT (2003) Relationships between insufficient potassium and crop maturity in cotton. Agron J 95:1323–1329
Ranieri A, Petacco F, Castagna A, Soldatini GF (2000) Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci 159:159–167
Rengel Z, Damon PM (2008) Crops and genotypes differ in efficiency of potassium uptake and use. Physiol Plant 133:624–636
Rubio F, Santa-Maria GE, Rodriguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Plant Physiol 109:34–43
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley belongs to a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9:2281–2289
Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101:8827–8832
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Intl Conf Intell Syst Mol Biol 6:175–182
Su H, Golldack D, Zhao CS, Bohnert HJ (2002) The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol 129:1482–1493
Su Q, Feng SY, An LJ, Zhang GH (2007) Cloning and functional expression in Saccharomyces cerevisiae of a K+ transporter, AlHAK1, from the graminaceous halophyte, Aeluropus littoralis. Biotechnol Lett 29:1959–1963
Tian XL, Wang GW, Yang FQ, Yang PZ, Duan LS, Li ZH (2008) Differences in tolerance to low-potassium supply among different types of cultivars in cotton (Gossypium hirsutum L.). Acta Agron Sin 34:1770–1780
Tomoaki H, Mitsuo S, Tomoyuki O, Koichiro T, Pulla KN, Maki K, Atsuhiko S, Hideki N (2011) Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioeng 111:346–356
Wang N, Hua HB, Egriny A, Eneji ZH, Duan LS, Tian XL (2012) Genotypic variations in photosynthetic and physiological adjustment to potassium deficiency in cotton (Gossypium hirsutum L.). J Photochem Photobiol B 110:1–8
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8:617–627
Xia Y, Jiang CC, Chen F, Lu JW, Wang YH (2011) Differences in growth and potassium-use efficiency of two cotton genotypes. Commun Soil Sci Plant 42:132–143
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183
Xu J, Tian XL, Egriny A, Eneji ZH (2014) Functional characterization of GhAKT1, a novel Shaker-like K+ channel gene involved in K+ uptake from cotton (Gossypium hirsutum). Gene 545:61–71
Yang Z, Gao Q, Sun C, Li W, Gu S, Xu C (2009) Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). J Genet Genom 36:161–172
Yang FQ, Wang GW, Zhang ZY, Egrinya Eneji A, Egrinya Eneji LS, Li ZH, Tian XL (2011) Genotypic variations in potassium uptake and utilization in cotton. J Plant Nutr 34:83–97
Yao X, Horie I, Xue S, Leung HY, Katsuhara M, Brodsky DE, Wu Y, Schroeder JI (2010) Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol 152:341–355
Zhang ZY, Tian XL, Duan LS, Wang BM, He ZP, Li ZH (2007) Differential responses of conventional and Bt-transgenic cotton to potassium deficiency. J Plant Nutr 30:659–670
Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y (2009a) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149:916–928
Zhang ZY, Wang QL, Li ZH, Duan LS, Tian XL (2009b) Effects of potassium deficiency on root growth of cotton seedlings and its physiological mechanisms. Acta Agron Sin 35:718–723
Zhu JK (2001) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4:401–406
Acknowledgments
This research was supported by the Natural Science Foundation of Hebei Province (No. C C2013201219), the Introduce Talents Start Scientific Research Projects of Hebei University (2014-277), Cutting-edge and Characteristic Disciplines of Biology (Botany) and Key subject of Biochemistry and Molecular Biology. The authors are grateful to Priscilla Licht for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. F. Liu and S. L. Zhang have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, J.F., Zhang, S.L., Tang, H.L. et al. Overexpression of an Aeluropus littoralis Parl. potassium transporter gene, AlHAK1, in cotton enhances potassium uptake and salt tolerance. Euphytica 203, 197–209 (2015). https://doi.org/10.1007/s10681-014-1310-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1310-2