Abstract
Greenbug is a major damaging insect to sorghum production in the United States. Among various virulent greenbug biotypes, biotype I is the most predominant and severe for sorghum. To combat with the damaging pest, greenbug resistant sources were obtained from screening sorghum germplasm collection. This experiment was conducted to identify the genomic regions contributing resistance to greenbug biotype I in a sorghum accession, PI 607900. An F2 mapping population consisting of 371 individuals developed from a cross of the resistant line with an elite cultivar, BTx623 (susceptible) were tested and scored for their response to greenbug feeding in the greenhouse. Significant differences in resistance were observed between the two parental lines and among their F2 progeny in response to greenbug feeding at 7, 10, 14 and 21 days after infestation. A linkage map spanning a total length of 729.5 cM across the genome was constructed with 102 polymorphic SSR markers (69 genomic and 33 EST SSRs). Of those microsatellite markers, 48 were newly developed during this study, which are a useful addition for sorghum genotyping and genome mapping. Single marker analysis revealed 29 markers to be significantly associated with the plant response to greenbug feeding damage. The results from interval mapping, composite interval mapping and multiple interval mapping analyses identified four major QTLs for greenbug resistance on chromosome 9. These QTLs collectively accounted for 34–82 % of the phenotypic variance in greenbug resistance. Minor QTLs located on chromosome 3 explained 1 % of the phenotypic variance in greenbug resistance. The major allele for greenbug resistance was on chromosome 9 close to receptor-like kinase Xa21-binding protein 3. These markers are useful to screen more resistant genotypes. Furthermore, the markers tagged to QTL regions can be used to enhance the sorghum breeding program for greenbug resistance through marker-assisted selection and map-based cloning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorghum is the fifth most important cereal crop worldwide with its multifaceted uses such as food, fiber and biofuel. The crop also has large commercial value in cattle, poultry and dairy industries as it is mainly used as feed for animals in the United States. Insect attack is one major factor limiting sorghum production. Sorghum is attacked by more than 150 insect pests, of which aphids are the most prominent (Young and Teetes 1977; Sharma 1993). Greenbug, Schizaphis graminum (Rondani), a sap-sucking aphid, is one of the most devastating to sorghum productivity (Teetes 1980). Greenbugs feed on the leaves of grasses and cereals, absorbing nutrients present in the sieve elements while incorporating phytotoxins resulting in visible symptoms in the plants, including chlorosis and red necrotic spots (Van-Emden and Harrington 2007). The loss of sorghum crop incurred due to greenbug damage was estimated for $274 million annually (Eddleman et al. 1999). To date, different biotypes of greenbug were discovered chronologically on sorghum, among them biotype I, identified and designated in 1990, is of economic importance because it causes huge losses in sorghum yield (Harvey et al. 1991; Kofoid et al. 1991; Teetes and Pendleton 2000).
Host plant resistance is perceived as an eco-friendly, socially acceptable and effective component of integrated pest management in deploying resistant cultivars against insect damage (Bramel-Cox et al. 1986; Andrews et al. 1993; Sharma 1993; Sharma and Ortiz 2002; Smith 2004). Therefore, host plant resistance is of prime importance in the arsenal of aphid-plant interaction. Different transcriptomic studies with microarray profiling have focused on the greenbug-sorghum interaction and emphasized the role of signaling compounds and defense-activated genes (Huang 2007). Suppression subtractive hybridization revealed the down-regulation of cysteine proteinase inhibitors and the up-regulation of genes such as Xa1, antimicrobial proteins and other signaling compounds in response to greenbug damage on sorghum plants (Park et al. 2006). Another transcriptomic study also identified the differential expression of 82 greenbug responsive genes, including a LRR-containing glycoprotein sequence and other defense related proteins in plants infested with greenbug (Zhu-Salzman et al. 2004). These studies have shown the prominent role of plant R genes in defense response to greenbug attack through signal transduction pathway.
Molecular markers have diverse utility in dissecting genes for greenbug resistance and in better understanding the genetic basis and mechanism of resistance (Yencho et al. 2000). Recent progress in sorghum genomics has availed the genome sequence to the public to aid in the development of several different types of molecular markers, including gene-based markers. Furthermore, post-genomic progress has accelerated linkage mapping experiments for all economically important traits including greenbug resistance (Paterson 2008; Paterson et al. 2009).
In the past decade, the use of molecular markers has helped to identify suitable greenbug resistant sorghum, which has fostered marker-assisted breeding programs for greenbug resistant crops. To date, five independent QTL mapping experiments have been conducted in sorghum to identify greenbug resistance to four different greenbug biotypes (Agrama et al. 2002; Katsar et al. 2002; Nagaraj et al. 2005; Wu et al. 2007; Wu and Huang 2008). In these studies, seven disparate genetic sources of resistance were used, which revealed multiple genomic regions accounting for resistance to greenbug biotypes C, E, I, and K. Among these studies, Katsar et al. (2002) identified at least three loci present on chromosome SBI05, SBI06 and SBI07 conferring resistance to greenbug biotype I. Agrama et al. (2002) quantified chlorophyll loss caused by greenbug injury and detected nine genomic regions affecting both biotype-specific and biotype non-specific resistance and tolerance to Biotype I and K. Of the seven QTLs detected by Agrama et al. (2002) that were associated with biotype-specific resistance and tolerance to greenbug damage, three markers present on chromosomes SBI02, SBI05 and SBI09 were linked with biotype I-specific resistance and tolerance. A similar study by Nagaraj et al. (2005) using chlorophyll loss as an indicator to greenbug damage identified three QTLs present on the sorghum chromosome SBI01 and SBI04 for biotype I resistance and tolerance. The recent study conducted by Wu and Huang (2008) have shown a major QTL located on sorghum chromosome SBI09 conditioning resistance to biotype I. It is obvious from these studies that resistance and tolerance to greenbug damage originates from multiple regions of the genome depending on the resistance source contributed by various genotypes and that some of the alleles are biotype-specific, and others are biotype non-specific.
Despite the economic importance of continuous breeding efforts to develop resistant sorghum cultivars, progress has been slow in identification of greenbug biotype I resistance sources for incorporation into existing greenbug resistance management practices. The resistance to aphid attack is governed by very few resistance loci and alleles, considering the meager sources of resistance (Dogimont et al. 2010). Previous work indicated that sorghum line PI 607900 contained strong resistance to biotype I and was genetically distinct from other known major resistant genotypes (Tuinstra et al. 2001; Wu et al. 2006). Tuinstra et al. (2001) reported PI 607900 (KS 97) had superior general combining ability of greenbug biotype I resistance compared to PI 550610, which carried one major and one minor QTL on SBI09 resistant to greenbug biotype I (Wu and Huang 2008). A worldwide collection of 40,000 sorghum germplasm accessions was evaluated for greenbug resistance, which resulted in identifying 21 resistant sources. Among these 21 lines, PI 607900 outperformed other lines with a damage rating of 1.1 (Huang 2011). Therefore, new resistant sources are always sought for their natural variation for resistance towards insects (Broekgaarden et al. 2011).
Our objective in the present research was to identify genomic regions associated with greenbug biotype I resistance in sorghum accession, PI 607900 using SSR markers. Microsatellite markers, diagnostic to biotype I resistance, developed in this study will be a useful tool for identifying resistant genotypes from the sorghum germplasm pool and facilitating greenbug resistance improvement in sorghum through marker-assisted selection.
Materials and methods
Selection of resistant source for development of the mapping population
The parental lines of sorghum for our QTL study were BTx623 (susceptible parent) and PI 607900 (resistant parent). BTx623 is the cultivar utilized in the sorghum genome sequencing project (Kresovich et al. 2005) and is susceptible to greenbug biotype I. PI 607900, also known as KS 97, is highly resistant to greenbug biotype I (Tuinstra et al. 2001). PI 607900 was developed by Dr. Gerald Wilde at Kansas State University using IS 2388 as a heterogenous seed source from South Africa (Wilde and Tuinstra 2000). In our preliminary screening for greenbug biotype I resistance, involving three major resistant sources (PI 550607, PI 550610, PI 607900) and two susceptible checks, we confirmed the PI 607900 accession as an outstanding greenbug biotype I-resistant line among currently available sorghum lines (Wu and Huang 2006 unpublished data). We used 371 F2 individuals for genotyping and their F2:3 families for phenotyping experiment.
Marker development and genotyping analysis
All the markers utilized in this study were microsatellite markers. The information of nuclear SSR primers was obtained from publically available sorghum linkage maps (Brown et al. 1996; Taramino et al. 1997; Dean et al. 1999; Bhattramakki et al. 2000; Kong et al. 2000; Cordeiro et al. 2001; Schloss et al. 2002). In addition, we developed additional nuclear SSRs and genic SSRs in the present investigation. Genic microsatellites were developed either in silico (gene-based SSR) using the sequence data base from the Phytozome website (http://www.phytozome.net/sorghum) or using collective sequence information from various EST databases (EST SSR). SSRIT, a SSR identification tool (http://www.gramene.org/db/markers/ssrtool) (Temnykh et al. 2001) was used to search the presence of microsatellites among these sequences. The search criteria to mine the core repeat motif was set to identify the maximum repeat motif length group with the five repeats as the minimum threshold repeats in the sequence. These sequences containing the SSR were further utilized in designing primers to amplify the repeat motifs with flanking sequences. Primer 3.0 software, v 0.4.0 was used with default parameters to obtain both forward and reverse flanking primer sequences (Rozen and Skaletsky 2000). The expected PCR product size was set to 100–300 bp, 40–60 % GC content with optimum of 50 %, and an annealing temperature of 55–58 °C. The forward primer was extended with the M13 primer sequence (5′-CACGACGTTGTAAAACGACG-3′) before the 5′ end of the sequence. The standard naming system for the newly developed marker was followed as proposed by De Vicente et al. (2004).
DNA was extracted from 1.5-month-old seedlings of F2 plants grown in the greenhouse using a modified CTAB (cetyl-trimethyl-ammonium bromide) procedure as described by Murray and Thompson (1980) except that we used a different method for grinding tissue samples (drill with a blunt 1 ml tip). The final concentration of DNA was diluted to 10 ng/μl as a working stock of PCR DNA template. We first screened all 401 available SSR markers with DNA from the parents, which resulted in identification of 107 polymorphic markers for the genotyping experiment. The PCR reaction volume and amplification procedure were followed as described by Wu and Huang (2008). We conducted the PCR reactions for genotyping all 371 F2 individuals along with the parental lines in a PTC-220 Dyad Thermal Cycler (MJ Research Inc, MA, USA) and 2720 thermal cycler (Applied Biosystems, CA, USA). The PCR reactions were performed with an initial denaturation step of 94 °C for 5 min proceeded by 13 cycles of denaturation at 94 °C for 20 s, primer annealing at 58 °C for 1 min, primer extension at 72 °C for 30 s, followed by 27 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 1 min, extension at 72 °C for 30 s, and a final primer extension at 72 °C for 10 min. The PCR products were separated in 6.5 % polyacrylamide gels mounted using a LI-COR 4300 DNA Analyzer (LI-COR Biosciences, Lincoln, NE, USA). The electrophoretic conditions were as follows: 1,500 V, 40 mA, 40 W, and 45 °C for 2.00 h. The two parents were added as controls either in the beginning of the 700 dye gel or at the end wells in the 800 dye gel. The DNA banding pattern in the gel was scored manually. For each marker, we recorded whether the individual was homozygous for the BTx623 or PI 607900 allele, heterozygous (both BTx623 and PI 607900 allele present), or was missing the marker amplification.
Phenotyping and data analysis
Phenotyping for greenbug resistance was conducted with the F2-derived F3 sorghum families. Two phenotyping experiments were performed, each arranged in a randomized complete block design consisting of three blocks. Within a block, each of the 371 F2-derived F3 families (F2–3) was represented with two seedlings along with two parental lines. Two F2–3 seeds from a single genetic family were planted together in a cell present in a growing tray of 12″ X 20″ X 1.75″ size filled with Redi-Earth soil. The two experiments were conducted in different section of the greenhouse with a gap of 3 days in sowing time.
Greenbug biotype I cultures were reared on barley (cultivar ‘Schuyler’) seedlings in the greenhouse of USDA-ARS, Stillwater until ready for use. For infestation, barley seedlings co-cultivated with greenbugs were cut and placed immediately between the rows of 12-day-old sorghum seedlings, equally and effectively infesting all the sorghum seedlings. F2-derived F3 families (F2–3) were phenotyped for greenbug feeding response at 7, 10, 14, and 21 days post-infestation. There were 12 plants across two experiments assayed for greenbug resistance which represented one F2 family. Evaluation of the response of sorghum seedlings to greenbug feeding was conducted using a visual estimation to class damage to seedling foliage with a discrete scale that ranged from 1 to 6, where 0 = no damage, 1 = < 20 % damage, 2 = 20–40 % damage, 3 = 40–60 % damage, 4 = 60–80 % damage, 5 = > 80 % damage, and 6 = dead (Starks and Burton 1977). The greenhouse conditions were maintained with constant temperature (28 ± 2 °C) and constant photoperiod (14 L:10 D) throughout the experiment (Wu et al. 2007; Wu and Huang 2008).
SAS software version 9.2 (SAS Institute 2008) was used to estimate heritability and variance components. The means and standard errors for parental lines and F2-derived F3 families were calculated using PROC MEANS. For all analyses, we calculated the mean of greenbug response of the two seedlings within an F2-derived F3 family raised within a single growing cell. To determine whether the F2-derived F3 families differed in greenbug resistance, we performed analysis of variance (ANOVA) with family and block as random factors (PROC MIXED). Similarly, to determine whether the two parental lines differed in greenbug resistance at the 4 time points, we performed an ANOVA with family as a fixed factor (PROC GLM). REML estimates of variance components were obtained using SAS/MIXED. We calculated the heritability of greenbug resistance at 7, 10, 14, and 21 days post-infection on a plot (block) basis and family mean basis following the REML univariate mixed-model analysis described by Holland et al. (2003). The phenotypic correlations among greenbug resistance at the four time points were estimated using multivariate REML module.
Linkage analysis and QTL mapping
The genetic map was constructed using MAPMAKER/EXP 3.0 (Lander et al. 1987) with a logarithm of odds score (LOD) of 3.0 and maximum linkage threshold of 40 cM. First, a few known markers were used as anchoring markers to ascertain the number of linkage groups that were formed in the present mapping population using the ‘Group’ command. Assignment of linkage group to a specific chromosome was defined based on the previous map information and the current number of linkage groups obtained in this experiment. The linkage groups were assembled, ordered and named based on the nomenclature given by Kim et al. (2005). The best order among the markers was chosen after using the ‘Compare’ command. The rest of the markers were added using the ‘Try’ command. Finally, the ‘Ripple’ command was executed to confirm the best possible order for constructing the framework map with log-likelihood threshold value of 2.0. The relative map distances between the markers were estimated by translating the recombination fractions into genetic mapping distances using the Kosambi mapping function (Kosambi 1944).
The output files were fed into QTL Cartographer version 2.5 (Wang et al. 2010) for QTL analysis. The mean value of all twelve plant’s greenbug resistance scores associated with each trait was used in QTL analysis. The empirical LOD threshold significant values for declaring QTLs associated with each trait was determined by conducting a 1000 permutation test (Churchill and Doerge 1994). Single Marker Analysis, Simple Interval Mapping and Composite Interval Mapping were performed prior to multiple interval mapping (MIM) analyses to have a glimpse of the significant associated markers and the variation explained by these markers. We selected a new model in the MIM module by selecting QTLs that had high LOD values in Composite Interval Mapping, Interval Mapping and Single Marker Analysis. The optimum position of QTLs and significant QTLs were tested in an iterative manner. The P value chosen for declaring a significant QTL was 0.05 with LOD values obtained from 1,000 permutation tests.
Results
Resistance source and phenotypic analysis
Based on the phenotyping data, plant response to greenbug feeding varied significantly in resistance between the two parents, BTx623 and PI 607900 as well as among the F2-derived F3 families (Table 1). Heritability values were high, ranging from 71 to 83 % for all four greenbug resistance traits which are derived on a family mean basis of plant responses. These values are reported along with various components of variation (Table 2). All four greenbug resistance traits were positively correlated, with coefficients from 0.89 to 0.97 (Table 3).
Genotyping and QTL detection
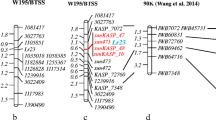
Of the 401 SSR markers, 33 % of nuclear SSRs and 20–25 % of ESTs and gene-based SSRs were polymorphic. We obtained 107 polymorphic markers for genotyping among F2 individuals, which consisted of 73 genomic SSRs, 30 EST-based SSRs, and 4 gene-based SSRs. In this research, we developed and used 48 novel SSR markers that represent additional tools for sorghum genomic studies. These newly developed markers include 34 genic markers and 14 nuclear SSRs (Table 4). There were 18 markers that deviated from the expected segregation ratio in this mapping experiment. Of these, 13 markers belong to chromosome 2, which deviated towards PI 607900. The linkage map included 13 groups with 102 markers, which spanned a total length of 729.5 cM (Fig. 1). The linkage map covered nine chromosomes, except chromosome SBI05. Five markers (sb6_036-SBI03, Xtxp303, Xtxp299-SBI05, Xtxp224-SBI07, and Starssbem94-SBI09) were unlinked and two of these markers were assigned to chromosome 5 in previous sorghum maps. The marker order and map distances were in consensus with previously published maps (Wu and Huang 2006; Mace et al. 2009).
Single marker analysis identified 29 markers linking to four greenbug resistance traits with R 2 (%) values explaining from 1 to 72 % of the phenotypic variation within these traits. All the markers present on chromosome 9 were significantly associated with the greenbug resistance traits measured. Interval mapping and composite interval mapping results were corroborative in identification of the major interval influencing greenbug resistance, Starssbnm 78–Starssbnm 102.
Initial MIM results indicated a consistent QTL for the four different time points of the study in the intervals of Starssbnm 78–Starssbnm 81, Starssbnm 81–Starssbnm 102, and one minor QTL near Starssbnm 47–Strassbnm 64. Additional microsatellite markers were developed within these regions for candidate genes of insect resistance using the information from the phytozome sorghum database. At least three markers were polymorphic for the homologue similar to the receptor-like kinase Xa21-binding protein 3 gene, but one of them was not consistent and failed to produce sufficient information, hence only two markers were used in the mapping experiment. In addition, we developed one marker each for the chitinase gene and the jasmonate precursor, OPDA (12-oxo-phytodienoic acid) gene.
The identified QTLs with their corresponding genetic effects are summarized in Table 5. A major QTL for greenbug resistance response from plants for all time points was evident between the interval Starssbnm 81–Starssbnm 102 located on chromosome 9 after incorporating gene-based markers (Fig. 2). The locus associated with the interval Starssbnm 93–Starssbem 296 was consistent across all traits, but explained different phenotypic variation. Nine significant QTLs were detected for all traits together with LOD values from 2.5 to 138.3 and some of these overlapped for different traits. Of the nine QTLs, two minor QTLs were located on chromosome 3b, between Starssbem 162 and Starssbem 265, and between Xtxp16 and Starssbem 162, which explained 1.3 and 1.0 % of greenbug resistance at day 14 and 21, respectively. These minor QTLs are reported here owing to the high LOD values associated with them. The markers Starssbnm 93 and Starssbem 296 were tagged to greenbug resistance across three time points post-infestation, and hence a major QTL resides at this region, which we designated Qstsgr-sbi09i (Q-QTL, st-Stillwater-ARS, sgr-Schizaphis graminum resistance, sbi09i- sorghum bicolor chromosome 9 first QTL). This region involving Starssbnm93 and Starssbem 296 was responsible for 82.4 % of phenotypic variation at day 21 post-infestation. Two more QTLs were designated as Qstsgr-sbi09ii and Qstsgr-sbi09iii in the intervals Starssbem 286–Starssbnm 93 and Starssbem 298–Starssbnm 102, which together accounted for 74.5 % of phenotypic variation for the trait day 7. There were two more QTLs consistently found in the region flanked by GSR 78–GSR 81 explaining 17.6 and 17.3 % of the greenbug resistance for the trait day 10 and 14, respectively. Hence, we designated QTL found in this region as Qstsgr-sbi09iv. Although the QTL Qstsgr sbi09iv is associated with LOD values less than 3.0, it is reported here as it was responsible for high phenotypic variation. In total, we identified four major QTLs on chromosome 9 of which region flanked by GSR 93–GSR 296 explained highest phenotypic variation. Hence, the major allele responsible for greenbug resistance was closer to receptor-like kinase Xa21-binding protein 3. The final model at day 14 post-infestation explained the highest phenotypic variation among all traits and this model consisted of alleles present on chromosomes 9 and 3b accounting for 85.3 % phenotypic variation. Additive and partial dominance effects were associated with all of the QTLs identified in this study. The negative sign associated with additive effects indicated that increasing allelic effect was derived from the resistant source (PI 607900) for all the identified loci. However, we also observed a few more putative QTLs on sorghum chromosome SBI 3b (Xtxp285-Xtxp34), SBI 6b (Xtxp57-Xcup37) and SBI 10 (Xtxp320-ESR78), which explained either less phenotypic variation (less than 1 %) or were associated with low LOD values. QTLs with an increased effect from the susceptible parent (BTx623) were also observed to be associated with the chitinase gene, but were not reported here due to a lack of consistency and low LOD values. No significant QTLs were associated with the marker Starssbem 274 developed for the OPDA (12-oxo-phytodienoic acid) gene, a precursor for jasmonate synthesis. Hence, among the newly developed markers, genic markers developed for sequences similar to receptor-like kinase Xa21-binding protein 3 and chitinase were useful in detecting significant QTLs.
The major QTLs identified in this study. The LOD score peak profile using MIM analysis for four greenbug resistance traits identified on chromosome 9. GDR07, GDR10, GDR14, and GDR21, represents greenbug damage response scored at 7, 10, 14, and 21 days after infestation. The identified QTLs for four traits are designated with symbols given in legend and distances between markers is in centimorgan (cM), filled circle GDR07, filled triangle GDR10, filled cresent GDR14, filled star GDR21
Discussion
Since the outset of extensive sorghum greenbug attack in 1968, varieties resistant to various biotypes had been developed but were frequently overtaken by newly emerging greenbug biotypes. Screening efforts to identify new sources of resistance have been a vital component of sustainable practices for greenbug management. Genomics-assisted techniques have hastened sorghum breeding efforts by facilitating marker-assisted selection for developing greenbug resistant varieties. The availability of the sorghum genome sequence has aided in developing novel markers for use in the current study. To our understanding, this is the first published map to tag genic SSRs for greenbug resistance in sorghum, which has further relevance to gene expression in response to aphid feeding because the genic microsatellites developed from expressed sequences have some advantages over genomic SSRs (Varshney et al. 2005; Yonemaru et al. 2009).
Resistance to greenbug biotype I was governed by a complimentary gene action between two major dominant genes (Tuinstra et al. 2001). PI 607900 is a genetically distinct source of resistance against greenbug biotype I (Wu et al. 2006). Resistance is manifested in the distinctive categories; antibiosis, antixenosis and tolerance (Painter 1951). The resistance categories for this source were classified as antibiosis and tolerance (Wilde and Tuinstra 2000). However, later studies have shown that resistance to greenbug biotype I is controlled by polygenes (Agrama et al. 2002; Katsar et al. 2002; Nagaraj et al. 2005; Wu and Huang 2008). Our main objective was to use sorghum accession, PI 607900, which proved to be distinct from several previous studies and to identify QTL regions controlling greenbug resistance. In this study several genomic regions contribute to greenbug resistance in resistant sorghum line PI 607900 and it contained several genes for insect resistance which were identified in the major QTL region. The identification of major QTLs for resistance to greenbug biotype I on chromosome 9 corroborates earlier mapping efforts for greenbug resistance (Wu and Huang 2008). But, the location of QTL regions was different compared to the early study conducted by Wu and Huang (2008). Therefore, the present investigation provided novel alleles for greenbug resistance using a different resistant source. Moreover, sorghum chromosome SBI09 also harbored genes for resistance to different greenbug biotypes, including C and E (Agrama et al. 2002; Katsar et al. 2002; Wu et al. 2007). Furthermore, the major QTL accounting for the highest phenotypic variation was consistently observed in the interval of Starssbnm 93–Starssbem 296 or near Starssbnm 93 at all four post-infestation times. Equally important, all the major QTLs are located in the interval of Starssbnm 78–Starssbnm 102. The high phenotypic variation can be attributed to a bigger mapping population size, more closely spaced markers and a clear phenotypic response. The selection of plants with a combination of the above flanking markers for Qstsgr-sbi09i would better assist in precise selection of a greenbug resistant variety compared to using a single marker alone. The region between Starssbnm 78–Starssbnm 102, which roughly corresponds to 1.02 Mb of physical distance on sorghum chromosome SBI09, contained several potential putative candidate genes. Most genes prominent and relevant to disease and insect resistance were the homologues similar to receptor-like kinase Xa21-binding protein 3 (Song et al. 1995), the chitinase gene, cysteine protease and amino acid selective channel protein. Among all the genic markers developed, the markers developed for Xa21-binding protein 3 was tightly linked to greenbug resistance traits measured in this study. The QTLs identified for correlated traits for greenbug resistance resided in the same region of the chromosome; similar results were observed in the earlier mapping experiments due to the phenomenon of pleiotropy of a single gene or tight linkage of a few genes affecting the trait (Aastveit and Aastveit 1993; Agrama 1996; Agrama et al. 2002; Wu et al. 2007; Wu and Huang 2008; Satish et al. 2009).
Genetic and molecular basis of aphid resistance has been reviewed and deciphered more recently with an emphasis on the involvement of R gene products in aphid resistance among agriculturally important crops (Thompson and Goggin 2006; Smith and Boyko 2007; Howe and Jander 2008; Dogimont et al. 2010). The genetic diversity of the sorghum gene pool from various parts of the world was assessed to identify resistance genes associated with greenbug attack from different resistant donors (Radchenko and Zubov 2007). Resistance to greenbugs was often conferred by either a few genes on a gene-for-gene basis or controlled by polygenes (Puterka and Peters 1995; Tuinstra et al. 2001; Dogimont et al. 2010). Aphid resistant plants are characterized with specific responses involving a gene-for-gene interaction and resistance in such a case involves loci containing NBS-LRR sequences (Smith and Boyko 2007; Dogimont et al. 2010). In the present findings, alleles accounting for a major proportion of variation were more closely linked to a homologue similar to receptor-like kinase Xa21-binding protein 3 than to a chitinase gene found in the nearby interval. Moreover, this project particularly suggests the involvement of Xa21 gene in a defensive response mounted by the plant. The up-regulation of Xa1 gene in greenbug-infested sorghum plants supports our finding that similar but slightly different genes are involved in greenbug response (Park et al. 2006). However, it remains to be determined whether the Xa21 plays a direct or indirect role in modifying response of the plant to greenbug damage.
In conclusion, this study identified major QTLs in the marker interval Starssbnm 78–Starssbnm 102 on SBI-09 for greenbug resistance in sorghum. The resistant parent used in this study contains novel alleles for greenbug resistance. This project aided in the development of molecular markers and in the identification of the location of these markers on the chromosomes for future map-based cloning experiments. The efforts to improve sorghum breeding programs for greenbug resistance management can be accelerated by using these tagged molecular markers.
References
Aastveit AH, Aastveit K (1993) Effects of genotype-environment interactions on genetic correlations. Theor Appl Genet 86:1007–1013
Agrama HAS (1996) Sequential path analysis of grain yield and its components in maize. Plant Breed 115:343–346
Agrama HA, Wilde GE, Reese JC, Campbell LR, Tuinstra MR (2002) Genetic mapping of QTLs associated with greenbug resistance and tolerance in Sorghum bicolor. Theor Appl Genet 104:1371–1378
Andrews DJ, Bramel-Cox PJ, Wilde GE (1993) New sources of resistance to greenbug, biotype I, in sorghum. Crop Sci 33:198–199
Bhattramakki D, Dong J, Chhabra AK, Hart GE (2000) An integrated SSR and RFLP linkage map of Sorghum bicolor (L.) Moench. Genome 43:988–1002
Bramel-Cox PJ, Olunju Dixon AG, Reese JC, and Harvey TL (1986) New approaches to the identification and development of sorghum germplasm resistant to the biotype E greenbug. In: 41st Annual Corn and Sorghum Research Conference, vol. 41, pp 1–16
Broekgaarden C, Snoeren TAL, Dicke M, Vosman B (2011) Exploiting natural variation to identify insect-resistance genes. Plant Biotechnol J 9:819–825
Brown SM, Hopkins MS, Mitchell SE, Senior ML, Wang TY, Duncan RR, Gonzalez-Candelas F, Kresovich S (1996) Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum (Sorghum bicolor (L.) Moench). Theor Appl Genet 93:190–198
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
De Vicente C, Metz T, Alercia A (2004) Descriptors for genetic markers technologies. International Plant Genetic Resources Institute, pp 1–24
Dean RE, Dahlberg JA, Hopkins MS, Mitchell SE, Kresovich S (1999) Genetic redundancy and diversity among ‘orange’ accessions in the U.S. National sorghum collection as assessed with simple sequence repeat (SSR) markers. Crop Sci 39:1215–1221
Dogimont C, Bendahmane A, Chovelon V, Boissot N (2010) Host plant resistance to aphids in cultivated crops: genetic and molecular bases, and interactions with aphid populations. CR Biol 333:566–573
Eddleman BR, Chang CC, McCarl BA (1999) Economic benefits from grain sorghum variety improvement in the United States. In: Wiseman BR, Webster JA (eds) Economic, environmental, and social benefits of resistance in field crops. Entomological Society of America, Lanham
Harvey TL, Kofoid KD, Martin TJ, Sloderbeck PE (1991) A new greenbug virulent to E-biotype resistant sorghum. Crop Sci 31:1689–1691
Holland JB, Nyquist WE, Cervantes-Martinez CT (2003) Estimating and interpreting heritability for plant breeding. Plant Breed Rev 22:9–112
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66
Huang Y (2007) Phloem feeding regulates the plant defense pathways responding to both aphid infestation and pathogen infection. In: Xu Z, Li J, Xue Y, Yang W (eds) Biotechnology and sustainable agriculture 2006 and beyond. Springer, Amsterdam, pp 215–219
Huang Y (2011) Improvement of crop protection against greenbug using the worldwide sorghum germplasm collection and genomics-based approaches. Plant Genetic Resour 9(2):317–320
Katsar CS, Paterson AH, Teetes GL, Peterson GC (2002) Molecular analysis of sorghum resistance to the greenbug (homoptera: aphididae). J Econ Entomol 95:448–457
Kim JS, Klein PE, Klein RR, Price HJ, Mullet JE, Stelly DM (2005) Chromosome identification and nomenclature of Sorghum bicolor. Genetics 169:1169–1173
Kofoid KD, Harvey TL, Sloderbeck PE (1991) A new greenbug, biotype I, damaging sorghum. Proceedings of the 46th Annual Corn and Sorghum Research Conference. American Seed Trade Association, Washington, DC
Kong L, Dong J, Hart GE (2000) Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs). Theor Appl Genet 101:438–448
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kresovich S, Barbazuk B, Bedell J, Borrell A, Buell R, Burke JJ, Clifton S, Cordonnier-Pratt M, Cox S, Dahlberg J, Erpelding JE, Fulton TM, Fulton B, Fulton L, Gingle A, Goff S, Hash C, Huang Y, Jordan D, Klein P, Klein RR, Magalhaes J, Mccombie R, Moore PH, Mullet JE, Ozias-Akins P, Paterson AH, Porter K, Pratt L, Roe B, Rooney W, Schnable P, Steely DM, Tuinstra M, Ware D, Warek U (2005) Toward sequencing the sorghum genome. A U.S. National Science Foundation-sponsored workshop report. Plant Physiol 138:1898–1902
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Mace ES, Rami J-F, Bouchet S, Klein PE, Klein RR, Kilian A, Wenzl P, Xia L, Halloran K, Jordan DR (2009) A consensus genetic map of sorghum that integrates multiple component maps and high-throughput diversity array technology (DArT) markers. BMC Plant Biol 9:13
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagaraj N, Reese JC, Tuinstra MR, Smith CM, Amand P, Kirkham MB, Kofoid KD, Campbell LR, Wilde GE (2005) Molecular mapping of sorghum genes expressing tolerance to damage by greenbug (homoptera: aphididae). J Econ Entomol 98:595–602
Painter RH (1951) Insect resistance in crop plants. MacMillan, New York
Park S-J, Huang Y, Ayoubi P (2006) Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 223:932–947
Paterson AH (2008) Genomics of sorghum. Int J Plant Genomics 2008:6. doi:10.1155/2008/362451 (Article ID 362451)
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboobur R, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Puterka GJ, Peters DC (1995) Genetics of greenbug (homoptera: aphididae) virulence to resistance in sorghum. J Econ Entomol 88(2):421–429
Radchenko EE, Zubov AA (2007) Genetic diversity of sorghum in greenbug resistance. Russ Agric Sci 33(4):223–225
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana, Totowa, pp 365–386
SAS Institute (2008) SAS Proprietary Software version 9.2. SAS Inst Cary NC
Satish K, Srinivas G, Madhusudhana R, Padmaja PG, Nagaraj Reddy R, Murali Mohan S, Seetharama N (2009) Identification of quantitative trait loci (QTL) for resistance to shoot fly in sorghum [Sorghum bicolor (L.) Monech]. Theor Appl Genet 119:1425–1439
Schloss SJ, Mitchell SE, White GM, Kukatla R, Bowers RE, Paterson AH, Kresovich S (2002) Characterization of RFLP probe sequences for gene discovery and SSR development in sorghum [Sorghum bicolor (L.) Moench]. Theor Appl Genet 105:912–920
Sharma HC (1993) Host-plant resistance to insects in sorghum and its role in integrated pest management. Crop Prot 12:11–34
Sharma HC, Ortiz R (2002) Host plant resistance to insects: an eco-friendly approach for pest management and environment conservation. J Environ Biol 23:111–135
Smith CM (2004) Insect/host plant resistance in crops. Encyclopedia of Plant and Crop Science, pp 605–608
Smith CM, Boyko EV (2007) The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol Exp Appl 122:1–16
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Starks KJ, Burton RL (1977) Greenbugs: determining biotypes, culturing, and screening for plant resistance. Technical Bulletin No. 1556. USDA-ARS, Washington, DC, pp 1–12
Taramino G, Tarchini R, Ferrario S, Lee M, Pe’ ME (1997) Characterization and mapping of simple sequence repeats (SSRs) in Sorghum bicolor. Theor Appl Genet 95:66–72
Teetes, GL. (1980) Breeding sorghums resistant to insects. In: FG Maxwell, PR Jennings (eds) Breeding plants resistant to insects. Wiley, New York, pp 457–485
Teetes GL, Pendleton BB (2000) Insect pests of sorghum. In: Smith CW, Frederiksen RA (eds) Sorghum: origin, history, technology, and production. Wiley, New York, pp 463–466
Temnykh S, Declerck G, Luashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57:755–766
Tuinstra MR, Wilde GE, Kriegshauser T (2001) Genetic analysis of biotype I greenbug resistance in sorghum. Euphytica 121:87–91
Van-Emden HF, Harrington R (2007) In: van Emden HF, and Harrington R (eds) Aphids as crop pests. CAB International, Oxford
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Wang S, Basten CJ, Zeng ZB (2010) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wilde GE, Tuinstra MR (2000) Registration of KS 97 sorghum. Crop Sci 40:866
Wu YQ, Huang Y (2006) An SSR genetic map of Sorghum bicolor (L.) Moench and its comparison to a published genetic map. Genome 50:84–89
Wu YQ, Huang Y (2008) Molecular mapping of QTLs for resistance to the greenbug Schizaphis graminum; (Rondani) in Sorghum bicolor, L. (Moench). Theor Appl Genet 117:117–124
Wu YQ, Huang Y, Tauer CG, Porter DR (2006) Genetic diversity of sorghum accessions resistant to greenbugs as assessed with AFLP markers. Genome 49:143–149
Wu YQ, Huang Y, Porter DR, Tauer CG, Hollaway L (2007) Identification of a major quantitative trait locus conditioning resistance to greenbug biotype E in sorghum PI 550610 using simple sequence repeat markers. J Econ Entomol 100:1672–1678
Yencho GC, Cohen MB, Byrne PF (2000) Applications of tagging and mapping insect resistance loci in plants. Annu Rev Entomol 45:393–422
Yonemaru J, Ando T, Mizubayashi T, Kasuga S, Matsumoto T, Yano M (2009) Development of genome-wide simple sequence repeat markers using whole-genome shotgun sequences of sorghum (Sorghum bicolor (L.) Moench). DNA Res 16:187–193
Young WR, Teetes GL (1977) Sorghum entomology. Annu Rev Entomol 22:193–218
Zhu-Salzman K, Salzman RA, Ahn J-E, Koiwa H (2004) Transcriptional regulation of Sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134:420–431
Acknowledgments
The author would like to thank Dr. Kathleen Yeaters, Area Statistician, ARS, College Station, TX for her assistance in statistical analysis. The author would also thank Ms. Angela Phillips for technical support during the project. The author would like to acknowledge the funding support received from Oklahoma Sorghum Commission for the project.
Disclaimer
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of a product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products that may also be suitable.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Punnuri, S., Huang, Y., Steets, J. et al. Developing new markers and QTL mapping for greenbug resistance in sorghum [Sorghum bicolor (L.) Moench]. Euphytica 191, 191–203 (2013). https://doi.org/10.1007/s10681-012-0755-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0755-4