Abstract
Diploid and triploid intergeneric hybrids obtained by crosses among Gloriosa superba ‘Lutea’ (2n = 2x = 22), G. ‘Marron Gold’ (2n = 4x = 44), Littonia modesta (2n = 2x = 22), and Sandersonia aurantiaca (2n = 2x = 24) were analyzed for their meiotic chromosome pairing in pollen mother cells by genomic in situ hybridization (GISH) with digoxigenin-labeled total DNA of one parent as probe. Chromosomes from each parent could be clearly distinguished in pollen mother cells of all the five intergeneric hybrids by GISH. For three diploid hybrids, L. modesta × G. superba ‘Lutea’ (2n = 2x = 22), L. modesta × S. aurantiaca (2n = 2x = 23) and S. aurantiaca × G. superba ‘Lutea’ (2n = 2x = 23), 0.04−0.27 autosyndetic bivalents (intragenomic pairing of non-homologous chromosomes) and 0.13−0.36 allosyndetic bivalents (intergenomic chromosome pairing) were observed per pollen mother cell, indicating that there are some homologous chromosomal regions within each genome and among the genomes of Gloriosa, Littonia and Sandersonia. Differences in the average number of allosyndetic bivalents per pollen mother cell among different genome combinations may reflect the evolutionary distances among the three genera, and Gloriosa and Littonia may be closely related to each other, while Sandersonia may have relatively distant relationships with Gloriosa and Littonia. For two triploid hybrids, L. modesta × G. ‘Marron Gold’ (2n = 3x = 33) and S. aurantiaca × G. ‘Marron Gold’ (2n = 3x = 34), no allosyndetic bivalents were observed. Based on the results obtained in the present study, possible utilization of the diploid and triploid intergeneric hybrids for further breeding of colchicaceous ornamentals is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
GISH analysis is a powerful tool for discriminating chromosomes from different genomes and allows identification of the genome constitution in allopolyploids (Lim et al. 2003; Pendinen et al. 2012) and wide hybrids (Ji et al. 2004; Marasek et al. 2004). In addition, GISH analysis is one of the most effective means to study intra- or intergenomic relationships by visualizing intra- or intergenomic chromosome pairing during meiosis of allopolyploids and wide hybrids. Cao et al. (2000) observed a high chromosome pairing affinity between Lolium perenne and Festuca mairei by GISH analysis and demonstrated that chromosomes of Lolium and Festuca may be genetically equivalent and reciprocal mixing of the genomes may be possible. Ge and Li (2007) investigated intragenomic chromosome homology in the B genome of Brassica nigra and their homoeology with chromosomes of the A genome of B. rapa and the C genome of B. oleracea in trigenomic triploid hybrids of different origins by GISH analysis and provided an evidence for the hypothesis that the three basic diploid genomes of the cultivated Brassica species evolved from one common ancestral genome with a lower chromosome number. Furthermore, Yao et al. (2010) analyzed trigenomic triploid hybrids among B. juncea, B. carinata and B. maurorum by dual-color GISH and concluded that intergenomic homoeology was higher than intragenomic homology in Brassica.
Gloriosa spp., Littonia modesta and Sandersonia aurantiaca are tuberous plants belonging to the family Colchicaceae and cultivated as ornamental plants because of their beautiful, unique flowers and good vase life (Nakamura et al. 2005). In order to obtain wide variability in horticultural traits and to develop novel cultivars, we have tried intergeneric hybridization among these plants and produced a number of hybrid plants in various combinations via ovule culture (Kuwayama et al. 2005; Nakamura et al. 2005; Amano et al. 2007, 2008, 2009). All the intergeneric hybrids were clearly distinguishable from the corresponding parents and had novel morphological characteristics, some of which were horticulturally attractive.
In a preliminary study, we examined chromosome behavior during meiosis in pollen mother cells (PMCs) of some diploid intergeneric hybrids among G. superba, L. modesta and S. aurantiaca by acetic-orcein staining (Amano 2008). Although a few bivalents were observed at metaphase I in all the hybrids, it is unclear whether these bivalents are autosyndetic (intragenomic pairing of non-homologous chromosomes) or allosyndetic (intergenomic chromosome pairing). Recently, Nakazawa et al. (2011) successfully applied GISH analysis for identifying the genome constitution of some intergeneric hybrids of colchicaceous ornamentals. In the present study, therefore, we analyzed meiotic chromosome pairing in diploid and triploid intergeneric hybrids by GISH in order to obtain some information on the intergenomic relationships among Gloriosa, Littonia and Sandersonia.
Materials and methods
Plant materials
The parents of intergeneric hybrids, G. superba‘Lutea’ (Gsu; 2n = 2x = 22), G. ‘Marron Gold’ (Gma; 2n = 4x = 44), L. modesta (Lit; 2n = 2x = 22) and S. aurantiaca (Sau; 2n = 2x = 24), diploid intergeneric hybrids, L. modesta × G. superba ‘Lutea’ (Lit × Gsu-2; 2n = 2x = 22), L. modesta × S. aurantiaca (Lit × Sau-1; 2n = 2x = 23) and S. aurantiaca × G. superba ‘Lutea’ (Sau × Gsu-1; 2n = 2x = 23), and triploid intergeneric hybrids, L. modesta × G. ‘Marron Gold’ (Lit × Gma-1; 2n = 3x = 33) and S. aurantiaca × G. ‘Marron Gold’ (Sau × Gma-6; 2n = 2x = 34) (Kuwayama et al. 2005; Amano et al. 2007, 2008, 2009), were used in the present study. All plants were cultivated in the greenhouse according to Amano et al. (2008).
Chromosome preparation
Young anthers were collected one week before anthesis. They were pre-treated with 0.5 % (w/v) amiprophos-methyl (Hayashi Pure Chemical Industries, Osaka, Japan) in water for 3 h at room temperature (20−25 °C), fixed in a 3:1 (v/v) mixture of absolute alcohol and glacial acetic acid, and stored at −20 °C. Chromosome preparations for GISH were made according to Nakazawa et al. (2011). Anthers were washed with distilled water for 10 min, cut into ca. 5 mm square pieces, and digested in an enzyme mixture containing 0.6 % (w/v) Cellulase Onozuka RS (Yakult Honsha, Co., Tokyo, Japan), 0.3 % (w/v) Pectolyase Y-23 (Seishin, Co., Tokyo, Japan) and 0.5 % (w/v) Macerozyme R-200 (Yakult Honsha, Co., Tokyo, Japan) for 80 min at 37 °C. PMCs were spread out in a drop of a 3:1 (v/v) mixture of absolute alcohol and glacial acetic acid. Prepared slides were air dried and stored at −20 °C.
Probe preparation and GISH analysis
Total genomic DNA was isolated from young leaves by the cetyltrimethylammonium bromide (CTAB) method (Rogers and Bendich 1985). Genomic DNA of Lit and Sau was labeled with digoxigenin-dUTPs by using the DIG Nick Translation Mix (Roche Diagnostics GmbH, Mannheim, Germany) and used as a probe.
In situ hybridization was carried out according to Nakazawa et al. (2011). Standard stringency conditions in GISH were applied to distinguish chromosomes of each genome according to Ji et al. (2004). Total DNA of one parent was used as a probe and 50-fold excess of salmon sperm DNA (Funakoshi, Co. Ltd., Tokyo, Japan) was used instead of blocking DNA. Hybridization signals of the probe were detected using anti-digoxigenin-rhodamine (Roche Diagnostics GmbH, Mannheim, Germany). Chromosomes were counterstained with 1 % (w/v) 4′-6-diamino-2-phenylindole (DAPI) in the antifade solution (Vector Laboratories, Inc., CA, USA). Chromosomes were examined under a fluorescent microscope (OPTIPHOT-2 9 2F EFD2, Nikon Corp., Tokyo, Japan) equipped with a CCD camera (VB-6010, Keyence Corp., Osaka, Japan). One hundred and 50 PMCs were observed for diploid and triploid intergeneric hybrids, respectively. Images were processed by the Photoshop Element 10 (Adobe Systems Inc., CA, USA). Differences in the average number of allosyndetic bivalents among different diploid intergeneric hybrids were determined by t test according to Molnár and Molnár (2010).
Results
Chromosomes from each parent could be clearly distinguished in PMCs at metaphase I of meiosis by GISH for all the five intergeneric hybrids analyzed in the present study.
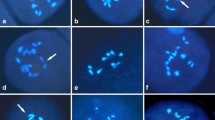
For three diploid hybrids, Lit × Gsu-2, Lit × Sau-1 and Sau × Gsu-1, most PMCs contained only univalents, but both allosyndetic and autosyndetic bivalents were sometimes observed in each hybrids. In Lit × Gsu-2, allosyndetic bivalents of Gloriosa (G)−Littonia (L) genomes (Fig. 1A) were formed with an average of 0.36 per PMC corresponding to 1.70 % of the total number of chromosome conformations, while autosyndetic bivalents of G and L genomes were formed with average numbers of 0.26 and 0.27 per PMC, respectively (Table 1). In Lit × Sau-1, an average number of allosyndetic bivalents between L−Sandersonia (S) genomes (Fig. 1B) was 0.13 per PMC corresponding to 0.57 % of the total number of chromosome conformations, while average numbers of autosyndetic bivalents of L and S genomes were 0.05 and 0.22 per PMC, respectively (Table 1). In Sau × Gsu-1, an average number of allosyndetic bivalents of G−S genomes (Fig. 1C) was 0.18 per PMC corresponding to 0.79 % of the total number of chromosome conformations, while average numbers of autosyndetic bivalents of G and S genomes were 0.04 and 0.23 per PMC, respectively (Table 1).
GISH analysis of PMCs during meiosis of intergeneric hybrids of colchicaceous ornamentals. A Lit × Gsu-2: univalents of Littonia probed with digoxigenin-dUTPs and detected with anti-digoxigenin-rhodamine, were red (arrowheads). Double-arrowhead indicates an allosyndetic bivalent of Littonia−Gloriosa genomes. B Lit × Sau-1: univalents of Sandersonia probed with digoxigenin-dUTPs and detected with anti-digoxigenin-rhodamine, were red (arrowheads). Double-arrowhead indicates an allosyndetic bivalent of Littonia−Sandersonia genomes. C Sau × Gsu-1: univalents of Sandersonia probed with digoxigenin-dUTPs and detected with anti-digoxigenin-rhodamine, were red (arrowheads). Double-arrowhead indicates an allosyndetic bivalent of Gloriosa−Sandersonia genomes. D Lit × Gma-1: univalents of Littonia probed with digoxigenin-dUTPs and detected with anti-digoxigenin-rhodamine, were red (arrowheads). E Sau × Gma-6: univalents of Sandersonia probed with digoxigenin-dUTPs and detected with anti-digoxigenin-rhodamine, were red (arrowheads). Bars 10 µm. (Color figure online)
Table 2 shows the results of t test describing differences in the average number of allosyndetic bivalents among different diploid hybrids. The average number of allosyndetic bivalents in Lit × Gsu-2 (G−L genomes) was significantly higher than those in Sau × Gsu-1 (G−S genomes) and in Lit × Sau-1 (L−S genomes) at P = 0.05 and P = 0.01 levels, respectively. No significant difference in the average numbers of allosyndetic bivalent was observed between Lit × Sau-1 (L−S genomes) and Sau × Gsu-1 (G−S genomes).
For two triploid hybrids, Lit × Gma-1 and Sau × Gma-6, no allosyndetic bivalents were observed in PMCs at metaphase I of meiosis. Most PMCs of Lit × Gma-1 contained only 11 univalents of L genome and 11 bivalents of G genome (Table 3; Fig. 1D). Similarly, only 12 univalents of S genome and 11 bivalents of G genome were observed in most PMCs of Sau × Gma-6 (Table 3; Fig. 1E). In both hybrids, univalents of G genome were sometimes observed.
Discussion
The success to distinguish different genomes by GISH in allopolyploids and wide hybrids largely depends on the sequence homology (Ji et al. 2004). Genomes sharing 80–85 % or less sequence homology can generally be discriminated by standard GISH conditions (Schwarzacher et al. 1989). On the other hand, increased stringency conditions in combination with an excess of blocking DNA are required to discriminate genomes sharing up to 90−95 % of sequence homology (Parokonny et al. 1997). In our previous study on GISH analysis of mitotic chromosomes, parental chromosomes in colchicaceous intergeneric hybrids could be clearly discriminated under standard stringency conditions without an excess of blocking DNA (Nakazawa et al. 2011). Similar GISH conditions also allowed clear discrimination of parental chromosomes in PMCs at metaphase I of meiosis in the present study, indicating that the sequence homology among G, L and S genomes is relatively low.
On the other hand, allosyndetic bivalents as well as autosyndetic bivalents were observed in all the three diploid intergeneric hybrids investigated in the present study, although their average numbers per PMC were low (0.13−0.36 for allosyndetic bivalents and 0.04−0.27 for autosyndetic bivalents). These results indicate that there are some homologous chromosomal regions within each genome and among the genomes of Gloriosa, Littonia and Sandersonia. For allosyndetic bivalents, the average number of IIG-L was significantly higher than those of IIG-S and IIL-S, indicating that the genome affinity between G−L genomes may be higher than G−S and L−S genomes. An informal classification of Colchicaceae incorporating the classifications by Nordenstam (1998) and Dahlgren et al. (1985) includes all the three genera, Gloriosa, Littonia and Sandersonia, into the tribe Iphigenieae. However, a phylogenetic analysis by sequencing three plastid regions revealed that Gloriosa and Littonia form a clade, whereas Sandersonia forms the different clade with Ornithoglossum in this tribe (Vinnersten and Reeves 2003). Therefore, the difference in the average number of allosyndetic bivalents among different genome combinations may reflect the evolutionally distances among Gloriosa, Littonia and Sandersonia. Gloriosa and Littonia may be closely related to each other, while Sandersonia may have relatively distant relationships with these two genera.
In the present study, all the three diploid intergeneric hybrids produced allosyndetic bivalents during meiosis of PMCs, indicating that intergenomic recombination may occur among Gloriosa, Littonia and Sandersonia. Gametes with intergenomic recombination could contribute to introgression breeding, although the gametes should have fertility for this purpose. Actually, these diploid hybrids showed very low or no pollen fertility (0−2.4 %) as assessed with acetocarmine staining (Amano et al. 2008, 2009). Although chromosome doubling has generally been used for fertility restoration of wide hybrids (Van Tuyl 1989; Van Tuyl et al. 1992; Isshiki and Taura 2003; Dunn and Lindstrom 2007), this approach could not contribute much to introgression breeding, because amphidiploids obtained via chromosome doubling usually produce 2x-gametes without intergenomic recombination (Ramanna and Jacobsen 2003; Van Tuyl and Lim 2003). On the other hand, it has been reported for Alstromeria (Kamstra et al. 1999) and Lilium (Lim et al. 2003; Zhou et al. 2008) that some interspecific hybrids spontaneously produced fertile 2x-gametes with intergenomic recombination via the first division restitution and these 2x-gametes were used for introgression breeding. In addition, artificial induction of 2x-gametes by nitrous oxide gas or high temperature treatments during meiosis has been reported for Populus adenopoda (Lu et al. 2013) and interspecific hybrids in Lilium (Barba-Gonzalez et al. 2006; Hasegawa et al. 2013). Therefore, we are now examining such treatments for inducing unreduced 2x-gametes in the diploid hybrids investigated in the present study.
For the two triploid intergeneric hybrids, allosyndetic bivalents were never observed in PMCs during meiosis. Most PMCs contained only univalents of L or S genome and bivalents of G genome, which might be derived from homologous chromosome pairing. Because of very low homology among three genomes, homologous chromosomes of G genome might paired prior to form allosyndesis. Thus, the triploid hybrids are inadequate as materials for introgression breeding via intergenomic recombination. On the other hand, fertile allotriploid hybrids have been used to produce alien chromosome addition lines in several genera such as Allium (Vu et al. 2012) and Lilium (Lim et al. 2003). Amano (2008) and Amano et al. (2009) reported that the triploid intergeneric hybrids in Colchicaceae showed 7.0−8.3 % of pollen fertility as assessed with acetocarmine staining and produced varied sizes of pollen grains, which might be resulted from abnormal chromosome segregation and cytokinesis. Therefore, the triploid hybrids investigated in the present study are possible materials for producing intergeneric chromosome addition lines in Colchicaceae. Backcross pollination using the triploid hybrids and subsequent ovule culture are now in progress.
References
Amano J (2008) Studies on intergeneric hybridization among colchicaceous ornamentals. PhD Thesis, Niigata University, Japan (in Japanese with English summary)
Amano J, Kuwayama S, Mizuta Y, Oomiya T, Nakamura T, Nakano M (2007) Early identification of intra- and intergeneric hybrids among colchicaceous ornamentals, Gloriosa spp., Littonia modesta Hook. and Sandersonia aurantiaca Hook., by flow cytometry and random amplified polymorphic DNA analyses. J Japan Soc Hort Sci 76:73–78
Amano J, Kuwayama S, Mizuta Y, Godo T, Okuno H, Nakano M (2008) Morphological characterization of three intergeneric hybrids among Gloriosa superba ‘Lutea’, Littonia modesta and Sandersonia aurantiaca (Colchicaceae). Hort Sci 43:115–118
Amano J, Nakazawa D, Kuwayama S, Mizuta Y, Okuno H, Watanabe Y, Godo T, Han D-S, Nakano M (2009) Intergeneric hybridization among colchicaceous ornamentals, Gloriosa spp., Littonia modesta and Sandersonia aurantiaca via ovule culture. Plant Biotechnol 26:535–541
Barba-Gonzalez R, Miller CT, Ramanna MS, Van Tuyl JM (2006) Nitrous oxide (N2O) induces 2n gametes in sterile F1 hybrids between Oriental × Asiatic lily (Lilium) hybrids and leads to intergenomic recombination. Euphytica 148:303–309
Cao M, Sleper DA, Dong F, Jiang J (2000) Genomic in situ hybridization (GISH) reveals high chromosome pairing affinity between Lolium perenne and Festuca mairei. Genome 43:398–403
Dahlgren RMT, Clifford HT, Yeo PF (1985) The families of monocotyledons. Springer-Verlag, Berlin
Dunn BL, Lindstrom JT (2007) Oryzalin-induced chromosome doubling in Buddleja to facilitate interspecific hybridization. Hort Sci 42:1326–1328
Ge X-H, Li Z-Y (2007) Intra- and intergenomic homology of B-genome chromosomes in trigenomic combinations of the cultivated Brassica species revealed by GISH analysis. Chromosom Res 15:849–861
Hasegawa Y, Nukui S, Okazaki K (2013) Production of unreduced male and female gametes in hybrid lilies by high temperature treatment. Breed Sci 15(suppl. 2):114 (in Japanese)
Isshiki A, Taura T (2003) Fertility restoration of hybrids between Solanum melongena L. and S. aethiopicum L. Gilo Group by chromosome doubling and cytoplasmic effect on pollen fertility. Euphytica 134:195–201
Ji Y, Pertuze R, Chetelat RT (2004) Genome differenciation by GISH in interspecific and intergeneric hybrids of tomato and related nightshades. Chromosom Res 12:107–116
Kamstra SA, Kuipers AGJ, De Jeu MJ, Ramanna MS, Jacobsen E (1999) The extent and position of homoeologous recombination in a distant hybrid of Alstromeria: a molecular cytogenetic assessment of first generation backcross progenies. Chromosoma 108:52–63
Kuwayama S, Nakamura T, Mizuta Y, Oomiya T, Nakano M (2005) Cross-compatibility in interspecific and intergeneric hybridization among the Colchicaceous ornamentals. Gloriosa spp., Littonia modesta and Sandersonia aurantiaca. Acta Hort 673:421–427
Lim KB, Ramanna MS, Jacobsen E, Van Tuyl JM (2003) Evaluation of BC2 progenies derived from 3x−2x and 3x−4x crosses of Lilium hybrids: a GISH analysis. Theor Appl Genet 106:568–574
Lu M, Zhang P, Kang X (2013) Induction of 2n female gametes in Populus adenopoda Maxim by high temperature exposure during female gametophyte development. Breed Sci 63:96–103
Marasek A, Hasterok R, Wiejacha K, Orlikowska T (2004) Determination by GISH and FISH of hybrid status in Lilium. Hereditas 140:1–7
Molnár I, Molnár-Láng M (2010) GISH reveals different levels of meiotic pairing with wheat for individual Aegilops biuncialis chromosomes. Biol Plant 54:259–264
Nakamura T, Kuwayama S, Tanaka S, Oomiya T, Saito H, Nakano M (2005) Production of intergeneric hybrid plants between Sandersonia aurantiaca and Gloriosa rothchildiana via ovule culture (Colchicaceae). Euphytica 142:283–289
Nakazawa D, Kishimoto T, Sato T, Saito T, Amano J, Kuwayama S, Okuno H, Godo T, Watanabe Y, Han D-S, Nakano M (2011) Genomic in situ hybridization (GISH) analysis of intergeneric hybrids in Colchicaceae. Euphytica 181:197–202
Nordenstam B (1998) Colchicaceae. In: Kubitzki K (ed) The families and genera of vascular plants, vol 3. Springer-Verlag, Berlin, pp 175–185
Parokonny AS, Marshall JA, Bennett MD, Cocking EC, Davey MR, Power JB (1997) Homoeologous paring and recombination in backcross derivatives of tomato somatic hybrids [Lycopersicon esculentum (+) L. peruvianum]. Theor Appl Genet 94:713–723
Pendinen G, Spooner DM, Jiang J, Gavrilenko T (2012) Genomic in situ hybridization reveals both auto- and allopolyploid origins of different North and Central American hexaploid potato (Solanum sect. Petota) species. Genome 55:407–415
Ramanna MS, Jacobsen E (2003) Relevance of sexual polyploidization for crop improvement—a review. Euphytica 133:3–8
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wide hybrid. Ann Bot 64:315–324
Van Tuyl JM (1989) Research on mitotic and meiotic polyploidization in lily breeding. Herbertia 45:97–103
Van Tuyl JM, Lim KB (2003) Interspecific hybridization and polyploidizition as tools in ornamental plant breeding. Acta Hort 612:13–22
Van Tuyl JM, Meijer H, Van Diën MP (1992) The use of oryzalin as an alternative for colchicine in in vitro chromosome doubling of Lilium and Nerine. Acta Hort 325:625–630
Vinnersten A, Reeves G (2003) Phylogenetic relationships within Colchicaceae. Am J Bot 90:1455–1462
Vu HQ, Yoshimatsu Y, Khrustaleva LI, Yamauchi N, Shigyo M (2012) Alien genes introgression and development of alien monosomic addition lines from a threatened species, Allium roylei Stearn, to Allium cepa L. Theor Appl Genet 124:1241–1257
Yao X-C, Ge X-H, Chen J-P, Li Z-Y (2010) Intra- and intergenomic chromosome pairings revealed by dual-color GISH in trigenomic hybrids of Brassica juncea and B. carinata with B. maurorum. Genome 53:14–22
Zhou S, Ramanna MS, Visser RGF, Van Tuyl JM (2008) Genome composition of triploid lily cultivars derived from sexual polyploidization of Longiflorum × Asiatic hybrids (Lilium). Euphytica 160:207–215
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (No. 20580023) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kishimoto, T., Yamakawa, M., Nakazawa, D. et al. Meiotic chromosome pairing in intergeneric hybrids of colchicaceous ornamentals revealed by genomic in situ hybridization (GISH). Euphytica 200, 251–257 (2014). https://doi.org/10.1007/s10681-014-1152-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1152-y