Abstract
The Pi-z gene in rice confers resistance to a wide range of races of the rice blast fungus, Magnaporthe oryzae. The objective of this study was to characterize Pi-z in 111 rice germplasm accessions using DNA markers and pathogenicity assays. The existence of Pi-z in rice germplasm was detected by using four simple sequence repeat (SSR) markers (RM527, AP4791, AP5659-1, AP5659-5) closely linked to Pi-z, and was verified using pathogenicity assays with an avirulent strain (IE1k) and two virulent races (IB33 and IB49). Among 111 germplasm accessions evaluated, 73 were found to contain the Pi-z gene using both SSR markers and pathogenicity assays. The remaining 38 germplasm accessions were found to be inconsistent in their responses to the blast races IB33, IEIk and IB49 with expected SSR marker alleles, suggesting the presence of unexpected SSR alleles and additional R gene(s). These characterized germplasm can be used for genetic studies and marker-assisted breeding for improving blast resistance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice blast disease, caused by the filamentous ascomycete fungus Magnaporthe oryzae (formerly Magnaporthe grisea), is a major fungal disease threatening rice production worldwide. Genetic resistance in rice to M. oryzae belongs to a classic gene-for-gene system where a resistance (R) gene is effective in preventing infection by the races of M. oryzae containing the corresponding avirulence (AVR) gene (Silue et al. 1992). Resistant cultivars, fungicides, suitable planting dates, optimum fertilizer applications and adequate flood depth are useful tools to manage the disease (Bonman 1992; Lee 1994). Among them, the utilization of R genes is the most economical and environmentally-benign method for control of this disease. Thus far, more than 80 race-specific R genes to M. oryzae have been identified and some of them have been molecularly cloned and tagged for marker-assisted selection (MAS) (Yu et al. 1991; McCouch et al. 1994; Godwa et al. 2003; Ballini et al. 2008; Jia et al. 2009).
Among the tagged blast R genes, the Pi-z gene, first identified by Kiyosawa (1967) in the medium grain cultivar ‘Zenith’, has been effectively introgressed into numerous rice cultivars around the globe to prevent infection by a wide range of races of the rice blast pathogen. In the U.S., Pi-z is usually found in tropical japonica medium grain rice cultivars (Fjellstrom et al. 2006). This gene has been shown to confer resistance to five U.S. races of blast (IH-1, IG-1, IC-17, IE-1 and IE1k), and susceptibility to two races (IB49 and IB33) Historically, IE1k as an avirulent race and IB49 and IB33 as virulent races have been used for predicting the presence of Pi-z in a rice germplasm (Marchetti et al. 1987; Conaway-Bormans et al. 2003). The Pi9/2/zt complex mapped at the same chromosomal location of Pi-z(t) has been recently characterized (Liu et al. 2002; Zhou et al. 2006). Therefore, t is removed from Pi-z(t) throughout this manuscript. Four simple sequence repeat (SSR) markers associated with the Pi-z gene have been identified and recommended for use in germplasm characterization and MAS in the U.S. (Fjellstrom et al. 2006).
Marker-assisted selection (MAS) is a recent tool in breeding for improved resistance to rice blast (Jia 2003). For MAS, selection is made based on DNA markers closely linked to a blast R gene that confers resistance to a particular race of the pathogen. MAS can be used to screen seeds or seedlings under laboratory conditions, which is much faster than traditional pathogenicity assays where accurate selection can only be made during the later stages of plant growth. In addition, MAS can avoid the overlapping effects of other matched pairs of R and AVR genes. However, disagreements between markers and disease reactions can occur in some breeding lines due to different genetic backgrounds and/or potential recombination events between markers and trait. Accurate identification of a particular R gene in diverse elite germplasm using DNA markers and differential blast races is an essential step for ensuring the accuracy of R gene utilization in using MAS for different rice breeding programs.
The objectives of this study were to identify the Pi-z gene in 111 rice germplasm using SSR markers closely linked to the Pi-z gene, to determine disease reactions of these germplasm to differential U.S. blast races.
Materials and methods
Plant materials and growth
The USDA core collection consisting of 1,790 accessions from 113 countries representing 70% of genetic diversity of the entire USDA collection (Yan et al. 2007) was screened with AP5659-1, the marker most closely linked to the Pi-z gene prior to the purification of the collection by single seed decent (Agrama et al. 2010). One gram of seed from each accession from the purified core was provided by the Genetic Stock Collections of Oryza at Dale Bumpers National Rice Research Center (DB NRRC) for pathogenicity assays. Four rice cultivars from the Molecular Plant Pathology Program at DB NRRC, Bengal (PI 561735), Jefferson (PI 593892) [+Pi-z], Wells (PI 612439), and Zhe733 (PI 629016) [−Pi-z], were used as the controls. Eight seeds of each accession were germinated in 96 well inserts (10 × 20 × 2 cm) (Hummert International, Missouri USA). Prior to seeding the inserts were placed in trays [26.67 × 53.34 × 6.35 (in cm), Model # INT0804, Hummert International, Missouri, USA] and filled with silt loam soil (pH 5.5–5.8) fertilized with Osmocote Pro 15-9-12 (Scotts-Sierra Horticultural Products Company, OH), autoclaved and stored at −20°C for three days. The trays were completely filled with water. Rice plants were grown for 3 weeks in the greenhouse maintained at 23–29°C during the day in winter (November–April) and 29–32°C in summer (May–October) and 22–25°C during the night all year long until the 3–4 leaf stage, for pathogenicity assay and subsequent DNA extraction.
Pathogenicity assay
Pathogenicity assays were performed on 131 experimental germplasm and four control germplasm. M. oryzae isolates, an avirulent (AVR) race IB49 (isolate ZN61), virulent (VIR) races, race IE1k (isolate TM2), and race IB33 (isolate FL9) were selected for pathogenicity tests. There were four replicates for each germplasm. The Pi-z gene could be verified by the pattern of resistance or susceptibility to a pair of AVR and VIR races. Pathogen inoculation was performed using a modified procedure based on Valent et al. (1991). Briefly, plants were inoculated with 40 ml of a spore suspension (5 × 105 spores/ml, 0.25% gelatin) using a hand atomizer (100 kPa) connected to an air compressor. Inoculated plants were maintained at approximately 95% relative humidity in a clear polyethylene autoclave bag 24 × 36 (in cm) and 1.5 mm thick at room temperature (Product code 018143 Fisher Scientific, USA). Approximately 24 h after inoculation, plants were moved to the greenhouse for an additional 6 days. Disease reactions were assessed 7 days after inoculation using a rating scale (Fig. 1). For each accession, 7–8 seedlings were evaluated and each pathogenicity assay was conducted three times.
Improved evaluation standard for determining disease reactions of rice germplasm. Resistant (0–2): No lesion formation 0; Lesions covering less than 5% of total leaf area, lesions restricted at the site of infection 1; Lesions covering between 5 to 10% of the total leaf area; restricted spindle lesions at diameter less than 2 mm 2; Susceptible (3–5): Lesions in several locations on the leaf to form a large eye-shaped brown area (diameter greater than 2 mm) 3; Lesions covering greater than 50% of the leaf area, diseased area with lesion greater than 30% of the total leaf area 4; Lesions covering greater than 70% of the total leaf area 5. Note: Improvement was based on disease reactions of both indica and japonica cultivars to blast. Plants at the three to four leaf stages were inoculated and the second youngest leaf was evaluated one week after inoculation
DNA extraction
DNA was extracted using a rapid DNA extraction procedure (Xin et al. 2003). DNA was extracted from bulked leaves from each of four replicates for further analysis. After extraction, sample DNAs were prepared for PCR through a Biomek 2000 Lab Automation Work Station (Beckman and Coulter, Brea, CA) using manufacturer protocols.
SSR marker selection
Five SSR markers from which data are already available from the USDA purified core collection (Agrama et al. 2009) were used to screen current germplasm to preclude any seed mixtures or experimental error. The markers selected were RM224, RM208, RM231, RM447, and RM171 because all five markers are robust and have high PIC (polymorphism information content) value. Four SSR markers, RM527, AP4791, AP5659-1 and AP5659-5, from the five markers identified by Fjellstrom et al. (2006) as associated with the presence of the Piz gene were selected for the present study because: (1) AP5659-1 was used for the initial screening of the core prior to purification and Fjellstrom et al. (2006) found this marker displays a unique 220 nt allele in germplasm carrying the Pi-z gene; (2) Fjellstrom et al. (2006) indicated AP5659-5 has a 279 nt allele for all germplasm with Pi-z, although this also was found in an accession carrying Pi-9, another R allele at the Pi-z locus (Liu et al. 2002). Fjellstrom et al. (2006) found that RM 527 had a 217 nt allele in all Pi-z germplasm but this allele was also found in germplasm not carrying Pi-z; (3) AP4791 is another marker that can be used to detect association with Pi-z; and (4) AP5659-3 was reported to co-segregate with the Pi-z resistance, and considered the most closely linked to Pi-z of those identified to date. However, this marker has a null allele in some medium and long grain cultivars and therefore the marker was not selected (Fjellstrom et al. 2006, Fig. 2).
Genetic and physical maps of the Pi-z gene as defined by SSR markers. Genetic map showing indicated SSR markers spanning the Pi-z locus (a) and physical map of the Pi-z locus as delimited by indicated SSR markers (b). Modified from Fjellstrom et al. (2006)
SSR marker analysis
Fluorescently labeled SSR markers were analyzed by capillary electrophoresis. For each marker, forward primers were labeled with fluorescent dyes (6FAM, NED, and Hex) from Applied Biosystems (Foster City, CA, USA) or Integrated DNA Technologies (Coralville, IA, USA). Reverse primers were not labeled. DNA was amplified with MJ Research Tetrad thermocyclers (Waltham, MA, USA) under the following PCR conditions: (1) initial denaturation at 94°C for 5 min; (2) 35 cycles of 94°C for 1 min, 55–67°C (marker dependent) for 1 min, and 72°C for 2 min; (3) 5 min final extension at 72°C. Two–three PCR products were pooled based on color and size range of the amplified PCR products using a Mini Prep75 (Tecan Group Ltd., Männedorf, Switzerland) instrument based on the manufacturer protocols. PCR products were diluted between 500 and 1,000×, and 2 μl of the diluted product were added to 9 μl of formamide-containing ROX-labeled size standards (Applied Biosystems, Foster City, CA). DNA was denatured by heating at 94°C for 5 min. The reaction was run on an ABI Prism 3730 DNA Analyzer (Applied Biosystems) following the manufacturer instructions. Fragment size and SSR marker genotype analysis were performed with Gene Mapper® software version 3.7 (Applied Biosystems). Analyzed alleles were exported into a Microsoft Excel spreadsheet.
Statistical analysis
To verify accuracy of Fjellstrom et al. (2006) previous observed association of AP5659-1 220 nt allele with Piz resistance, χ2 analysis was conducted assuming Hardy–Weinberg equilibrium of IE1k resistance in general rice lines using software GraphPad χ2 calculator at http://www.graphpad.com.
Results and discussion
Identification and verification of germplasm with potential Pi-z resistance
An initial screen of the core collection with marker AP5659-1 identified 143 germplasm with the 220 nt allele that Fjellstrom et al. (2006) indicated was associated with the presence of the Pi-z gene. When seed from the purified core was obtained 12 germplasm failed to germinate in time to perform pathogenicity assays. In the remaining 131 germplasm, 114 retained the 220 nt allele, while the remaining 17 germplasm either contained a different allele or failed to amplify although PCR was attempted three times, thus indicating a possible null allele. The 17 germplasm with different AP5659-1 results between the purified and unpurified core are likely due to changes in the germplasm during purification. To preclude seed mixtures and/or erroneous germplasm we screened the 131 germplasm with six SSR’s previously run on the purified core collection (five high PIC value SSR’s and RM527 from the Pi-z region). Fourteen germplasm were either mixtures or failed to match previous marker data on the purified core.
To confirm previously published AP5659-1 association with IE1k resistance presumably from the Pi-z locus we conducted χ2 analyses on the purified germplasm that retained the 220 nt AP5659-1 allele assuming that IE1k resistance in the Core Collection is in Hardy–Weinberg Equilibrium. Under that assumption, if we select germplasm using a marker that lacks an association with IE1k resistance 50% of the germplasm should be resistant to IE1k and 50% should be susceptible to IE1k. The assumption of Hardy–Weinberg equilibrium for IE1k resistance was verified by performing χ2 analysis on data published by Wang et al. (2010) in which germplasm were screened for the presence of blast resistance gene Pi-ta, and pathogenicity assays were performed with both IE1k and IB49. Pi-ta confers resistance to IB49 but not IE1k. 50% of this germplasm were IE1k resistant, χ2 0.53. However our current germplasm did not fit a model of Hardy–Weinberg equilibrium indicating marker AP5659-1 is selecting for IE1K resistance (Table 1). This analysis could not be conducted on the other markers because we do not have data from the entire core collection with marker’s AP5659-5 and AP4791 and we did not analyze all core collection germplasm that contained the RM527 217 nt allele. Standard association mapping could not be performed on this germplasm because there is inadequate representation of both IE1k susceptible germplasm and the alleles Fjellstrom et al. (2006) identified as associated with Piz resistance are extremely overrepresented in the current germplasm.
The Pi-z resistance
In the present study, we relied on previously identified markers spanning the Pi-z locus and differential blast races to identify germplasm accessions with Pi-z and with additional R genes from 1,790 rice germplasm (Yan et al. 2007). In addition we verified accuracy of our germplasm using SSRs for which data was available from the purified core collection. We excluded the fourteen germplasm that were either mixtures or failed to contain the same alleles observed in previous core collection data (Agrama et al. 2009) at any of the six marker loci used for germplasm verification from further analysis or presentation. Six germplasm gave inconsistent pathogenicity results and were also excluded from further analysis or presentation. The remaining 111 germplasm were used to identify the presence of Pi-z and additional R genes.
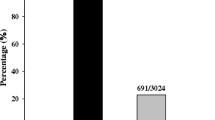
The gene-for-gene theory predicts that a germplasm is resistant due to Pi-z when this germplasm is (i) resistant to an AVR race IE1k (ii) and susceptible to a virulent (VIR) race, IB33 or IB49. As expected, the cultivar Bengal carrying Pi-z was found to be resistant to IE1k and susceptible to both IB33 and IB49. The cultivar Wells lacking Pi-z was found to be susceptible to all three races. The cultivar Zhe733 carrying Pi42(t) and Pi43(t) was found to be resistant to all three M. oryzae races (Lee et al. 2009). The cultivar Jefferson carrying Pi-z was found to be susceptible to IB33 and IB49 but resistant to IE1k (Table 2). Using these M. oryzae differential races the current study found that 77 germplasm accessions were resistant to IE1k and susceptible to IB33 and IB49, indicating the possible presence of Pi-z in these accessions (Fig. 3). Out of the 77 accessions with Pi-z as detected by pathogenicity assays, 40 had identical marker alleles for all four SSR markers (Table 2). The presence of the same marker alleles in these germplasm suggests that they contain a single Pi-z haplotype. This finding is important because these 40 germplasm were collected from several geographic regions of the world: the United States, South America, Europe, Asia and Africa (Table 2). The most likely reason for this haplotype similarity is that the original donor parent for the Pi-z gene may contain the same genomic fragment for all these cultivars. In contrast, 33 germplasm accessions showed 1–3 of the Pi-z allele (haplotype) markers, suggesting that these rice germplasm contain different Pi-z haplotypes, presumably either inherited from different donors or the result of recombination during the breeding process. Although pathogenicity data supported the presence of Pi-z, no expected maker (null) alleles were found in the remaining four germplasm accessions. The existence of the Pi-z gene in these four germplasm cannot be verified using our present markers and differential blast races.
If observed resistance in these germplasm is due to Pi-z only, susceptibility to IB49 and IB33 is expected. By pathogenicity assays, a total of 16 germplasm accessions were found to be resistant to all three races (Fig. 3; Table 2). These findings suggest the presence of additional R genes in these germplasm. There were nine germplasm that were susceptible to all isolates tested, eight of which showed the presence of 2–4 expected marker alleles, however, Montakcl from Egypt was the only cultivar which was susceptible to all isolates evaluated and did not show any expected marker allele. There were five germplasm that were resistant to IB49 and three that were resistant to IB33 in addition to being resistant to IE1k indicating the presence of additional R genes. There was one germplasm that was resistant to both IB33 and IB 49 but susceptible to IE1k suggesting Pi-z was absent or non functional.
Race IB49 versus IB33
In the rice blast system, a pair of blast races is adequate to identify the corresponding R gene (Silue et al. 1992). An additional blast race will increase the complexity for R gene identification because it may contain a different avirulence gene because each AVR gene is sufficient to trigger the corresponding R gene mediated resistance. IB33 was a laboratory-generated strain (F. Lee, unpublished data); and IB49 was a field isolate; both of which were highly similar in fingerprinting (Correll et al. 2000; Zhou et al. 2007). If a pair of AVR/VIR cannot detect an R gene, it may suggest that there are R genes in rice that interfere with expected disease reactions. In the present study, 16 accessions were found to be resistant to all three blast races. The presence of the Pi-z gene in these germplasm can not be verified although 12 of them contain one to four expected alleles using differential blast races. Under these situations, MAS is a better choice because it can detect the presence of a particular R gene in rice (Jia 2003).
R gene modifier
Historically, it has been commonly observed that an R gene has different phenotypical effects in different germplasm and/or under different environmental conditions. These differences are often conditioned by R gene modifiers. Some of these modifiers are critical for complete resistance. In the present study, we also found nine germplasm accessions that may contain 0–4 Pi-z marker alleles that were susceptible to all three races tested. Although it is possible that mutations in the coding region of Pi-z can result in the loss of resistant function, these findings suggest that some of these germplasm may have at least one nonfunctional critical modifier rendering susceptibility.
Identification of additional R genes
In our study, five germplasm accessions, Chao Puak Deng and Assaw from China, Biribra from Ghana, CA902/b/2/2 from Chad, and Agami Mont-1 from Egypt, were found to be resistant to both IE1k and IB49 and susceptible to IB33, suggesting these germplasm accessions contain additional R genes. In addition, three germplasm accessions, Perititovo 1417 from Madagascar R 100/2 from Zaire and Ku Mun Do No. 84 from Korea, were also determined to carry additional R genes to IB33. The cultivar Wanni Dahanala from Sri Lanka was known to be resistant to IB49 and IB33 but susceptible to IE1K, indicating the absence of Pi-z yet conferring the presence of additional R genes. Three accessions (PI184675-4 from Iran; Ken Yen from China, and GP No. 22232 from Germany) did not show any Pi-z haplotype alleles for the SSR markers, yet conferred resistance to all three races. Despite the nature of R genes it is unknown if these materials could be useful as resistant donors.
Geographic distribution of germplasm carrying Pi-z
Rice germplasm with Pi-z was found in 42 countries from six continents, Asia, Europe, North America, South America, Africa and Australia (Table 2). The most Pi-z containing germplasm was found in the U.S. and Puerto Rico, 5 + 4 of 77, respectively. Cote D’ Ivoire of West Africa was the next to the US and Puerto Rico where seven of 77 germplasm with Pi-z in Cote D’ Ivoire were verified in the present study. However, we have not found any germplasm with Pi-z from India and China where most of the rice is being grown in the world.
In conclusion, we not only verified the Pi-z gene in 73 of 77 rice germplasm using previously identified DNA markers but also demonstrated the usefulness of DNA markers and pathogenicity assays with differential blast races for germplasm characterization. We verified differential blast races; IE1k, IB33 and IB49 should be useful for predicting the existence of the Pi-z gene for conventional breeding for blast resistance. All of these new findings presented by this work were summarized in Table 2. For germplasm requests, please visit (http://www.ars.usda.gov/spa/dbnrrc/gsor) at GSOR of DB NRRC.
Abbreviations
- AVIR:
-
Avirulent
- D:
-
Day
- MAS:
-
Marker assisted selection
- R:
-
Resistance
- SSR:
-
Simple sequence repeat
- VIR:
-
Virulent
- PIC:
-
Polymorphism information content
References
Agrama HA, Yan W, Lee F, Fjellstrom R, Chen MH, McClung A (2009) Genetic assessment of a mini-core subset developed from the USDA rice genebank. Crop Sci 49:1336–1346
Agrama HA, Yan W, Jia MH, Fjellstrom RG, McClung AM (2010) Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Natural Sci 2:247–291
Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D (2008) A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant-Microbe Interact 21:859–868
Bonman JM (1992) Blast. In: Webster RK, Gunnell PS (eds) Compendium of rice diseases. APS Press, St. Paul, pp 14–17
Conaway-Bormans CA, Marchetti MA, Johnson CW, McClung AM, Park WD (2003) Molecular markers linked to the blast resistance gene Pi-z in rice for use in marker-assisted selection. Theor Appl Genet 107:1014–1020
Correll JC, Harp TL, Guerber JC, Lee FN (2000) Differential changes in host specificity among MRG586 DNA fingerprint groups of Pyricularia grisea. In: Tharreau D et al (eds) Advances in rice blast research. Kluwer Academic Publishers, Dordrecht, pp 234–242
Fjellstrom R, McClung AM, Shank AR (2006) SSR markers closely linked to the Pi-z locus are useful for selection of blast resistance in a broad array of rice germplasm. Mol Breed 17:149–157
Godwa M, Venu RC, Roopalakshmi K, Sreerekha MV, Kulkarni RS (2003) Advances in rice breeding, genetics and genomics. Mol Breed 11:337–352
Jia Y (2003) Marker assisted selection for the control of rice blast disease. Pesticide Outlook 14:150–152
Jia Y, Liu G, Costanzo S, Lee S, Dai Y (2009) Current progress on genetic interactions of rice with rice blast and sheath blight fungi. Front Agric China 3:231–239
Kiyosawa S (1967) The inheritance of resistance of the Zenith type varieties of rice to the blast fungus. Jap J Breed 17:99–107
Lee FN (1994) Rice breeding programs, blast epidemics and blast management in the United States. In: Zeigler RS, Leong SA, Teng PS (eds) Rice Blast Disease. CAB International, Wallingford, pp 489–500
Lee S, Wamishe Y, Jia Y, Liu G (2009) Identification of two major resistance genes against race IE1k of Magnaporthe oryzae in the indica rice cultivar Zhe733. Mol Breed 24:127–134
Liu G, Lu G, Zeng L, Wang GL (2002) Two broad-spectrum blast resistance genes, Pi9(t) Pi2(t) are physically linked on rice chromosome 6. Mol Genet Gen 267:472–480
Marchetti MA, Lai X, Bollich CN (1987) Inheritance of resistance to Pyricularia oryzae in rice cultivars grown in the United States. Phytopathology 77:799–804
McCouch SR, Nelson RJ, Tohme J, Zeigler RS (1994) Mapping of blast resistance genes in rice. In: Zeigler RS, Leong SA, Teng PS (eds) Rice blast disease. CAB International, Wallingford, pp 167–186
Silue D, Notteghem JL, Tharreau D (1992) Evidence of a gene-for gene relationship in the Oryza sativa–Magnaporthe grisea pathosystem. Phytopathology 82:577–580
Valent B, Farrall L, Chumley FG (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127:87–101
Wang X, Fjellstrom RG, Jia Y, Yan W, Jia MH, Scheffler BE, Wu D, Shu Q, McClung AM (2010) Characterization of Pi-ta Blast resistance gene in an international rice core collection. Plant Breed 129:491–501
Xin Z, Velten JP, Oliver MJ, Burke JJ (2003) High-throughput DNA extraction method suitable for PCR. Biotechniques 34:820–825
Yan W, Rutger JN, Bockelman HE, Fjellstrom RG, Chen MH, Tai T, McClung AM (2007) Development and evaluation of a core subset of the USDA rice (Oryza sativa L.) germplasm collection. Crop Sci 47:869–878
Yu ZH, Mackill DJ, Bonman JM, Tanksley SD (1991) Tagging genes for blast resistance in rice via linkage to RFLP markers. Theor Appl Genet 81:471–476
Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang G-L (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. MPMI 19:1216–1228
Zhou E, Jia Y, Singh P, Correll JC, Lee FN (2007) Instability of the Magnaporthe Oryzae avirulence gene AVR-Pita alters virulence. Fungal Genet Biol 44:1024–1034
Acknowledgments
The authors would like to thank Michael Lin and Lorie Bernhardt, Ellen McWhirter, and Dr. Kathy Yeater, USDA-SPA area statistician and other staff members of DB NRRC for their excellent technical assistance. This research was conducted at the DB NRRC partially supported by the Molecular Plant Pathology program of DB NRRC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
RoyChowdhury, M., Jia, Y., Jackson, A. et al. Analysis of rice blast resistance gene Pi-z in rice germplasm using pathogenicity assays and DNA markers. Euphytica 184, 35–46 (2012). https://doi.org/10.1007/s10681-011-0481-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-011-0481-3