Abstract
Genetic distance analysis among quality protein maize (QPM) inbred lines and the correlation of genetic distance with heterosis would help to design breeding strategy and predict hybrid performance. This study was carried out to determine the amount of genetic diversity among QPM inbred lines using SSR markers and morphological distances; to classify the inbred lines according to their relationships; and to estimate the correlations of SSR markers and morphological distances with hybrid performance, heterosis and specific combining ability (SCA). One-hundred and five hybrids generated by diallel crossing of 15 QPM inbred lines were evaluated with the 15 parents for 17 morphological traits at Harare, Zimbabwe and Bako, Ethiopia and also examined for DNA polymorphism using 40 SSR markers. SSR markers and morphological methods of genetic distance estimates showed moderately high genetic distance among the inbred lines studied. Cluster analysis based on the two distance measures grouped the 15 parental lines differently. The SSR marker-based genetic distance was positively and highly significantly correlated with grain yield (r = 0.37), and negatively and highly significantly with days to anthesis (r = −0.40) and days to silking (r = −0.42). These relationships suggest that high grain yield and earliness of QPM hybrids can be predicted from SSR marker determined distances of the parents, although the correlation values were not very high. The correlations of SSR marker distance with heterosis were too low to be of predictive value except for the case of plant height. Morphological distances were of less importance in predicting hybrid performance and SCA effects of hybrids.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In maize, hybrid breeding remains the method of choice for attaining maximum genetic gain from the effects of heterosis. Nevertheless, identification of lines with superior cross performance is the most crucial, costly and time consuming phase in hybrid development programs (Melchinger et al. 1990) due to the need for extensive yield trails. If inbred lines could be screened, and superior crosses predicted before field evaluation, this would greatly enhance the efficiency of hybrid breeding programs. However, per se performance of maize inbred lines does not predict the performance of maize hybrids for grain yield and agronomic traits (Hallauer and Miranda 1988). In addition, trait expression is often significantly influenced by environmental factors. The efficiency of hybrid breeding programs could be increased if the inbred lines could be screened for genetic diversity using molecular markers and superior crosses accurately predicted prior to field evaluation (Melchinger et al. 1991).
Genetic diversity in relation to hybrid performance has been studied extensively in maize. Moll et al. (1965) reported positive correlations between morphological markers based genetic diversity of the parents with heterosis in maize hybrids. The amount of heterosis measured between crosses of maize populations and races from different geographical regions increased as the genetic distance between populations increased (Moll et al. 1965). Studies with isozymes demonstrated that genetic distance estimates were in most cases positively associated with F 1 performance for grain yield (Stuber 1994). Several studies have attempted to use isozyme variation to measure genetic diversity among inbred lines and to examine the relationship between genetic diversity and grain yield (Smith and Smith 1989). In general, diversity or genetic distance, as measured by isozyme differences, has not been a good predictor of grain yield or grain yield heterosis.
DNA markers have been used to analyze the genetic relationships among maize inbred lines and to examine the relationship between DNA marker-based genetic distance and single-cross grain yields and heterosis in maize. Several reports have demonstrated the high correlation between RFLP distance and hybrid performance in maize (Ajmone-Marsan et al. 1998). Lanza et al. (1997) observed positive correlations between RAPD-based genetic distances and maize single-cross hybrid grain yield whereas Parentoni et al. (2001) reported that correlation between RAPD marker estimated genetic distance and SCA for yield was too low to be of predictive value. Ajmone-Marsan et al. (1998) and Kiula et al. (2008) reported that the correlation between AFLP markers, hybrid performance and SCA estimates may have a practical use in predicting hybrid performance. Legesse et al. (2008) reported low correlation of AFLP measured genetic distance with hybrid performance. SSR markers are reliable, reproducible, good for discrimination and standardization, and cost effective compared to other marker types (Smith et al. 1997; Melchinger 1999). SSR markers provide an effective method for predicting hybrid performance (Drinic et al. 2002). Genetic distance based on SSR markers was found to be significantly correlated with hybrid yield in maize (Xu et al. 2004).
The degree of heterosis depends on the relative performance of inbred parents and the corresponding hybrids. Environment can have different effects on the performance of hybrids, altering the relationship between genetic distance and heterosis. Little information is available on genetic distances and their relationship to yield characteristics in QPM germplasm. Therefore, the objectives of this study were to determine SSR marker and morphological traits based genetic distances among QPM inbred lines; to classify the inbred lines according to their relationship based on these markers, and to estimate the correlations of SSR markers and morphological distances with hybrid performance, heterosis and SCA in QPM in two environments.
Materials and Methods
Locations
Trials were evaluated under optimal management conditions at Harare, Zimbabwe and Bako, Ethiopia. Field experiments were planted in the main seasons at CIMMYT-Harare Research Station, Zimbabwe (2006/07) and at Bako Agricultural Research Centre, Ethiopia (2007). The locations represent sub-humid mid-altitude maize growing mega-environments of sub-Saharan Africa (Hartkamp et al. 2000). Harare lies at 17o08′S latitude, 31o05′E longitude at an altitude of 1489 m above sea level (masl). The soil is reddish brown clay (nitosol) with a pH (CaCl2) of 5.2 and 5.4 for top soil (0–30 cm) and sub-soil (30–60 cm), respectively (data not shown). Harare received a total precipitation of 630 mm, and an average maximum and minimum temperature of 27.7 and 16.1 °C, respectively, during the growing season (November 2006 to March 2007). The site at Bako is at 9o06′N latitude and 37o09′E longitude and at an altitude of 1,650 masl. The soil of the centre is reddish brown clay (nitosol) with pH (H2O) of 6.0 and 5.9 for top soil (0–30 cm) and sub-soil (30–60 cm), respectively (data not shown). The total precipitation during the growing season (May–November 2007) was 975 mm, and the mean maximum and minimum temperatures were 28.1 and 14.0 °C, respectively.
Optimum growing conditions were maintained by applying the recommended rates of fertilizer for the respective locations, managed by crop rotation and residue incorporation. Prior to sowing, 28 kg ha−1 nitrogen was incorporated, and 69 kg ha−1 nitrogen was side-dressed at both 28 and 56 days after emergence at Harare while 100 kg ha−1 nitrogen was applied in two splits, half at planting and the rest at 37 days after emergence at Bako. At Harare, phosphate (P2O5) and potassium (K) were basally applied before planting at rates of 56 and 24 kg ha−1. Pre-emergence herbicides, Atrazine (4.0 l ha−1) and Dual (1.8 l ha−1), were applied at planting to control weeds. The later emerging weeds were controlled manually. At 6 weeks after emergence, Dipterex granules (3.5 kg ha−1) were applied to control stalk borer (Busseola fusca). The experiments at Bako received 100 kg P2O5 ha−1 at planting. Pre-emergence herbicide, Lasso–Atrazine (5.0 l ha−1) was applied at planting to control weeds; and then the weeds were controlled by hand weeding. The experiments at Harare were irrigated to field capacity at planting using sprinkler irrigation. Thereafter, the trials were rain fed. Experiments at Bako were grown exclusively under rain fed conditions.
Germplasm, experimental design and field measurements
Fifteen white grained QPM inbred lines used for the current study were obtained from CIMMYT-Zimbabwe (Table 1). The lines were developed by pedigree breeding and had good combining ability in top-cross and per se performance across a range of tropical and subtropical environments (data not shown). Most of the lines were resistant/tolerant to major foliar diseases of the tropics (CIMMYT 2004). The inbred lines were planted in separate trial using alpha-lattice design with two replicates at each location. Grain yield (t ha−1), days to anthesis, anthesis-silking interval (days), plant height (cm), ear height (cm), plant aspect (overall phenotypic appearance of the plant where 1 = excellent and 5 = poor), ear diameter (cm), ear length (cm), thousand kernel weight (g), number of rows per ear, number of kernels per row, leaf length (cm), leaf width (cm), leaf area (cm2), tassel size [(recorded after milk stage as 3 (small), 5 (medium) and 7 (large)], leaf orientation [recorded after flowering as 1 (erect), 2 (intermediate) and 3 (pendant)] and foliage rating [rating of total leaf surface after milk stage as 3 (small), 5 (intermediate) and 7 (large)] were used for morphological characterization of the 15 parental inbred lines. These 17 characteristics were used to determine morphological genetic distances between the inbreds.
Diallel crosses were made among the 15 inbred lines in the winter of 2006 at Muzarabani, Zimbabwe. Seeds from crosses and reciprocal crosses were bulked to form a set of 105 F 1 hybrids. The F 1 hybrids were evaluated along with two QPM (SC527Q and CML144/CML159//CML176) and one non-QPM (SC633) hybrid check at Harare, and with two non-QPM (BH540 and BH541) checks and one QPM (BHQP542) hybrid check at Bako. The 108 hybrids (105 F 1s and three checks) were planted adjacent to the inbred line trials at both locations as 9 × 12 alpha-lattice design with two replicates in each environment. The plots were over sown with two seeds per hill every 25 cm at Harare and every 30 cm at Bako. Each plot consisted two rows of 4.0 (Harare) and 4.8 m (Bako) in length spaced at 75 cm apart. The plots were later thinned to the desired plant densities of 53,333 and 44,444 plants ha−1 at Harare and Bako, respectively, according to local production guidelines. Measurements for the hybrid trials were taken on grain yield, days to anthesis, days to silking, plant height, ear height and ears per plant of the hybrids.
SSR analysis
Leaves of each of the inbred lines were harvested from 15 three-week old plants and freeze-dried. Equal amounts of freeze-dried leaf material from 15 plants of the same inbred line were bulked and ground to a fine powder using a Tissue-lyzer (QIAGEN, GmbH, Hilden, Germany). Genomic DNA was isolated using the CTAB (hexadecyltrimethylammonium bromide) procedure according to the Applied Biotechnology Center’s Manual of Laboratory Protocols (CIMMYT 2005). The concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, Delaware, USA) and the DNA was diluted to working concentrations of 25 ng/μl. The 40 SSR markers used in this study were selected from previous studies (Smith et al. 1997; Warburton et al. 2002) and from the public maizeGDB database (http://www.agron.missouri.edu/ssr_probes/ssr.htm) based on repeat unit and bin location to provide a uniform coverage of the entire maize genome. Fluorescently labeled SSR primers were multiplexed in polymerase chain reactions (PCRs) for maximum efficiency. PCR reactions were set up in a total volume of 10 μl containing 25 ng template DNA, 0.05–0.2 μM each of forward and reverse primers (amount varied according to activity of the primer), and 2× Taq Mastermix (Promega Corporation). The 2× Taq Master Mix is a premixed, ready-to-use solution containing Taq DNA polymerase, dNTPs, MgCl2 and reaction buffers at optimal concentrations for efficient amplification of DNA templates by PCR. The PCR Master Mix has been optimized for use in routine PCR for amplifying DNA templates.
The PCR amplification was performed in a MBS 384-well thermal cycler (Thermo Electron Corporation, Somerset, NJ). The amplification protocol consisted of an initial denaturation cycle at 94 °C for 1 min, followed by 10 cycles at 94 °C for 1 min, at 65 °C to 55 °C by decreasing 1 °C each cycle for 1 min, and at 72 °C for 90 s. This was followed by 30 cycles of 1 min at 94 °C, 1 min at 55 °C and 72 °C for 90 s. A final extension of 72 °C for 5 min was followed by a hold step at 4 °C. The PCR products were diluted in sterile water at a ratio of 1:25 and 1 μl of diluted PCR products were added to a mix of 9 μl Hi-Di Formamide and 0.15 μl size standard (Genescan LIZ 500) in the wells of a 384-well microtitre plate. The samples were denatured at 94 °C for 3 min in a PCR machine, and then immediately cooled on ice. PCR products were subsequently fractionated on an ABI 3130×l Genetic Analyser (PE Applied Biosystems, Foster City, Calif.). Capillary electrophoresis was carried out at an oven temperature of 60 °C in POP-7 polymer using dye set G5. Samples were injected at 1.2 kV for 23 s and run at 15 kV for 20 min. The electropherograms were analysed with Genemapper 4.0 software (PE Applied Biosystems, Foster City, Calif.). Bands were coded as present (1) or absent (0) for all the genotypes. A matrix of binary data was constructed with rows equal to number of inbred 60 lines and columns equal to SSR marker alleles. The body of the matrix was filled in with zeros and ones, for absence and presence of the fragments for each inbred line, respectively.
Statistical analysis
Mid-parent heterosis was calculated as: \( {\text{MPH}} = \frac{{\left( {F_{1} - {\text{MPV}}} \right)}}{{\text{MPV}}} \times 100 \), where F 1 is the mean performance of the cross and MPV is mean of the two inbred parents. High-parent heterosis was calculated as: \( {\text{HPH}} = \frac{{\left( {F_{1} - {\text{HPV}}} \right)}}{{\text{HPV}}} \times 100 \), where HPV is the mean value of the highest performing parent. SCA was analyzed using a modification of the DIALLEL-SAS program (Zhang and Kang 1997) and SCA effects of the crosses were estimated following Griffing’s Method IV (crosses only) and Model I (fixed) of diallel analysis (Griffing 1956). Euclidean distance was computed from 17 morphological traits after the mean observation for each line was standardized (subtracting from each observation the mean value of the trait and dividing by its respective standard deviation). Band profiles generated by SSR markers as 1 or 0 for presence or absence of a specific band, respectively, were used to estimate the Euclidian distances among all possible pairs of the 15 inbred lines. The distances were computed using the Number Cruncher Statistical System, NCSS 2000 (Hintze 1998). A dendrogram was constructed from the distance matrix using the unweighted pair group method with the arithmetic averages (UPGMA) method of cluster analysis. Pearson correlation coefficients were computed to estimate the association of SSR marker- and morphological distances with F 1 hybrid performance, mid-parent heterosis (MPH), higher-parent heterosis (HPH) and SCA.
Results
There was significant inbred (data not shown) and hybrid with environment interaction for all measured characteristics. There was also significant SCA with environment interaction for almost all measured characteristics (Table 2). For this reason, the SCA values and the MPH and HPH were separated for the two environments and all correlations were calculated separately for the two environments. MPH and HPH was consistently higher at Bako than Harare. MPH for grain yield was, on average, very high for Harare and Bako (Table 3). Mean HPH over the two locations for grain yield ranged from −30.4 to 221.4 % with a mean of 89.8 %. Crosses VL052 × VL054178, VL05468 × VL054178 and CML511 × CML491 showed higher MPH and HPH (above 200 %) for grain yield. Generally, about 38 % of the crosses showed above 100 % HPH values for grain yield (Table 4). All the hybrids showed negative MPH and HPH for days to anthesis and silking for both locations. MPH for plant height and for ear height was much lower than the grain yield MPH at both locations, but was still an average of 50.2 and 62.6 % for the two locations. MPH for ears per plant was on average 9.1 % for the two locations. HPH was again very high for grain yield (almost 90 % on average). Plant height and ear height HPH was lower than the MPH, but the average was still 39.8 and 49.8 % respectively. There was almost no positive HPH for ears per plant. Statistically significant positive or negative SCA effects for grain yield and agronomic traits shows that the crosses performed better or poorer than what would be expected from the GCA effects of their respective parents (Tables 2 and 4). The SCA effects for grain yield were high for the crosses VL054178 × CML144, VL05483 × CML144, VL05483 × CML491 and CML511 × CML491 across locations (Table 4).
Analysis of 40 SSR markers yielded a total of 169 alleles among the 15 QPM inbred lines. The number of amplified alleles per primer varied from two to seven with a mean value of 4.2. Primers with a low number of alleles (two per primer) were phi006, phi029, phi032, phi050, phi089 and umc1993 whereas primers phi041, phi053, phi96100 and umc1757 showed a higher number of alleles (seven alleles per primer). A total of 37 unique alleles were detected among the 15 inbred lines (data not shown).
The pairwise genetic dissimilarity values determined using Euclidean distance based on SSR markers ranged from 0.25 (between VL0523 and VL0524) to 0.67 (between VL05200 and VL05561) with a mean of 0.57 (Table 5). Euclidean distance based on morphological evaluation ranged from 0.73 (between VL052 and VL0524) to 2.17 (between VL052887 and VL05482) with a mean of 1.39. Higher genetic distances for SSR markers were observed between VL05200 and VL05561, VL05482 and CML144, VL05561 and CML159, VL05483 and CML144, and CML511 and CML159. Higher morphological distance was observed between VL054178 and VL052887, VL054178 and CML144, and VL052887 and VL05482. Parental inbred lines VL05561 and CML144 showed the highest mean SSR measured genetic distance while VL052 had the lowest. On the other hand, mean morphological distance was higher for VL054178, VL052887 and VL05482. The correlation coefficient between the two distance measures was non-significant (data not shown).
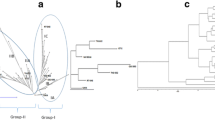
Cluster analyses based on SSR marker and morphological distances classified the parental inbred lines into different groups (Figs. 1 and 2). However, the two distance measures grouped the 15 inbred lines quite differently. The SSR dendrogram formed four groups and the morphological dendrogram five groups. Cluster I of the SSR marker distance contained eight of the 15 inbred lines (Fig. 1). In this cluster the most closely related inbred lines were VL052, VL052887 and CML491, and VL0523 and VL0524. The inbred lines were not related by pedigree, however, VL052, VL052887 and CML491 shared some common phenotypic characteristics such as late anthesis and silking, taller plants and ear heights. VL0523 and VL0524 had similar performance in almost all morphological traits. Inbred lines in cluster II had a closer pedigree relationship; two of them are the reselected versions of CML176. Cluster III contained three inbred lines among which CML159 and VL054178 had a very close pedigree relationship. Inbred lines VL05561 and CML144 were outliers, which clustered distantly from the other inbred lines. In a few cases, morphological distance based clustering of the inbred lines followed the pedigree relationship among the inbreds (Fig. 2). On the basis of this distance measure, inbred lines with closely related pedigrees such as CML144 and VL05200 (cluster III), and CML511 and VL05483 (cluster II) were tightly clustered. Inbred line VL052887 was clustered separately from the others.
SSR marker distance had a strong correlation with hybrid grain yield in both environments (Table 6). SSR distance was inversely correlated with days to anthesis and silking in both environments. The correlation values of genetic distance measured by SSR markers with plant height were highly significant. Morphological distance was significantly correlated with hybrid performance for grain yield at Harare and with days to anthesis and silking. SCA values were also significantly correlated with grain yield for both SSR and morphological data distances. Other significant correlations with SCA were only present at Bako, and were not consistent for the two environments.
The MPH for grain yield showed positive association with morphological distance at Harare and with SSR distance at Bako (Table 7). Across environments, the two distance measures showed positive and significant correlation with MPH. HPH for grain yield had positive and significant correlation with SSR marker distance at Harare and across both locations; but no strong association was observed between morphological distance and HPH. The two distance measures had negative correlations with MPH and HPH of days to anthesis and silking. In some cases, the distance measures had positive and significant correlations with plant and ear height.
Discussion
Highly significant differences observed among the hybrids and inbred lines for various traits indicated the existence of considerable variation among these sets of genotypes, which allows selection of preferred inbred lines and hybrids.
The mean (4.2) and range (2–7) of alleles amplified per primer in the current study confirmed results of previous studies using SSRs in maize inbred lines (Xu et al. 2004; Legesse et al. 2007). Xu et al. (2004) reported a mean of 4.4 and a range of 2–9 alleles per locus for 43 SSR primers in 15 key inbred lines of Chinese maize. Legesse et al. (2007) genotyped 56 highland and mid-altitude maize inbred lines from CIMMYT programs in Ethiopia and Zimbabwe using 27 SSR loci and reported a mean of 3.85 and range of 2–7 alleles per SSR locus. Parental inbred lines VL05482, VL05561, CML144 and VL05200 with abundant unique alleles could easily be differentiated from the other inbred lines. Similar findings were reported previously (Bantte and Prasanna 2003; Choukan et al. 2006).
The ability to provide distance measures between the inbred lines that reflect pedigree relationship ensures a more stringent evaluation of the adequacy of a marker profile data. The fact that maximum distance revealed by both SSR markers and morphological distance between inbred lines with unrelated pedigree is a good indication conferring the ability of the two distance measures to distinguish between maize inbred lines. The efficiency of SSR markers in genetic diversity analysis of maize inbred lines has been proven valuable (Smith et al. 1997; Warburton et al. 2002; Bantte and Prasanna 2003; Legesse et al. 2007). Morphological traits are also useful for preliminary evaluation because it is fast, simple, and can be used as a general approach for assessing genetic diversity among morphologically distinguishable accessions (Yoseph et al. 2006). DNA markers are generally seen as more reliable as morphological markers have shortcomings to detect differences among closely related genotypes and are influenced by prevailing environmental conditions (Smith and Smith 1989).
Most crosses showed positive heterosis for grain yield in both locations, indicating the existence of substantial heterosis in the hybrids. Among all the traits studied, both MPH and HPH were the highest for grain yield, which is consistent with other reports in maize (Saleh et al. 2002; Legesse et al. 2008). The level of mean MPH (112.8 %) and HPH (89.8 %) for grain yield observed in the current study is higher than that reported by Xu et al. (2004) and Legesse et al. (2008), but lower than that reported by Saleh et al. (2002) and Betran et al. (2003). All hybrids showed desirable negative MPH and HPH for days to anthesis and silking, showing that the hybrids were earlier in anthesis and silking than their parental inbred lines. Moderately high positive MPH and HPH for plant and ear height indicates the preponderance of dominance effects among the parental inbred lines for taller plant stature. Similar results were reported previously (Saleh et al. 2002; Legesse et al. 2008). Significant SCA effect indicates the deviation of specific crosses from the average performance of their parents (Hallauer and Miranda 1988). Vasal et al. (1992) argued that positive SCA effects indicate that lines are in opposite heterotic groups while negative SCA effects indicate that lines are in the same heterotic group.
In maize, breeding for hybrid varieties is a well recognized approach for yield increment through the exploitation of heterosis. The role of genetically divergent germplasm is of primary importance for the phenomenon of heterosis to occur. Such phenomenon has been reported repeatedly in maize where genetically unrelated parents will have better cross performance (Hallauer and Miranda 1988; Saleh et al. 2002). In the current study, highly significant positive correlations (although the values were not very high) were seen between SSR marker-based genetic distance and hybrid performance for grain yield, indicating the potential of molecular markers for prediction of hybrid performance. Such significant correlations between genetic distance and grain yield, and suitability of molecular distance for predicting the maize single-cross performance were reported by several investigators (Betran et al. 2003; Barbosa et al. 2003; Xu et al. 2004; Legesse et al. 2008; Kiula et al. 2008). Highly significant negative correlation of SSR marker distance with days to anthesis and silking indicated the dominance for earliness. Consistent with the current study, Legesse et al. (2008) reported positive and significant correlation between plant height and molecular genetic distance. The correlation coefficients of morphological distance with grain yield and agronomic traits were mostly non-significant; except with days to anthesis and silking, where correlation was highly significant.
The magnitude of correlation coefficients of SSR marker and morphological distances with MPH, HPH and SCA of grain yield and agronomic traits were mostly not significant except for SSR genetic distance with heterosis (MP and HP) for plant height at Harare and Bako and morphological distance which was negatively correlated with MP and HP for days to silking at Harare. This indicates that the two distance measures may not be very effective predictors of heterosis and SCA effects in the set of materials studied. Similar results were previously reported (Melchinger et al. 1990; Ajmone-Marsan et al. 1998; Parentoni et al. 2001; Legesse et al. 2008). Contrary to the current finding, Betran et al. (2003) reported a highly significant correlation (r = 0.80) between genetic distance and SCA effects in tropical maize inbreds grown under stress and non-stress environments. Melchinger (1999) and Betran et al. (2003) suggested that hybrid performance and heterosis can better be predicted when genetic distance is smaller than a certain threshold, depending on the germplasm under consideration. Melchinger et al. (1990) noted that the level of correlations between genetic distance, and hybrid performance and heterosis depend on the germplasm used.
Several reasons have been suggested for the low correlation of genetic distance with heterosis and SCA. These include lack of linkage between genes controlling the traits measured, unequal genome coverage, random marker distribution and diversified effect of dominance (Melchinger et al. 1990; Charcosset et al. 1991; Kwon et al. 2002). Effective prediction of heterosis using molecular markers would be feasible when a significant proportion (50 %) of the selected markers used in the calculation of genetic distance are linked with quantitative trait loci (QTL) affecting performance and heterosis of the target trait in the germplasm under study (Melchinger et al. 1990; Charcosset et al. 1991).
To conclude, grain yield showed the highest MPH and HPH among all the traits studied. Negative MPH and HPH for days to anthesis and silking showed that hybrids were earlier in anthesis and silking than their parental inbred lines. The desirable heterosis observed in this study for grain yield and earliness indicates the potential of the inbred lines for hybrid development. SSR markers and morphological distances estimated between all possible pair-wise comparisons indicated the presence of considerable variations among the inbred lines studied, which can systematically be exploited in the breeding program. SSR marker-based genetic distances showed higher correlation values with grain yield and some agronomic traits than morphological distances, indicating the potential of SSR markers in predicting hybrid performance. Even though the correlations of SSR marker and morphological distances with heterosis and SCA effects were significant in some cases, the magnitudes were relatively low. In general, the two distance measures can be useful for the identification of genetically similar/different genotypes.
References
Ajmone-Marsan P, Castiglioni P, Fusari F, Kuiper M, Motto M (1998) Genetic diversity and its relationship to hybrid performance in maize as revealed by RFLP and AFLP markers. Theor Appl Genet 96:219–227
Bantte K, Prasanna BM (2003) Simple sequence repeat polymorphism in quality protein maize (QPM) lines. Euphytica 129:337–344
Barbosa AMM, Geraldi IO, Benchimol LL, Garcia AAF, Souza JR, Souza AP (2003) Relationship of intra- and interpopulation tropical maize single cross hybrid performance and genetic distances computed from AFLP and SSR markers. Euphytica 130:87–99
Betran FJ, Ribaut JM, Beck DL, de Leon DG (2003) Genetic diversity, specific combining ability, and heterosis in tropical maize under stress and non stress environments. Crop Sci 43:797–806
Charcosset A, Lefort-Busen MM, Gallais A (1991) Relationship between herosis and heterozgosity at marker loci: a theoretical computation. Theor Appl Genet 81:571–575
Choukan R, Hossainzadeh A, Ghannadha MR, Warburton ML, Talei AR, Mohammadi SA (2006) Use of SSR data to determine relationships and potential heterotic groupings within medium to late maturing Iranian maize inbred lines. Field Crop Res 95:212–222
CIMMYT (2004) Maize inbred lines release by CIMMYT: a compilation of 497 CIMMYT maize lines (CMLs), CML1–CML497. CIMMYT, Mexico, DF
CIMMYT (2005) Laboratory protocols: CIMMYT applied molecular genetics laboratory, 3rd edn. CIMMYT, Mexico, DF
Drinic SM, Trifunovic S, Drinic G, Konstantinov K (2002) Genetic divergence and its correlation to heterosis in maize as revealed by SSR-based markers. Maydica 4:1–8
Griffing B (1956) Concept of general and specific combining ability in relation to diallel crossing system. Aust J Biol Sci 9:463–493
Hallauer AR, Miranda JB (1988) Quantitative genetics in maize breeding, 2nd edn. Iowa State University Press, Ames, IA
Hartkamp AD, White JW, Rodríguez AA, Bänziger M, Srinivasan G, Granados G, Crossa J (2000) Maize production environments revisited: a GIS-based approach. CIMMYT, Mexico, DF
Hintze JL (1998) NCSS-2000 Statistical System for Windows. Number Cruncher Statistical Systems, Keysville, Utah 84037, Canada
Kiula BA, Lyimo NG, Botha AM (2008) Association between AFLP-based genetic distance and hybrid performance in tropical maize. Plant Breed 127:140–144
Kwon SJ, Ahn SN, Jeong EG, Hwang HG, Choi HC, Moon HP (2002) Relationship between genetic divergence and hybrid performance in Japonica rice grown in a cold water irrigated field. Euphytica 128:389–396
Lanza LLB, de Souza Jr CL, Ottoboni LMM, Vieira MLC, Souza AP (1997) Genetic distance of inbred lines and prediction of maize single-cross performance using RAPD markers. Theor Appl Genet 94:1023–1030
Legesse BW, Myburg AA, Pixley KV, Botha AM (2007) Genetic diversity of African maize inbred lines revealed by SSR markers. Hereditas 144:10–17
Legesse BW, Myburg AA, Pixley KV, Twumasi-afriye S, Botha AM (2008) Relationship between hybrid performance and AFLP based genetic distance in highland maize inbred lines. Euphytica 162:313–323
Melchinger AE (1999) Genetic diversity and heterosis. In: Coors JG, Pandey S (eds) The genetics and exploitation of heterosis in crops. ASA, CSS, and SSSA, Madison, WI, pp 99–118
Melchinger AE, Lee M, Lamkey KR, Woodman WL (1990) Genetic diversity for restriction fragment length polymorphisms: relation to estimated genetic effects in maize inbreds. Crop Sci 30:1033–1040
Melchinger AE, Messmer MM, Lee M, Woodman WL, Lamkey KR (1991) Diversity and relationships among U.S. maize inbreds revealed by restriction length polymorphisms. Crop Sci 31:669–678
Moll RH, Lonquist JH, Foreuno JV, Johnson EC (1965) The relationship of heterosis and genetic divergence in maize. Genetics 52:139–144
Parentoni SN, Magalhaes JV, Pacheco CAP, Santos MX, Abadie T, Gama EEG, Guimaraes PEO, Meirelles WF, Lopes MA, Vasconcelos MJV, Paiva E (2001) Heterotic groups based on yield-specific combining ability data and phylogenetic relationship determined by RAPD markers for 28 tropical maize open pollinated varieties. Euphytica 121:197–208
Saleh G, Abdullah D, Anuar AR (2002) Performance, heterosis and heritability in selected tropical maize single, double and three-way cross hybrids. J Agr Sci 2002:21–28
Smith JSC, Smith OS (1989) The description and assessment of distances between inbred lines of maize: II. The utility of morphological, biochemical and genetic descriptors and a scheme of testing of distinctiveness between inbred lines. Maydica 34:151–161
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparison with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Stuber CW (1994) Heterosis in plant breeding. Plant Breed Rev 12:227–251
Vasal SK, Srinivasan G, Pandey S, Cordova HS, Han GC, Gonzalez F (1992) Heterotic patterns of ninety-two white tropical CIMMYT maize lines. Maydica 37:259–270
Warburton ML, Xianchun X, Crossa J, Franco J, Melchinger AE, Frisch M, Bohn M, Hoisington D (2002) Genetic characterization of CIMMYT inbred maize lines and open pollinated populations using large scale fingerprinting methods. Crop Sci 42:1832–1840
Xu S, Liu J, Liu G (2004) The use of SSRs for predicting the hybrid yield and yield heterosis in 15 key inbred lines of Chinese maize. Hereditas 141:207–215
Yoseph B, Botha AM, Myburg AA (2006) Genetic diversity in traditional Ethiopian highland maize accessions assessed by AFLP markers and morphological traits. Biodivers Conserv 15:2655–2671
Zhang Y, Kang MS (1997) DIALLEL-SAS: a SAS program for Griffing’s diallel analyses. Agron J 89:176–182
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wegary, D., Vivek, B. & Labuschagne, M. Association of parental genetic distance with heterosis and specific combining ability in quality protein maize. Euphytica 191, 205–216 (2013). https://doi.org/10.1007/s10681-012-0757-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0757-2