Abstract
Members of the APETALA2/ethylene response factor (AP2/ERF) transcription factor superfamily are widely present in plants and play important roles in the plant cell cycle, growth and development, as well as the response to biotic and abiotic stresses. The maize genome project has been completed; therefore, it is possible to identify all of the AP2/ERF genes in the maize genome. In this study, 184 AP2/ERF genes were identified by an in silico cloning method, and were compared with AP2/ERF genes from Arabidopsis, rice, grape and poplar. The 184 AP2/ERF maize genes were classified into four subfamilies: DREB (51), ERF (107), AP2 (22) and RAV (3), as well as one soloist. The amino acid sequence composition, physical and chemical characteristics, phylogenetic trees, conserved domain sequences, functional domains, and chromosomal location of the genes were predicted and analyzed. The 184 AP2/ERF genes are distributed on maize chromosomes 1–10 (31, 21, 13, 19, 22, 18, 21, 16, 11 and 12 genes, respectively). Under 0, 1, 2, 4 h waterlogging stress, the expression of 184 AP2/ERF genes in root of Hz32 inbred line (tolerance to waterlogging) were performed using RNA-sequence, and the result showed that 38 genes were responsive to waterlogging stress. This study lays the foundation for subsequent cloning and investigation of the function of the AP2/ERF genes responding to waterlogging stress in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants experience various abiotic stresses during their life cycle, such as drought, salinity, and extreme temperatures. Abiotic stress is currently one of the major factors responsible for agricultural crop losses worldwide, and can reduce the yields of most major crop plants by up to 50 % (Boyer 1982; Bray et al. 2000). Plant adaptation to abiotic stress is regulated by a variety of molecular signaling cascades. Although relatively little is known about the molecular networks which regulate plant tolerance to abiotic stress, a large number of stress-associated genes and signal-transduction systems have been identified (Seki et al. 2003; Zhang et al. 2004). Some of these genes are induced by stress stimuli and encode products that confer tolerance to abiotic stress, whereas others encode upstream regulators that can regulate the signaling pathways induced by various abiotic stresses (Kizis et al. 2001). Previous research has shown that the APETALA2/ethylene response factor (AP2/ERF) genes, a large multigene superfamily of transcription factors, play an important role in the regulation of gene expression during abiotic stress, especially for waterlogging or flooding stress. (Fukao et al. 2006; Hattori et al. 2009; Hinz et al. 2010).
AP2/ERF genes encode either one or two AP2 domains, and can be classified into at least five subfamilies: AP2, DREB (dehydration responsive element binding), RAV (related to ABI3/VP), ERF, and others (Sakuma et al. 2002). The ERF and DREB subfamilies can be further divided into six subfamilies: the B1–6 subfamilies and A1–6 subfamilies, respectively (Sakuma et al. 2002). The first AP2 subfamily gene was isolated from Arabidopsis thaliana (Jofuku et al. 1994). Subsequently, an ERF subfamily gene was also cloned from tobacco (Ohme-Takagi and Shinshi 1995). It has been demonstrated that AP2/ERF proteins play important roles in the regulation of plant growth and development, as well as the response to environmental stress. For example, overexpression of SHN and WIN1, two ERF-related genes, increased leaf epidermal wax accumulation in transgenic Arabidopsis plants (Aharoni et al. 2004; Broun et al. 2004). The level of EREB1 expression in cotton was significantly elevated following treatment with Verticillium wilt toxin (Meng et al. 2010). Under flooding conditions, ethylene production in rice was inhibited due to expression of Sub1A, which led to leaf and internode elongation, inhibition of chlorophyll degradation, as well as the consumption of carbohydrates (Xu et al. 2006; Fukao et al. 2006). In contrast, internode elongation was significantly promoted due to overexpression of SNORKEL1 and SNORKEL2 in deep water rice (Hattori et al. 2009). The Arabidopsis gene RAP2.2 partly affects the low-oxygen response, by inducing genes involved in sugar metabolism and fermentation, as well as ethylene biosynthesis. Overexpression of RAP2.2 improved plant survival under hypoxic stress (Hinz et al. 2010). HRE1 and HRE2 genes play a partially redundant role in low oxygen signaling in Arabidopsis, and improve the tolerance to anoxia by enhancing anaerobic gene expression and ethanolic fermentation (Licausi et al. 2010). Collectively, these studies suggest that the AP2/ERF genes play a crucial role in the tolerance to waterlogging in plants. As an economically important crop, corn suffers from varying degrees of waterlogging, which leads to the loss of 25–30 % of the maize yield each year (Rathore et al. 1997; DMR 2001). Therefore, it is important to identify AP2/ERF genes in the maize genome, which are likely to contribute to stress tolerance, in order to provide new resources for genetically improving the waterlogging tolerance of maize.
Many plant genome projects have been completed, making it possible to identify the AP2/ERF genes in different species. There are 147, 164, 200, 132, and 116 AP2/ERF genes in the Arabidopsis (Sakuma et al. 2002; Nakano et al. 2006), rice (Riaño-Pachón et al. 2007), poplar (Zhuang et al. 2008), grapevine genomes (Zhuang et al. 2009), and Chinese plum (Du et al. 2012), respectively. However, few reports of the AP2/ERF superfamily are available in maize. In the present report, the establishment of a complete picture of the AP2/ERF genes in maize was attempted. Here, 184 AP2/ERF transcription factors were identified from either the MaizeSequence and NCBI databases using the AP2/ERF domain of the tobacco ERF2 protein as a query sequence, or downloaded from PlnTFDB (Riaño-Pachón et al. 2007; http://plntfdb.bio.uni-potsdam.de/v3.0/fam_mem.php?family_id=AP2-EREBP&sp_id=ZMA). The phylogenetic relationships between the maize AP2/ERF genes and their homologs in Arabidopsis were analyzed; and the maize AP2/ERF genes were classified into subfamilies and mapped onto maize linkage groups, as well as, the expression of maize AP2/ERF genes under waterlogged stress were performed. These analyses will be valuable to isolate and understand the molecular mechanism of AP2/ERF genes responded to waterlogging stress in maize.
Materials and methods
Database search
MaizeSequence (http://www.maizesequence.org/index.html) and the NCBI database (http://www.ncbi.nlm.nih.gov) were mined to identify members of the AP2/ERF family. The amino acid sequence of the AP2/ERF domain from tobacco ERF2 protein was used as a query sequence to search the databases using BLASTP. Position-specific iterated BLAST was also used to increase the extent of the database search results. The maize AP2/ERF database was also downloaded from PlnTFDB. The Arabidopsis AP2/ERF sequences were downloaded from the database of Arabidopsis transcription factors (DATF) website at http://datf.cbi.pku.edu.cn/ (Guo et al. 2005).
Sequence analysis and phylogenetic tree construction
Multiple sequence alignment analysis was performed using ClustalX (Chenna et al. 2003; http://bioinformatics.ubc.ca/resources/tools/clustalx). Phylogenetic trees of the aligned maize AP2/ERF protein sequences were constructed using MEGA version 5.0 (Tamura et al. 2007; http://www.megasoftware.net) via the neighbor-joining (NJ) method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1,000 replicates; random seed). The amino acid variation rates were also determined and motif detection was performed with MEME version 3.5.7 (Bailey et al. 2006; http://meme.sdsc.edu/meme/meme.html).
Bioinformatic analysis
Bioinformatic analysis of the AP2/ERF superfamily genes from maize and Arabidopsis, such as the gene and deduced amino acid sequences, composition, physical and chemical characterization, and conserved domain sequences, were performed using the Expert protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (http://cn.expasy.org). The solubility of the recombinant proteins when overexpressed in Escherichia coli was predicted using the statistical model from the University of Oklahoma (http://biotech.ou.edu) (Zhuang et al. 2008). The folding states of the AP2/ERF proteins from maize and A. thaliana were predicted using the FoldIndex program (http://bioportal.weizmann.ac.il) (Zhuang et al. 2008).
Location of AP2/ERF genes on maize chromosomes
Chromosomal locations were determined according to chromosomal location information gathered from maizeGDB (http://www.maizegdb.org).
RNA sequencing
Seeds of maize inbred line Hz32 were germinated for 2 days in the dark on moist filter paper at room temperature. Germinated seeds were transferred to silica sand, and uniform seedling with two visible leaves were selected and planted in a greenhouse for waterlogging treatment 0, 1, 2 and 4 h, respectively. Total RNA was isolated from the roots of above waterlogging treatment seedling, respectively, and RNA was sequenced in BGI-shenzhen.
Results
Identification and prediction of maize AP2/ERF transcription factors
A total of 330 maize AP2/ERF genes were downloaded from the PlnTFDB database, and 121 maize AP2/ERF genes were obtained from BLASTP searches of the NCBI and MaizeSequence databases. However, 69 of the 121 AP2/ERF genes identified using BLASTP searches were also downloaded from PlnTFDB. Therefore, a total of 382 AP2/ERF maize genes were obtained from PlnTFDB, NCBI and MaizeSequence. This number (382) is much larger than the number of AP2/ERF genes in other plants, such as rice (164), Arabidopsis (147), poplar (200) and grapevine (132). Thus, we concluded that there were a number of false-positive genes within the 382 putative AP2/ERF genes. In order to exclude false-positive genes, the deduced amino acid sequences of the 382 AP2/ERF genes were used as queries to search the maizeGDB database using BLAST. Finally, 198 false-positive AP2/ERF genes were excluded, and a total of 184 maize AP2/ERF genes were obtained.
We compared the AP2/ERF genes from maize, rice, grapevine, poplar and Arabidopsis (Table 1). In maize, 51 AP2/ERF genes were classified into the DREB subfamily, compared with 52, 36, 57 and 77 in rice, grapevine, Arabidopsis and poplar, respectively; 107 maize AP2/ERF genes were classified into the ERF subfamily, compared with 79, 73, 65 and 91 in rice, grapevine, Arabidopsis and poplar, respectively. Twenty-two maize AP2/ERF genes were predicted to encode proteins with two AP2/ERF domains, and were classified into the AP2 subfamily. In comparison, rice, grapevine, Arabidopsis and poplar contain 26, 18, 18 and 26, respectively. Three maize AP2/ERF genes were classified into the RAV subfamily, compared to 7, 4, 6 and 5 in rice, grapevine, Arabidopsis and poplar, respectively. The remaining maize AP2/ERF gene, GRMZM2G323172, was obtained by position-specific iterated BLAST using Arabidopsis At4g13040 as query sequence, and has a low homology with other AP2/ERF genes; therefore, this gene was designated as a soloist.
At the whole-genome level, the percentage of the AP2/ERF gene family in maize is 0.4640 %, which is more than in rice (0.4315 %), grapevine (0.4337 %) and poplar (0.4390 %), but less than in Arabidopsis (0.581 %). However, the average number of AP2/ERF genes per Mb in maize is 0.0736, less than in rice (0.3814), grapevine (0.271), poplar (0.4124) and Arabidopsis (1.176; Table 1).
Phylogenetic relationships between the AP2/ERF transcription factors in maize and Arabidopsis
Based on alignment of the AP2/ERF domain from maize and Arabidopsis, 184 maize AP2/ERF genes were classified into the DREB, ERF, AP2 and RAV subfamilies, and one soloist (Table 1). Phylogenetic trees of the DREB and ERF subfamilies in maize were constructed (Fig. 1a, b). A total of 51 DREB subfamily genes distributed into the A1–A6 groups in maize; A1, A2, A3, A4, A5 and A6 contain 105, 1, 11, 12 and 12 genes, respectively. Additionally, 107 genes belonging to the ERF subfamily in maize distributed into the B1–B6 groups; B1, B2, B3, B4, B5 and B6 contain 31, 20, 18, 10, 6 and 22 genes, respectively. Finally, 22 genes were classified into the AP2 subfamily and three genes into the RAV subfamily (Fig. 2a, b).
Sequence alignment and conserved motifs inside or outside the AP2/ERF domain
In order to thoroughly characterize the similarity and diversity of the AP2/ERF transcription factors in maize, multiple alignment analyses were performed using both the amino acid sequences of the AP2/ERF domain and the full amino acid sequences. The sequence alignments revealed that the DREB transcription factors in maize and Arabidopsis contain the residues G4, R6, R8, W14, V15, E17, R19, W29, G31, A39, A40, D44, A46 and N59 (The number was accorded with the GRMZM2G137341). All of the DREB transcription factors contain a WLG motif (W29, L30, G31), except for GRMZM2G175856 which contains V30. In the A2, A4 and A6 subfamilies, the residues G11, K12, W13, V14, E16, I17 and R18 are conserved. In the A4 and A6 subfamilies, the residues L58, N59, F60 and P61 are conserved, while L58, N59 and F60 are conserved in the A1 subfamily, and N59, F60 and P61 are conserved in the A5 subfamily (Supplementary Fig. 1). The full amino acid sequence alignments suggested that there is no conservation outside of the AP2/ERF domain in the A1–A6 subfamily (Supplementary Figs. 2–7).
In the ERF subfamily, the B6 subfamily contains more variation than the B1-B5 subfamilies. All ERF transcription factors in maize and Arabidopsis contain the residues G12 and G30 (The number was accorded with the GRMZM2G079825). In the B1–B5 subfamilies, R19, A3, Y42 and D43 are completely conserved; G51 and N58 are also conserved, except for the genes GRMZM2G379652 and GRMZM2G478965 which contain S51, and GRMZM2G105266 which contains G58. The WLG motif (W28, L29, G30) is also present in the B1–B6 subfamily; however, GRMZM2G703514 contains CLG; GRMZM2G379652, GRMZM2G478965, AC213666.3, GRMZM2G363052, GRMZM2G480434 and GRMZM2G016079 contain WIG; and GRMZM2G033656 contains WVG (Supplementary Fig. 8). Analysis of the multiple full length amino acid sequence alignments of the B1–B6 subfamilies revealed there was more variation outside the AP2/ERF domain in both maize and Arabidopsis (Supplementary Figs. 9–14).
Twenty-two maize AP2/ERF genes contain two AP2/ERF domains; these genes were classified into the AP2 subfamily. However, part of the AP2/ERF domain I is deleted in GRMZM2G131266, GRMZM2G141219, GRMZM2G008234, GRMZM2G317160, GRMZM2G700665 and GRMZM2G416701. AP2/ERF domain II is also deleted in GRMZM2G141219 and GRMZM2G399072 (Supplementary Figs. 15, 16). GRMZM2G059939, GRMZM2G159592 and GRMZM2G169654 contain one AP2/ERF domain and one B3 domain, and were classified into the RAV subfamily (Supplementary Figs. 17, 18).
Characterization of the maize AP2/ERF transcription factor proteins
The number of amino acids, theoretical Mw, theoretical pI and recombinant protein solubility were analyzed using ExPASy. Of the 184 maize AP2/ERF transcription factors, GRMZM2G141638 is the largest number with 706 amino acids (Supplementary Table 13). GRMZM2G060206 has the shortest amino acid sequence, with 124 amino acids (Supplementary Table 9). In the DREB-A1 subfamily and RAV subfamily of AP2/ERF transcription factors, the maize proteins contain more amino acids than their Arabidopsis homologs (Supplementary Table 1, 14).
More than 50 % of the AP2/ERF transcription factors from maize and Arabidopsis were predicted to be insoluble; however, less than 70 % of the Arabidopsis proteins in the DREB-A1 subfamily were predicted to be insoluble (Supplementary Table 1). In the ERF-B4 subfamily and DREB-A3 subfamily, over 90 % of the maize and Arabidopsis proteins were predicted to be insoluble (Supplementary Table 3, 10).
Over 30 % of the amino acids in the maize DREB, ERF and AP2 subfamilies are aliphatic, except for DREB-A2 and ERF-B6 subfamily, and over 40 % of the amino acids in the RAV subfamily are aliphatic (Supplementary Tables 1–14). For the DREB-A1 subfamily, the theoretical pI values in Arabidopsis and maize are less than 6 and 7 %, respectively (Supplementary Table 1). The theoretical pI values for the DREB-A4 subfamily in maize and Arabidopsis are less than 7 % (Supplementary Table 4). In the RAV subfamily, all of the theoretical pI values are over 9 %, expect for two Arabidopsis genes (AT1G50680.1, 6.62 %; AT1G51120.1, 6.77 %; Supplementary Table 14).
For the 184 maize AP2/ERF transcription factors, the percentage of aromatic amino acids is less than 10 %, except for GRMZM2G047999 (12 %), GRMZM2G079825 (12 %), GRMZM2G023708 (12 %) and GRMZM2G457562 (11 %; Supplementary Tables 1–14). In the DREB-A1 subfamily, DREB-A4 subfamily and RAV subfamily, the percentage of negative amino acids is over 15, 13 and 12 % in Arabidopsis and less than 15, 14 and 10 % in maize, respectively (Supplementary Table 1, 4, 14). The amino acid sequences, number of amino acids, theoretical Mw, theoretical pI and recombinant protein solubility of GRMZM2G120401 and GRMZM2G165257 in maize were identical (Supplementary Table 7).
Prediction of the folding state of AP2/ERF transcription factor proteins
Analysis of the folding states of the AP2/ERF transcription factors from maize and Arabidopsis was performed using FoldIndex software (http://bip.weizmann.ac.il/fldbin/findex). In the DREB-A1 subfamily, the percentage of disordered residues is over 60 % in Arabidopsis, except for AT1G63030.1 (46.96 %), compared to less than 60 % in maize (Supplementary Table 15). The percentage of disordered residues in the DREB-A4 subfamily from maize and Arabidopsis is less than 70 %, except for AT3G16280.1 (75.42 %; Supplementary Table 18). In the ERF-B2 subfamily, the percentage of disordered residues is over 80 % in Arabidopsis, and less than 80 % in maize (Supplementary Table 22). For all of the AP2/ERF transcription factors from maize and Arabidopsis, the highest of percentage of disordered residues were observed in GRMZM2G133168, in which 100 % of the residues were disordered (Fig. 3a). The lowest of percentage of disordered residues is in GRMZM2G169654 (4.11 %; Fig. 3b). In addition, a more than five-fold difference was observed between the percentage of disordered residues in GRMZM2G703514 and GRMZM2G132185, both in the ERF-B1 subfamily (89.50 and 17.01; Fig. 3c, d).
Chromosomal location of maize AP2/ERF genes
We identified 184 AP2/ERF genes in maize, located on 10 chromosomes. Chromosome 1 contains the highest number of AP2/ERF genes, 31/184 (16.8 %), and chromosome 9 contains the least, 11/184 (6.0 %); 21/184 (11.4 %) AP2/ERF genes are located on chromosome 2; 13/184 (7.1 %) on chromosome 3; 19/184 (10.3) on chromosome 4; 22/184 (12.0 %) on chromosome 5; 18/184 (9.8 %) on chromosome 6; 21/184 (11.4 %) on chromosome 7; 16/184 (8.7 %) on chromosome 8 and 12/184 (6.5 %) on chromosome 10 (Fig. 4). Maize AP2/ERF genes are mainly located on the ends of the chromosomes, especially on chromosome 2, 3, 4, 5 and 9. Many maize AP2/ERF genes in the same subfamily are clustered on one chromosome, for example GRMZM2G129674, GRMZM2G018984 and GRMZM2G061227 belonging to the ERF-B2 subfamily are clustered in a ~57 Kb segment (Chr.1); GRMZM2G016079, GRMZM2G480434 and GRMZM2G033656 belonging to the ERF-B6 subfamily are clustered in a ~63 Kb segment (Chr.1); GRMZM2G124037 and GRMZM2G124011 belonging to the DREB-A1 subfamily are clustered in a ~5 Kb segment (Chr.2); GRMZM2G020016 and GRMZM2G020150 belonging to the ERF-B1 subfamily are clustered in a ~12 Kb segment (Chr.6); GRMZM2G069082, GRMZM2G069126 and GRMZM2G069146 belonging to the DREB-A1 subfamily are clustered in a ~12 Kb segment (Chr.7), and GRMZM2G132223 and GRMZM2G132185 belonging to the ERF-B1 subfamily are clustered in a ~9 Kb segment (Chr.8; Fig. 4).
Expression of AP2/ERF family genes under waterlogging stress in maize
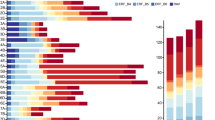
RNA-seq was performed in BGI-shenzhen, and the expression patterns of 184 AP2/ERF genes were analyzed. The results showed that the expression levels of 38 AP2/ERF genes were changed under waterlogging stress (Fig. 5). Above 38 AP2/ERF genes, DREB subfamily was 10, including five genes in A1 subgroups (GRMZM2G069146, GRMZM2G124011, GRMZM2G069126, GRMZM2G175856, and GRMZM2G006745), one gene in A2 subgroups (AC209257.4), one gene in A5 subgroups (GRMZM2G174917), and three genes in A6 subgroups (GRMZM2G055204, GRMZM2G061487, and GRMZM2G141679). 25 AP2/ERF genes in ERF subfamily were responsive to waterlogging stress, B1, B2, B3, and B6 subgroups were 9 (GRMZM2G120401, GRMZM2G438202, GRMZM2G068967, GRMZM2G132185, GRMZM2G310368, GRMZM2G089995, GRMZM2G 020054, GRMZM2G020150, and GRMZM2G174347), 9 (GRMZM2G018984, GRMZM2G169382, GRMZM2G053503, GRMZM2G025062, GRMZM2G171179, GRMZM2G052667, GRMZM2G131281, GRMZM2G129674, and AC206951.3), 5 (GRMZM2G055180, GRMZM2G080516, GRMZM2G123119, GRMZM2G466044, and GRMZM2G474326), and 2 (GRMZM2G081892 and AC187157), respectively. AP2 and RAV subfamily were 2 (GRMZM2G700665 and GRMZM2G416701), and 1 (GRMZM2G169654), respectively (Fig. 5).
Heatmap show expression level of 38 AP2/ERF genes in the root of Hz32. Under 0, 1, 2, and 4 h waterlogging stress, the expression of maize 184 AP2/ERF genes was measured by RNA-seq analysis using the root of Hz32 inbred line, respectively. Color scale represents reads per kilobase per million normalized log2 transformed counts where green indicates low level and red indicate high level. (Color figure online)
The expression levels of 38 AP2/ERF waterlogging stress responsive genes are diverse. There are seven genes (GRMZM2G120401, GRMZM2G174917, GRMZM2G131281, GRMZM2G020150, GRMZM2G055204, GRMZM2G061487, and GRMZM2G416701), and their expression levels are decreased under waterlogging condition. Under 1 h waterlogging stress, 11 AP2/ERF genes (GRMZM2G089995, GRMZM2G020054, GRMZM2G132185, GRMZM2G174347, GRMZM2G068967, GRMZM2G438202, GRMZM2G310368, GRMZM2G055180, GRMZM2G080516, GRMZM2G123119, and GRMZM2G474326) exhibit firstly decreased-expression levels, then are increased under 2, or 4 h waterlogging stress. Nine AP2/ERF genes (GRMZM2G006745, GRMZM2G141679, AC209257.4, GRMZM2G124011, GRMZM2G171179, GRMZM2G069146, GRMZM2G069126, AC187157, and GRMZM2G700665) show highest-expression level under 1 h waterlogging stress, then are decreased in 2, or 4 h. GRMZM2G169382 and GRMZM2G052667 genes are highest-expression levels under 2 h waterlogging. There are nine AP2/ERF genes (AC206951.3, GRMZM2G018984, GRMZM2G053503, GRMZM2G175856, GRMZM2G466044, GRMZM2G169654, GRMZM2G025062, GRMZM2G129674, and GRMZM2G081892), and their expression levels are higher under 4 h waterlogging than other treatments (Fig. 5).
Discussion
Plants are continuously exposed to inescapable environmental changes and various stresses, which may severely affect their growth and development. Plants have evolved a number of specific transcription factors to modulate the expression of numerous stress-responsive genes. Among these transcription factors, the AP2/ERF superfamily is one of the largest families in plants, and plays a prominent role in abiotic stresses (Xu et al. 2006; Fukao et al. 2006; Hattori et al. 2009; Licausi et al. 2010). The growth and productivity of the economically important crop maize are greatly affected by abiotic stresses, specifically drought and flooding. Therefore, it is important to identify and isolate the maize AP2/ERF genes, in order to characterize AP2/ERF-mediated regulation of the hormone biosynthetic and signaling pathways associated with enhanced abiotic stress, which may enable an understanding of the molecular mechanisms by which AP2/ERF genes regulate the response to abiotic stress in maize.
Previous research showed that the AP2/ERF genes in each subfamily share similar functions (Hinz et al. 2010; Licausi et al. 2010). ERF subfamily members, specifically the ERF-B2 subfamily members, are involved in the submergence stress response. SNORKEL genes encoding ERFs trigger internode elongation in deepwater rice (Hattori et al. 2009). Another water tolerance gene, SubA1, restricts rice elongation at the seedling stage during flash floods (Xu et al. 2006; Fukao et al. 2006). There are five ERF-B2 subfamily genes in Arabidopsis, namely RAP2.2 (At3g14230), RAP2.12 (At1g53910), RAP2.3 (At3g16770), HRE1 (At1g72360), and HRE2 (At2g47520). RAP2.2, RAP2.12, HRE1, and HRE2 genes have been documented that play important role in the response to hypoxia (Licausi et al. 2011; Hinz et al. 2010; Licausi et al. 2010). In present study, 184 maize AP2/ERF genes were identified, classified and mapped. RNA-seq analysis has confirmed that 38 of 184 genes are response to waterlogging stress. There are 20 ERF-B2 subfamily genes in maize genome (Table 1), and 9 of 20 are response to waterlogging stress (Fig. 5). Previous study revealed that genes in the ERF-B2 subfamily play a key role in waterlogging tolerance in rice and Arabidopsis. Therefore, it can be inferred that maize waterlogging tolerance gene may be existed among nine waterlogging-responsive genes in ERF-B2 subfamily. Further researches are carrying, and will be reported in the future.
References
Aharoni A, Dixit S, Jetter R, Thoenes E, Arkel GV, Pereira A (2004) The SHINE of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:369–373
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Bray EA, Bailey-Serres J, Weretilnyk E (2000) Response to abiotic stresses. In: Gruissem W, Buchannan B, Jones R (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rochville, pp 1158–1249
Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Nati Acad Sci USA 101:4706–4711
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res 31:3497–3500
DMR (2001). Research achievements. In: Azad CS (ed) Proceedings of 49th annual maize workshop. Directorate of maize research (DMR), University of Agriculture and Technology, Kanpur (UP)
Du DL, Hao RJ, Cheng TR, Pan HT, Yang WR, Wang J, Zhang QX (2012) Genome-wide analysis of the AP2/ERF gene family in prunus mume. Plant Mol Biol Rep. doi:10.1007/s11105-012-0531-6
Fukao T, Xu KN, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18:2021–2034
Guo A, He K, Liu D, Bai S, Gu X (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21:2568–2569
Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, Matsuoka M, Mori H, Ashikari M (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460:1026–1031
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153:757–772
Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Kizis D, Lumbreras V, Pagès M (2001) Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett 498:187–189
Licausi F, Dongen JTV, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HER1 and HER2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62:302–315
Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, Dongen TV (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479:419–422
Meng XP, Li FG, Liu CL, Zhang CJ, Wu ZX, Chen YJ (2010) Isolation and characterization of an ERF transcription factor gene from cotton (Gossypium barbadense L.). Plant Mol Biol Rep 28:176–183
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Ohme-Takagi M, Shinshi H (1995) Ethylene inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7(2):173–182
Rathore TR, Warsi MZK, Zaidi PH, Singh NN (1997) Waterlogging problem for maize production in Asian region. TAMNET News Lett 4:13–14
Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller RB (2007) PlnTFDB: an integrative plant transcription factor database. MBC Bioinform 8:42
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration and cold-inducible gene expression. Biochem Biophys Res Cummun 290:998–1009
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14(2):194–199
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Xu KN, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Zhang JZ, Creelman RA, Zhu JK (2004) From laboratory to field: using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XY, Tian YS, Zhao W, Qiao YS, Zhang Z, Xiong AS, Yao QH (2008) Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun 371:468–474
Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XF, Tian YS, Zhao W, Qiao YS, Zhang Z, Xiong AS, Yao QH (2009) Genome-wide analysis of the AP2/ERF gene family in Vitis vinifera. Sci Hortic 123:73–81
Acknowledgments
The work was supported by National Natural Science Foundation (31271741), the Hubei Province Natural Science Foundation (2011CDB006 and 2012FFA051), Youth Foundation of Hubei Province Department of Education (Q20111313), Hubei Key Laboratory of Economic Forest Germplasm Improvement and Resources Comprehensive Utilization (2011BLKF242), and Hubei Provincial Key Discipline, the Crop Science in Yangtze University.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hewei Du and Min Huang have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, H., Huang, M., Zhang, Z. et al. Genome-wide analysis of the AP2/ERF gene family in maize waterlogging stress response. Euphytica 198, 115–126 (2014). https://doi.org/10.1007/s10681-014-1088-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1088-2