Abstract
Gibberellic acid (GA) plays an essential role in many plant growth and developmental processes. Overexpression of rice (Oryza sativa L.) HOMEOBOX4 (OsHox4) gene in rice variety IR64 under the control of CaMV 35s promoter caused varying degrees of dwarfism and bushy tillers. Further investigation showed that over-expression of OsHox4 in indica rice induced semi-dwarf by repressing stems cell elongation. This repression could be eliminated by exogenously applied Gibberellin-4 (GA4) suggested that OsHox4 was involved in GA metabolism. Investigations of the expressions of rice DELLA-like subfamily genes, GA 3-oxidase family genes and GA 2-oxidase family genes by qRT-PCR showed that OsHox4 played an important role in GA deactivation and signaling by controlling the expression of rice DELLA-like subfamily genes, rice GA 2-oxidase family genes and rice GA 3-oxidase family genes. Our experiment provided an evidence that over-expression of OsHox4 in transgenic rice resulted in a semi-dwarf phenotype that could be fully rescued by application of exogenous GA4. The OsHox4 plays an important role in GA deactivation by controlling the expression of rice DELLA subfamily genes, rice GA 2-oxidase family genes and rice GA 3-oxidase family genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibberellic acid (GA) plays an essential role in many plant growth and developmental processes, including seed germination, hypocotyl and stem elongation, leaf expansion, reproductive development, and circadian and light regulation (Olszewski et al. 2002; Sun and Gubler 2004; Fleet and Sun 2005). Over the past decade, biochemical and genetic studies have identified most of the genes in GA-biosynthetic pathway and GA-deactivation enzymes (Sakamoto et al. 2004; Thomas et al. 2005). Nevertheless, the mechanisms of GA signaling are still poorly understood.
Derepression of the repressed state is currently considered to be the key step of GA action in the GA signaling pathway. In this model, DELLA subfamily proteins of the GRAS superfamily play an important role in the negative control of GA signaling (Gomi and Matsuoka 2003; Hartweck 2008). GRAS proteins are a recently discovered family of plant-specific proteins named after GAI, RGA and SCR, the first three of its members isolated. They are important regulatory components in a number of different cellular processes ranging from meristem maintenance, signal transduction to hormone signaling (Bolle 2004). It has been suggested that GA-dependent degradation of DELLA proteins is a key event for GA-signaling (Ueguchi-Tanaka et al. 2007). DELLA protein is rapidly degraded by GA through ubiquitination is reported in several organism, such as rice, Arabidopsis and barley (Sasaki et al. 2003; Silverstone et al. 2001; Itoh et al. 2002). The disappearance of SmDELLA1 protein after GA4 treatment is reported in selaginella moellendorffii also (Hirano et al. 2007). DELLA protein degradation causes derepression of the repression state of GA action. The processing of the GA signal in the nucleus depends directly on the presence or absence of DELLA proteins, which are therefore presently considered to be a ‘molecular switch’ for GA signaling (Gomi and Matsuoka 2003). The rice genome contains one DELLA protein ortholog, OsGAI/SLR1. The slr1 mutant is a recessive mutation and phenocopies a constitutive GA response (Ogawa et al. 2000; Ikeda et al. 2001). It has been postulated that DELLAs function as transcriptional repressors of genes involved in GA signaling (Olszewski et al. 2002). Taken together, bioactive GA promotes growth by turning on de-DELLA-repressing system. Conversely, if the bioactive GA is reduced, the degradation of DELLA protein will slow down or not at all, and the plant growth will be inhibited. This might be the reason why so many semi-dwarfs appear in the mutants where bioactive GA is reduced.
HD-Zip proteins constitute a large family of transcription factors and several lines of evidence have also shown that HD-Zip proteins play roles in regulating plant responses to hormones (Sawa et al. 2002; Johannesson et al. 2003; Rosin et al. 2003; Hay et al. 2004). Rice OsHox4 is a drought-repressed HD-Zip family I gene act as activators and express predominantly in vascular (Agalou et al. 2008, Meijer et al.2000).Loss-of-function analysis of OsHox4 reveals no specific phenotype, but over-expression of OsHox4 displayed a prolonged vegetative phase and reduced internodes elongation; the flowering time of OsHox4 overexpression lines is variant from as normal as wild type to no flowering (Agalou et al. 2008).
The objectives of this research are: (1) to figure out the mechanism of semi-dwarf caused by OsHox4 overexpression; (2) confirm the role of OsHox4 in GA metabolism pathway; and (3) to further explore the GA signaling pathway in rice if it is possible. Our results showed that OsHox4 involves in GA metabolism because the semi-dwarf phenotype in OsHox4 transgenic rice could be fully eliminated by applied GA. A further research provided evidence that OsHox4 regulate GA signaling and plant morphogenesis by interacting with DELLA-like subfamily genes and GA 2, 3-oxidasze family genes.
Materials and methods
Crop husbandry and sampling
Seeds of rice (Oryza sativa L. cv. IR64, IRTP A004) were obtained from the International Network for the Genetic Evaluation of Rice, International Rice Research Institute (IRRI), Philippines. Plants were grown individually in pots containing clay loam soil under glasshouse conditions with natural sun light and photoperiod. The tissue samples were frozen immediately in liquid nitrogen and stored at −80°C for future RNA extraction.
Rice transformation
The binary vector for over-expression was constructed based on pCAMIA-1301 (CAMBIA, Genbank accession number: AF234297) and pRTL2 (Mason et al. 1992). A HindIII fragment with a double CaMV 35S enhancer/promoter and the nopaline synthase gene terminator from pRTL2 was inserted into pCAMBIA-1301. The resulting vector was named as p1301DS. For the over-expression of OsHox4, the full-length cDNA was inserted into p1301DS digested with BamHI and KpnI. The PCR fragments were sequenced and inserted into pDS1301. The constructs were introduced into rice plants by Agrobacterium tumefaciens (strain EHA105)-mediated transformation following modified protocol of Hiei and Komari (2006).
Growth regulators treatments
An amount of 10 and 30 μM gibberellin acid (GA4, Sigma) was sprayed to transgenic rice at heading stage on 35 s::OsHox4 transgenic plants. The treatments were repeated every 2 days for 3 times. The length of the internodes was measured 7 days after the beginning of the treatments. For RNA extraction and quantitative RT-PCR analysis, the flag leaves were collected 24 h after GA4 spraying.
Light microscopy
2nd internodes (which are beneath peduncle) are used to compare the stem cell elongation. Stems were harvested from wild type, OsHox4 transgenic plant and 7 days after GA4 treatment OsHox4 transgenic plants. The microtome sections (10 μm) were mounted on glass slides for imaging.
Statistical analysis
Analyses of variances (ANOVA) were made with the General Linear Model Procedure of SPSS statistical software version 16.0 (IBM SPSS Statistics 2007). ANOVA was used to determine the treatment effects on each variable and confidence intervals derived from these analysis used to establish significant effects. Correlation analysis and regression analysis was performed by SPSS statistical software version 16.0 (IBM SPSS Statistics 2007) also.
RNA isolation and real-time quantitative PCR analysis
Total RNA was extracted from flag leaves of Null (wild type IR64) and transgenic plants (IR64-OsHox4-014, IR64-OsHox4-027, IR64-OsHox4-022) 24 h after different concentration GA4 applied using RNA extracting kit (TRIzol reagent, Invitrogen). 4 µg total RNA was pre-treated with RNase-free DNase I (Invitrogen), according to the manufacturer’s instruction. RT reaction was performed with SuperScriptTM II reverse transcriptase (Invitrogen) following the manufacturer’s instructions. Primers used for real-time PCR are listed in Supplementary 1. Real-time PCR was performed in an optical 96-well plate with an ABI PRISM® 7500 Real-time PCR System (Applied Biosystems) by using SYBR® Green to monitor dsDNA synthesis. All reactions contained 10 μl 2×SYBR® Green Master Mix Reagent (Applied Biosystems), 2.0 ng cDNA and 200nM of each gene-specific primer in a final volume of 20 μl. Thermal cycling was as follows: 50 °C for 2 min; 95 °C for 10 min; 50 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min. Relative expression level of reporter and target genes was determined based on the 2(-Delta Delta C(T)) method (Livak and Schmittgen 2001) by using rice GAPDH as internal control. Transcript levels from Null (wild type) were set at 1.
Result
Overexpression of OsHox4 induced semi-dwarf plants along with increased tiller number
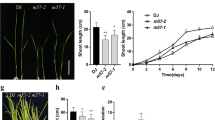
Rice HD-ZIP I family gene OsHox4 was overexpressed in IR64 under CaMV 35s promoter, which induced shorter plant height than Null (wild type of IR64, as a control) with ranging from a very strong dwarf phenotype (with essentially no stem) to semi-dwarf or wild type phenotype (Figs. 1, 2). The average plant height of Null was 100.96 cm; the average height of three OsHox4 overexpression lines, IR64-OsHox4-014, IR64-OsHox4-027 and IR64-OsHox4-022 was 48.667, 52.786 and 62.643 respectively (Table 1). ANOVA (Analyses of variances) analysis results showed that the plant height of three OsHox4 overexpression transgenic lines, IR64-OsHox4-027, IR64-OsHox4-022 and IR64-OsHox4-014 were significantly shorter than Null (Table 1). The difference of plant height between transgenic line IR64-OsHox4-027 and IR64-OsHox4-014 was not significant (Table 1; Fig. 2). The difference of plant height between transgenic line IR64-OsHox4-027 and IR64-OsHox4-022 was not significant also (Table 1; Fig. 2).The plant height of transgenic line IR64-OsHox4-014 was significant shorter than transgenic line IR64-OsHox4-022, the P value < 0.0001 (Table 1; Fig. 2).
Comparison of plant height and tiller number among Null (wild type) and OsHox4 transgenic plants. Comparison of plant height between OsHox4 transgenic plant and Null showed a repression of plant height in OsHox4 overexpression plants (a); comparison of tiller number between OsHox4 transgenic plant and Null (wild type) showed a increasing of tiller number in OsHox4 overexpression plants (b). Data are mean ± SD (standard error) (n ≥ 5 biological repeats)
Compare to wild type, over expression of rice OsHox4 gene induced more tillers also. The average tiller number of Null was 14.80; the average tiller number of three OsHox4 overexpression lines, IR64-OsHox4-014, IR64-OsHox4-027 and IR64-OsHox4-022 was 26.00, 22.357 and 21.571 respectively. ANOVA analysis results showed that the tiller numbers in 3 single copy OsHox4 overexpression transgenic lines, IR64-OsHox4-027, IR64-OsHox4-014 and IR64-OsHox4-022, were significantly higher than in wild type plants (Table 1; Fig. 2). The tiller numbers among the OsHox4 overexpression transgenic lines were not significant (Table 1).
Exogenous GA4 eliminates the semi-dwarf stature of OsHox4 transgenic plant
In order to investigate the causing of OsHox4 induced semi-dwarf and the role of OsHox4 in plant development, 10 and 30 μM exogenous GA4 was applied on OsHox4 transgenic rice at heading stage, the semi-dwarf phenotype which induced by the overexpression of OsHox4 was reversed in different extent (Fig. 3).
Exogenous GA4 eliminate the plant height repression of OsHox4 overepxression plant. Comparison of OsHox4-overexpressing plants treated with GA4 at different concentrations showed GA4 could eliminate the plant height repression of OsHox4 transgenic rice. OsHox4 overexpressing plants were treated with water (control) (a), 10 μM exogenous GA4 (b) and 30 μM exogenous GA4(c) showed different levels increase in plant height. This confirmed OsHox4 involved in GA deactivation in OsHox4 transgenic plant
Further investigations showed that the internodes of OsHox4 transgenic rice plant increased differently after exogenous GA4 applied. Compare to OsHox4 overexpression transgenic plants without GA4 treatment, 7 days after 30 μM exogenous treatment at heading stage, the 1st internodes (peduncle) was 2 times the length as those without GA4 treatment plants, which was the same as average peduncle length of Null; the average length of 2nd internodes (the internodes beneath peduncle) increased by 5 times long as those without GA4 treated OsHox4 overexpression transgenic plants, which was 2 times higher than Null. But the elongation of 3rd, 4th and 5th internodes was not significant faster than those in OsHox4 overexpression transgenic plants without GA4 treatment (Fig. 4). It showed GA4 could increase internodes elongation only if they were at elongation stage. These results confirmed exogenous GA4 could reverse the depression of stem elongation induced by OsHox4 overexpression and the elongation degree of internodes depends on their development stage while exogenous GA4 were applied.
Comparison of different internodes in Null (wild type) and OsHox4 transgenic plant. Comparison of internodes length among Null without GA4 treatment (blue column), OsHox4 overexpression plants without GA4 treatment (green column) and OsHox4 overexpression plant 7 days after 30 μM GA4 treatment (red column) showed the internodes elongation increasing by exogenous GA4 depending on plant growth stage. 1st internodes is peduncle, 2nd is the one beneath peduncle, so forth for 3rd, 4th, and 5th internodes. The average values were calculated from measures of at least ten plants ± SD (SE). (Color figure online)
A comparison of longitudinal section of 2nd internodes between Null and OsHox4 overexpressing plant without GA4 treatment showed overexpression of OsHox4 reduced plant height by inhibiting stem cell elongation (Fig. 5); and comparison between OsHox4 overexpression transgenic plants with 30 μM GA4 treatment and without GA4 treatment showed the exogenous GA4 eliminated the semi-dwarf stature of OsHox4 transgenic plant by increasing the stem cell elongation (Fig. 5). Therefore, over-expression of OsHox4 induced semi-dwarf by repressing stems cell elongation and application of exogenous GA4 could reverse dwarf phenomena by also eliminating this repression.
Longitudinal section comparison of Null (wild type), OsHox4 transgenic plant and GA4 applied OsHox4 transgenic plant. A comparison of longitudinal section of 2nd internodes beneath peduncle among Null (a), OsHox4 overexpressing plant treated with 50 ml 30 μM exogenous GA4 (b) and OsHox4 overexpressing plant without GA4 treatment (c) showed overexpression of OsHox4 reduced plant height by inhibiting stem cell elongation (c); application of exogenous GA4 could eliminate this repression (b). Null without GA4 treatment was used as control (a)
Expression analysis of rice GA 3-oxidase and GA 2-oxidase family genes
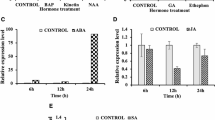
To investigate the molecular mechanism of OsHox4 on GA metabolism and characterize the role of OsHox4 in GA signaling, we checked the expression of 2 rice GA 3-oxidase family genes (OsGA3ox1 and OsGA3ox2), and 9 rice GA 2-oxidase family genes (OsGA2ox1, 2, 3, 4, 5, 6, 7, 8 and 9) in flag leaves of Null and OsHox4 overexpression transgenic plants which were treated by different concentration levels exogenous GA4 after 24 h by qRT-PCR. Our qRT-PCR results showed, compare to Null, the expressions of two rice GA 3-oxidase family genes, OsGA3ox1 and OsGA3ox2, were depressed in OsHox4 overexpression transgenic plants (Fig. 6). The average relative expression of OsGA3ox1 and OsGA3ox2 were lower to 16 and 29 % of their expression in Null respectively (Fig. 6). 24 h after different 50 ml 10 µm GA4 treatments, the expression of OsGA3ox1 upregulated to almost the same level as in null and the expression of OsGA3ox2 upregulated to 25 % higher than in Null (Fig. 6). After 30 μM GA4 treatment, the relative expression of OsGA3ox1 did not do change too much but the expression of OsGA3ox2 in 30 μM GA4 applied plant was lower to 45 % of its expression in 10 μM GA4 applied plants (Fig. 6). It showed the OsGA3ox2 was subjected to GA feedback.
Real-time PCR analysis of 2 rice GA 3-oxidase family genes. Transcripts of 2 rice GA 3-oxidase family genes, OsGA3ox2 and OsGA3ox2, in Null (wild type) and three OsHox4-over-expressing lines (IR64-OsHox4-027, IR64-OsHox4-022, IR64-OsHox4-017) were quantified by real time RT-PCR. cDNA samples were derived from flag leaves of Null and OsHox4 transgenic rice plants in different GA treatments. Data are mean ± SD (SE) (n = 3 biological repeats)
In rice GA 2-oxidase family, the expression of OsGA2ox4, 5, 6, 8, 9 in OsHox4 transgenic plant was differently lower than in Null, from 22 to 62 %. After 10 μM GA4 treatment, their expressions were highly up-regulated in OsHox4 transgenic plants. In 30 μM GA4 treated plant, the expressions of OsGA2ox3, 4, 5, 8 and 9 were decreased (Fig. 7).
Real-time PCR analysis of 7 rice GA 2-oxidase family genes. Transcripts of 7 GA 2-oxidase family genes in Null (wild type) and three OsHox4-over-expressing lines (IR64-OsHox4-027, IR64-OsHox4-022, IR64-OsHox4-006) were quantified by real time RT-PCR. cDNA samples were derived from flag leaves of Null and OsHox4 transgenic rice plants in different GA treatments. Data are mean ± SD (SE) (n = 3 biological repeats)
From our qRT-PCR analysis, the expressions of OsGA3ox1, OsGA3ox2, OsGA2ox4, 5, 6, 8 and 9 were down-regulated in OsHox4 transgenic rice; 10 μM GA4 treatment could induce their expression to a level higher than in Null, but 30 μM GA4 treatment could inhibit their expression in some extent (except OsGA2ox6, its expression still up-regulated after 30 μM GA treatment, that showed 30 μM GA4 treatment could not reach its responding threshold) (Figs. 6, 7). All of these showed the expressions of these genes were mainly controlled by active GA in vivo. Rice OsHox4 could regulate their expression indirectly by active GA level in plant.
The expression of OsGA2ox2 and OsGA2ox3 was 2.7 times and 1.6 times higher in OsHox4 overexpression transgenic plants than in Null respectively and was highly upregulted in 10 μM GA4 treated plants, 11 times and 42 times higher than in Null respectively. After 30 μM GA4 treatment, the expression of OsGA2ox3 was down-regulated a little bit, but the expression OsGA2ox2 was still up-regulated (Fig. 7). The expression analysis showed OsGA2ox2 and OsGA2ox3 were regulated by both of OsHox4 and active GA.
There was no expression of OsGA2ox1 and OsGA2ox7 in both of Null plants and OsHox4 transgenic plants.
Expression analysis of rice DELLA protein subfamily genes
The expressions of 5 rice DELLA protein subfamily genes OsGAI, OsSCR, OsSCRL, OsSCL14 and OsRGA1 were investigated in Null and GA4 applied transgenic plants. qRT-PCR results showed that the expression of rice DELLA protein gene OsGAI in OsHox4 transgenic plants was as low as 51 % in Null but up-regulated to 1.8 times as its in Null after 50 ml 10 μM GA4 applied. In 30 μM treated plants, the expression of OsGAI was down-regulated to lower than in 10 μM GA4 treated plants and Null (Fig. 8). It showed the expression of OsGAI was affected by active GA in plant. Compare to Null, the expression of OsSCR up-regulated in OsHox4 overexpression transgenic plants and was further increased in 10 μM GA4 treated transgenic plant (Fig. 8). It showed both of OsHox4 and exogenous GA4 had a positive role in its expression. Comparing to wild type, the expression of OsRGA1 and OsSCRL were down-regulated in OsHox4 transgenic plants but up-regulated in OsHox4 transgenic plants which were treated by 10 μM GA4 after 24 h. The expressions of all of these 4 genes (OsGAI, OsSCR, OsSCRL and OsRGA1) in 30 μM GA4 treated transgenic plant were lower than their expressions in 10 μM GA4 treated plants. It showed a GA feedback mechanism involved in their expression. Compare to Null, the expression of OsSCL14 was down-regualted in OsHox4 transgenic plant, application of GA4 intensified this down-regulation (Fig. 8). It showed Oshxo4 was a negative factor in expression of OsSCL14; GA4 could not eliminate the depression of its expression. The molecular mechanism needs a further investigation.
Real-time PCR analysis of 5 rice DELLA protein subfamily genes. Transcripts of 5 rice DELLA protein subfamily genes in Null (wild type) and three OsHox4-over-expressing lines (IR64-OsHox4-027, IR64-OsHox4-022, IR64-OsHox4-006) were quantified by real time RT-PCR. cDNA samples were derived from flag leaves of Null and OsHox4 transgenic rice plants in different GA treatments. Data are mean ± SD (SE) (n = 3 biological repeats)
Discussion
Rice OsHox4 gene regulate the GA metabolism and signaling
The semi-dwarf which is produced by OsHox4 over-expression indicates OsHox4 has a role of GA deactivation or/and GA signal response (Fig. 3). Expression analysis shows rice OsHox4 gene involves in repression GA response by rice GRAS family genes and regulates the GA metabolism by GA 2-oxidase family and GA 3-oxidase family genes (Figs. 6, 7, 8). Our results confirmed that GA is deactivated in OsHox4 transgenic plant because the exogenous GA4 can eliminate the semidwarf statures which are caused by OsHox4 over-expression (Fig. 3).
It is well know that rice GA3-oxidases are response for GA activation (Hartweck, 2008). From our expression analysis, the down-regulation of rice GA3-oxidase genes, OsGA3ox1 and OsGA3ox2, in OsHox4 transgenic plants implies the OsHox4 deactivate GA by inhibiting the expression of OsGA3-oxidases (Fig. 6). Except for rice GA3-oxidase genes, the expression analysis result shows OsGA2ox2 and OsGA2ox3 are regulated by OsHox4 also. The upregulation of GA deactivation factors OsGA2ox2 and OsGA2ox3, together with down regulation of GA biosynthesis factors OsGA3ox1 and OsGA3ox2, implies OsHox4 controls GA metabolism at biosynthesis and deactivation two directions.
It has been suggested that GA-dependent degradation of DELLA proteins is a key event for GA-signaling (Ueguchi-Tanaka et al. 2007). The processing of the GA signal in the nucleus depends directly on the presence or absence of DELLA proteins, which are therefore presently considered to be a ‘molecular switch’ for GA signaling (Gomi and Matsuoka 2003). But the direct targeted upstream and downstream components of DELLA proteins are still unknown. Promoter analysis based on PLACE database which is described by Higo et al. (1999) showed that except the OsGA3ox2, all of the genes analyzed in this paper have homeodomain binding sites (Supplementary 2). But only the expressions of OsGA2ox2 and OsGA2ox3 are upregulated in OsHox4 transgenic plant (Figs. 6, 7, 8), it implies OsHox4 directly/indirectly regulates a complex GA signaling network. Comparing to application of 10 μM GA4, the expressions of most genes are lower after 30 μM GA4 applied (Figs. 6, 7, 8), which hints a GA feed-back mechanism in controlling these genes at transcription level.
Directly/indirectly regulating the expression of rice GA3 oxidase family, GA2 oxidase family and GRAS family genes implies the complex role of OsHox4 in rice development. An extended biological and biochemical analysis will help to find a common mechanism of function, if it exists. Determining the specific roles of these proteins in response to developmental and environmental signals, as well as their cross-talk with other pathways, is an important future task.
Dai et al. (2008) report that the semidwarf induced by over-expression of OsHox4 could not be complemented by applied GA3 at seedling stage. Our result showed clearly that exogenous GA4 could reverse the semidwarf phenotype at different stem elongation stages (Figs. 3, 4, 5). It confirms our conclusion that the exogenous GA4 only works on the cells which are at elongation stage. All of these hints GA4 alone is not enough to increase cell elongation, it should work together with other factors which started the internodes cells elongation and reverse the semidwarf.
They also observed that the over-expression plants accumulated elevated levels of bioactive GA, while the GA catabolic gene OsGA2ox3 was upregulated in the transgenic plants (Dai et al. 2008). In some extent, the upregulation of OsGA2ox3 is contradicted with the elevation of bioactive GA in over-expression plants. Our results showed the OsHox4 control GA signaling in two directions, one was disturbing the bioactive GA metabolism by controlling the expression of GA 2-oxidase and GA 3-oxidase genes; another was disturbing the GA signaling through DELLA subfamily genes. The accumulation of bioactive GA in OsHox4 transgenic plants implies the OsHox4 mainly control GA activity by involving in GA signaling pathway. The related mechanism needs to be confirmed and unraveled by a further investigation.
Rice OsHox4 involves in rice morphogenesis by hormones coordination (372 words)
Dwarfism is one of the most valuable traits in crop breeding because semi-dwarf cultivars are more resistant to damage by wind and rain (lodging resistant) and are associated with stable increased yields (Evans 1993). Overexpression of OsHox4 can not only reduce the plant height but can increase the tiller number at same time (Fig. 1; Table 1). Further investigation shows exogenous GA4 can eliminate the semi-dwarf stature of OsHox4 transgenic plant that means the GA metabolism is deactivated in OsHox4 transgenic plant.
It has been long known that there is cross-talking between GA action and other hormones signaling to control the plant growth and development, but the mechanism remains unclear. It is reported that auxin can positively affect GA biosynthesis also, and GA signaling through DELLA proteins is mediated by auxin (Fu and Harberd 2003). Over 95 % of the transcriptional changes associated with GA application were included among the changes induced by IAA application in populus and arabidopsis (Covington and Harmer 2007, Bjorklund et al. 2007).IR64 OsHox4 transgenic plant has more tillers and short stature probably because the IAA which is repressed together with GA. Since the GA can positively affect auxin biosynthesis (Nemhauser et al. 2006; Desgagne-Penix and Sponsel 2008), the active GA is depressed in OsHox4 transgenic rice which causes a lower level of IAA in OsHox4 transgenic plant than in wild type plant. The lower level of active GA is the reason of semi-dwarf in OsHox4 transgenic plant; and the lower level of IAA is the reason of multiple tillers in OsHox4 transgenic plant because the elimination of apical dominance when IAA level is low. Fig. 9 is an illustration of discussion in above.
Conclusion
HD-ZIP family gene OsHox4 is over-expressed in IR64 under the control of CaMV 35 s promoter; 10uM and 30uM GA4 application result showed that the exogenous GA can rescue the semi-dwarf caused by OsHox4 over-expression; internodes section result showed that the cell length was increased after GA spraying. The expression of rice DELLA protein subfamily genes, rice GA 3-oxidase family genes and rice GA 2-oxidase family genes were checked by qRT-PCR; our result showed OsHox4 impaired the biosynthesis of active GA by down-regulating rice GA 3-oxidase family genes and deactive GA by upregulating rice GA 2-oxidase family genes. OsHox4 also involves in repression GA response by rice GRAS family genes.
References
Agalou A, Purwantomo S, Overnäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, Wang M, Xiong L, Meijer AH, Ouwerkerk PB (2008) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66:87–103
Bjorklund S, Antti H, Uddestrand I, Moritz T, Sundberg B (2007) Cross-talk between gibberellin and auxin in development of populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J 52:499–511
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5:1773–1784
Dai M, Hu Y, Ma Q, Zhao Y, Zhou DX (2008) Functional analysis of rice HOMEOBOX4 (OsHox4) gene reveals a negative function in gibberellin responses. Plant Mol Biol 66:289–301
Desgagne-Penix I, Sponsel VM (2008) Expression of gibberellin 20-oxidase1 (AtGA20ox1) in Arabidopsis seedlings with altered auxin status is regulated at multiple levels. J Exp Bot 59:2057–2070
Evans LT (1993) Crop evolution, adaptation and yield. Cambridge University Press, Cambridge
Fleet CM, Sun T (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8:77–85
Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421:740–743
Gomi K, Matsuoka M (2003) Gibberellin signalling pathway. Curr Opin Plant Biol 6:489–493
Hartweck LM (2008) Gibberellin signaling. Planta 229:1–13
Hay A, Craft J, Tsiantis M (2004) Plant hormones and homeoboxes: bridging the gap? Bioessays 26:395–404
Hiei Y, Komari T (2006) Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 85:271–283
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hirano K, Nakajima M, Asano K et al (2007) The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19:3058–3079
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell, 13:999–1010
Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin Ssignaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57–70
Johannesson H, Wang Y, Hanson J, Engström P (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol Biol 51:719–729
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Mason HS, Lam DM, Arntzen CJ (1992) Expression of hepatitis B surface antigen in transgenic plants. Proc Natl Acad Sci 89:11745–11749
Meijer AH, de Kam RJ, d’Erfurth I, Shen W, Hoge JH (2000) HD-Zip proteins of families I and II from rice: interactions and functional properties. Mol Gen Genet 263:12–21
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–477
Ogawa M, Kusano T, Katsumi M, Sano H (2000) Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245:21–29
Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism and response pathways. Plant Cell 14:61–80
Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ (2003) Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol 132:106–117
Sakamoto T, Miura K, Itoh H et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134:1642–1653
Sasaki A, Itoh H, Gomi K et al (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299:1896–1898
Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32:1011–1022
Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-P (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13:1555–1566
Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plant. Annu Rev Plant Biol 55:197–223
Thomas SG, Rieu I, Steber CM (2005) Gibberellin metabolism and signaling. Vitam Horm 72:289–338
Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58:183–198
Acknowledgments
The present research was financially supported by CGIAR (Consultative Group on International Agricultural Research) HarvestPlus Challenge Program (Project Number M013260-001). The authors are obliged to the anonymous reviewers and editors who we would also like to thank for their thoughts, comments and suggestions.
Conflict of interest
Authors declared that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, W., Malabanan, P.B. & Abrigo, E. OsHox4 regulates GA signaling by interacting with DELLA-like genes and GA oxidase genes in rice. Euphytica 201, 97–107 (2015). https://doi.org/10.1007/s10681-014-1191-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1191-4