Abstract
Reciprocal differences, mostly caused by cytoplasmic effects, are frequently observed in interspecific hybrids. Previously, we found that crosses onto Solanum demissum were much successful with the pollen of interspecific hybrids between S. tuberosum as female and S. demissum as male (TD hybrids) than the reciprocal ones (DT hybrids). To elucidate this reciprocally different crossability, we analyzed the pollen DNA of TD and DT using methylation-sensitive amplified polymorphism (MSAP) analysis. Using 126 primer combinations, MSAP analysis revealed 57 different bands between bulked pollen DNA samples of TD and DT. Individual examination of 16 TD and 9 DT plants disclosed eight bands uniformly different between TD and DT. Their sequencing results revealed two pairs of bands to be identical to each other, resulting in six distinct sequences. As expected, one band shared high homology with chloroplast DNA, and another one with mitochondrial DNA. However, one band that was apparently different at DNA sequence level and maternally transmitted from S. demissum, showed no homology with any known sequence. The remaining three bands were of DNA methylation level differences with no or uncertain homology to known sequences. To our knowledge, this is the first report detecting reciprocal differences in DNA sequence or DNA methylation other than those in cytoplasmic DNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reciprocal difference refers to differential phenotypic expression observed between reciprocal F1 hybrids, and the phenomenon is widely observed for various traits in various plant species (Burke et al. 1998; Campbell and Waser 2001; Tiffin et al. 2001; Rhode and Cruzan 2005; Gonzalo et al. 2007). In a perennial wild flower Penstemon davidsonii Greene, reciprocal F1 hybrids differed significantly in fruit set, seed number, seed weight, number of days to fruit maturity, vegetative growth and performance in field gardens (Kimball et al. 2008). In maize (Zea mays L.), inter-varietal F1 hybrids showed reciprocal differences in germination and traits such as whole-kernel growth rates (Groszmann and Sprague 1948) and embryo and endosperm dry weights (Bagnara and Daynard 1983). These differences occur mainly by the maternal effect as cytoplasmic factors of mitochondrial and chloroplast genomes, and xenia (refers to the effect of pollen on the endosperm phenotype in the same generation) (Roach and Wulff 1987).
A hexaploid Mexican wild species Solanum demissum Lindl. (2n = 6x = 72) has been extensively used in potato breeding as a source of resistance to potato late blight (Rudorf 1950; Ross 1986; Plaisted and Hoopes 1989). S. demissum is highly self-fertile, yet it shows unilateral incompatibility with the common potato (S. tuberosum L., 2n = 4x = 48). S. demissum can be easily crossed with the pollen of S. tuberosum and produces pentaploid hybrids which, and the back-cross progenies as well, are only crossable as female parents (Black 1943; Dionne 1961; Irikura 1968). In a previous study (Sanetomo et al., unpublished), we reconfirmed the unilateral incompatibility between S. demissum (D) and S. tuberosum (T). Further, we found that the obtained hybrid seeds were reciprocally very different in size: the average seed weights were 0.94 mg from D × T and 0.39 mg from T × D. The further crossing experiments in various combinations revealed reciprocal differences in crossability between T (female) × D (male) hybrids (TD hybrids) and the reciprocal ones (DT hybrids). Especially when TD and DT hybrids were crossed as pollen parents onto S. demissum, a significantly higher berry setting rate was obtained in TD (64.9%) compared with DT (24.2%). Based on these crossing results, we suggested at least three factors likely involved in hybrid seed development in these specific crosses: (1) a cytoplasmic, or maternally inherited factor, (2) a nuclear-encoded factor functioned in female gametophyte and (3) a nuclear-encoded factor functioned in pollen and affected by a balance between male and female gametes. Increasing the S. tuberosum nuclear germplasm and possessing S. tuberosum cytoplasm by backcrossing in both male and female gametophytes always resulted in superior berry setting rates (Sanetomo et al., unpublished). Due to the maternal inheritance of chloroplast and mitochondrial DNA, the genetic information was slightly different between TD and DT. However, most of genetic information was encoded in the nuclear DNA, which was composed of a half of S. demissum and a half of S. tuberosum genomes, and should be identical between TD and DT hybrids. Nevertheless, the pollen from TD and DT hybrids functioned differently.
It is widely recognized that DNA methylation affects gene activity mainly by repressing gene expression or transcription (reviewed in Bird and Wolffe 1999), whereas its demethylation promotes gene expression and activates silent genes (Cervera et al. 2002; Rutherford and Henikoff 2003). In plants, this mechanism likely occurs because methylation affects the accessibility of several plant proteins to their target DNA sequences (Gierl et al. 1988). Proteins with affinity for methylated sequences also have been isolated (Ehrlich 1993). Therefore, DNA methylation is one of most important epigenetic mechanisms for plant development and the regulation of fertilization in fungi and plants (Martienssen and Colot 2001).
The DNA cytosine methylation changes were observed among several plant inter-specific hybrids, allopolyploids and introgression lines (Comai et al. 2000; Madlung et al. 2002; Levy and Feldman 2004). The role of DNA methylation is well documented on seed development in Arabidopsis (reviewed in Kinoshita 2007). The endosperm was specifically hypomethylated (demethylated), lower than other tissues (Adams et al. 2000; Vinkenoog et al. 2000). And imprinted genes of female origin are activated and interacted with those of male origin (reviewed in Baroux et al. 2002). Such tissue-specific methylation differences between endosperm and leaf or other organ tissues have also been reported in maize (Lauria et al. 2004; Lu et al. 2008), rice (Xiong et al. 1999) and sorghum (Zhang et al. 2007). However, the methylation rate of male gametophyte, or pollen has been studied poorly.
In this paper, we investigated a global view of the DNA differences and cytosine methylated DNA differences between pollen DNA from reciprocal F1 hybrids TD and DT, and between pollen and leaf DNA using methylation-sensitive amplified polymorphism (MSAP) analysis (Reyna-López et al. 1997; Xiong et al. 1999). As expected, we found DNA level differences originated from chloroplast and mitochondrial DNA. In addition, we found other DNA sequence and DNA methylation level differences, which is to our knowledge the first report detecting difference at such levels between reciprocal hybrids.
Materials and methods
Plant materials
In this study, we used as parents a S. tuberosum breeding line Saikai 35 (referred to T), bred at Nagasaki Agricultural and Forestry Technical Development Center, Aino, Nagasaki, and seedlings of 6H36 and 7H16 families (collectively referred to D) derived by selfing from one of S. demissum PI 186551 plants. Since S. demissum is highly self-pollinated in nature and homogeneous within family as evidenced by random amplified polymorphic DNA analysis (unpublished data), we assumed all seedlings were genetically identical. The interspecific hybrid family 6H37 was obtained by the cross between S. demissum as female and S. tuberosum as male (DT), while 6H38 family was obtained by the reciprocal cross (TD). Most of S. tuberosum cultivars have chloroplast DNA of T type, as defined by Hosaka (1986), and mitochondrial DNA of β type, as defined by Lössl et al. (1999, 2000). However, since Saikai 35 descended maternally from S. phureja, it has S-type chloroplast DNA and ε-type mitochondrial DNA (Hosaka, unpublished). The parental S. demissum clone has W-type chloroplast DNA and α-type mitochondrial DNA (Hosaka, unpublished).
DNA extraction
Mature pollen grains were collected from many T individuals and seedlings of D. For interspecific hybrids, 16 TD and 9 DT genotypes were clonally propagated and grown in the field to obtain sufficient amount of pollen grains to extract DNA. Collected pollen grains were stored at −30°C until DNA extraction. Approximately 50–120 mg of pollen grains per genotype were used to extract DNA by the method of Hosaka and Hanneman (1998). Simultaneously, DNA from fresh leaves was extracted from T, D, 7 TD and 6 DT genotypes.
Methylation-sensitive amplified polymorphism (MSAP) analysis
The MSAP analysis was performed to detect differences in DNA sequences and DNA methylation status as well. The MSAP analysis is an adaptation of the amplified fragment length polymorphism (AFLP) technique (Vos et al. 1995), in which the isoschizomers HpaII and MspI were used instead of a usually used frequent cutter enzyme MseI (Reyna-López et al. 1997). HpaII and MspI recognize the same four-base sequence (CCGG) and cut with differential sensitivity to DNA methylation of internal or external cytosine. HpaII does not cut the recognition site if the internal cytosine is full-methylated, whereas MspI is insensitive and cuts it. If the external cytosine is hemi-methylated, MspI does not cut, whereas HpaII cuts the recognition site. Thus, if the presence/absence of an AFLP band was different between EcoRI + HpaII double-digest (E/H digest) and EcoRI + MspI double-digest (E/M digest) of the same DNA sample, it was regarded as a methylation-sensitive band caused by a difference of DNA methylation status.
Genomic DNA (500 ng) was digested with 12.5 units of MspI (Takara Bio, Japan) or HpaII (TOYOBO, Japan) separately by overnight incubation at 37°C. After the digestion reaction was stopped by incubating at 70°C for 10 min, digested DNA was precipitated by ethanol, dried, and re-suspended in 10 μl of distilled water. The second digestion was performed with 12.5 units of EcoRI (Takara Bio, Japan) in volumes of 25 μl by overnight incubation at 37°C. The double-digested DNA fragments were ligated to the adapters by adding 24 μl of ligation mixture [2× ligation buffer (Invitrogen), 0.1 pmol EcoRI adapter and 1 pmol HpaII/MspI adapter] and 1 μl of T4 DNA ligase (1 unit/μl, Invitrogen), and incubated overnight at 20°C. The adapter and pre-amplification primer sequences were the same as those described by Xiong et al. (1999). Pre-amplification was performed with 2.5 μl of the above DNA in volumes of 25 μl consisting of 0.3 μM pre-amplification primers, 12.5 μl of Ampdirect® Plus (Shimadzu, Japan) and 0.25 units of Taq DNA polymerase (Nova Taq™ Hot Start DNA Polymerase, Novagen®, USA). Pre-amplified products were adjusted to the concentration of 5 ng/μl. Selective amplification was performed with the same components in volumes of 10 μl with 2 μl of 5 ng/μl pre-amplified DNA. For selective amplification seven EcoRI primers and two sets of nine MspI/HpaII primers were used. EcoRI primers consisted of the core sequences of 5′-GACTGCGTACCAATTC and three selective nucleotides ANN (NN denotes CC, AG, CA, CG, CT, AC or GC). Two MspI/HpaII primer sets were used, both consisted of the same core sequences of 5′-ATCATGAGTCCTGCTCGG. The first primer set had three selective nucleotides ANN and the second one TNN (NN denotes TA, CG, GC, TG, CT, CC, TC, AC or GT). Thermal profiles for pre- and selective amplifications were those described in the original AFLP protocol (Vos et al. 1995). The amplification products were electrophoresed on 4% denaturing polyacrylamide gels and visualized by silver-staining (Bassam et al. 1991).
Bulked segregant analysis
Because potato is a highly heterozygous tetraploid crop, interspecific hybrids would become heterogeneous. Thus, bulked segregant analysis (Michelmore et al. 1991) was employed to compare TD and DT. Pollen DNA samples of 16 TD and 9 DT genotypes were pre-amplified separately, bulked with equal amounts as TD and DT bulked samples, and then, subjected to selective amplification. If different bands were found between TD and DT bulked pollen DNA samples, these bands were examined from individually pre-amplified samples. For bulked leaf DNA samples, 7 TD and 6 DT leaf DNA samples were respectively mixed prior to double-digestion.
Isolation and characterization of amplified bands
The target bands were cut out from polyacrylamide gels dried on the glass plate using a razor blade. The peeled gel fragment containing a target band was rehydrated in 10 μl of sterile water overnight, and centrifuged at 12,000×g for 10 min. The target DNA was amplified by PCR from 2 μl of the supernatant using the same primer pairs as those for the selective amplification. The thermal condition was 10 min at 95°C, then, 25 cycles of 1 min at 95°C, 1 min at 65°C and 1 min at 72°C, followed by final extension of 5 min at 72°C. After PCR amplification, unincorporated primers and deoxynucleotide triphosphates were removed by ethanol precipitation prior to sequencing. The PCR products were sequenced from both directions with forward and reverse primers separately using BigDye terminator version 3.1 on ABI 3100 (Applied Biosystems). The homology search of the obtained sequences was performed using the BLASTN program.
Results
Detection of methylation-sensitive bands
For MSAP analysis two sets of HpaII/MspI selective amplification primers were used. Primer sequences of the first and second sets consisted of the same core sequences plus three selective nucleotides ANN and TNN, respectively. Nine primers of each set had the same set of additional two selective nucleotides, and all combinations with the same seven EcoRI primers were used. Using a total of 126 pairs of selective amplification primers, bulked pollen and leaf DNA of inter-specific reciprocal F1 hybrids TD and DT were examined (Table 1). The total numbers of AFLP bands from bulked pollen DNA samples of TD and DT were 23,527 and 23,525, while those from bulked leaf DNA samples were 23,530 and 23,533, respectively. The total numbers of methylation-sensitive bands from bulked pollen DNA of TD and DT were 2,090 and 2,082, while those from bulked leaf DNA were 2,078 and 2,087, respectively. Thus, there was no significant difference in the total band numbers among four DNA samples, and in the methylation-sensitive band numbers as well. However, the numbers of methylation-sensitive bands detected from E/M-digested samples (1,776–1,780 bands) were much higher than those from E/H-digested samples (302–310 bands), the mean proportion being 85.3 and 14.7%, respectively, which also reflected on the total band numbers of E/M digests higher than those of E/H digests. When two primer sets were compared, the first set of primers generated a total of 45,175 bands from four samples (11,288–11,301 bands) and the second set of primers generated 48,940 bands (12,232–12,237 bands per sample), respectively, in which 3,437 and 4,900 bands were methylation-sensitive, respectively. Thus, compared with the first set primers, the second set primers increased total band numbers to 1.1 times and methylation-sensitive bands to 1.4 times, which increased the detection frequency of methylation-sensitive bands from 7.6 to 10.0%.
Rates of methylation-sensitive bands
When the DNA methylation rate was calculated as the percentage of the total number of methylation-sensitive bands over total band numbers detected in both E/H and E/M digests (Table 1), those for pollen DNA samples of TD and DT were 8.88 and 8.85%, and those for leaf DNA samples of TD and DT were 8.83 and 8.87%, respectively. Thus, average DNA methylation rates of pollen and leaf DNA samples were 8.87 and 8.85%, respectively.
Different banding types in TD and DT
Only for the first set primers, pollen and leaf DNA samples of TD and DT were electrophoresed together on the same gels. Thus, the presence/absence of each band among four samples for each of TD and DT could be compared (Table 2). A total of 6,085 bands were amplified from TD and the same number of bands from DT, which were categorized into 13 types. 5,197 bands (85.4%) of type 1 were present in all samples of TD, and similarly in DT. The bands of types 2 and 3 were methylation-sensitive bands, observed in E/M digests of pollen and leaf DNA, but not in the E/H digests (type 2), or vice versa (type 3). Types 4–13, counting 50 bands (0.82%) in TD and 49 bands (0.81%) in DT, were differentially amplified between leaf and pollen DNA. Among these tissue-specific bands, those of types 4 and 5 (12.0% in TD and 20.4% in DT) were detected as DNA sequence differences, while those of types 6–13 were regarded as DNA methylation differences (88.0% in TD and 79.6% in DT).
Differences in pollen DNA between TD and DT
TD and DT bulked DNA samples showed mostly the same AFLP banding patterns (Fig. 1). Yet, 57 bands from 43 primer pairs were different between TD and DT, of which 35 bands were specifically found in pollen DNA (Table 3).
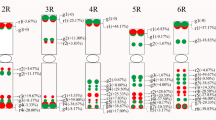
For these 57 bands, the pollen DNA samples of 16 TD and 9 DT plants were individually analyzed. Forty-nine bands segregated for presence/absence within each population. The presence or absence of eight bands (Bands 1–8) was uniform within each of TD and DT populations and consistently different between TD and DT plants (Fig. 2; Table 4). Irrespective of methylation-sensitive restriction enzymes, Bands 1–3 were detected in all DT but not in any TD plants, while Bands 4 and 5 were detected in all TD but not in any DT plants. Thus, these bands were regarded as the differences in DNA sequences. Bands 6–8 were detected in all E/M digests of TD and DT plants, whereas in E/H digests, Band 6 was present in all TD but absent in DT plants, and Bands 7 and 8 present in all DT but absent in TD plants. Presence or absence of these bands in the parental T and D was also analyzed (Table 4). Bands 1–5 of F1 hybrids were only present in the female parent, that is, Bands 1–3 of DT were shared with D, while Bands 4 and 5 of TD shared with T. Band 6 was present in D and absent in T in both E/H and E/M digests. Bands 7 and 8 were present in E/M digests of both parents, whereas in E/H digests, Band 7 was present only in D and Band 8 absent in both parents. These eight bands detected in pollen DNA were also examined in leaf DNA. Only the difference of Band 8 was pollen-specific, while the others were shown similarly in leaf DNA.
Comparison between EcoRI/HpaII-digested (E/H) and EcoRI/MspI-digested (E/M) bulked pollen DNA samples revealed a methylation sensitive AFLP band different between interspecific reciprocal F1 hybrids TD and DT (arrowed, Band 7), whose presence or absence in individual DNA samples was consistent within and between populations
Sequencing analysis
Bands 1–8 were eluted from the polyacrylamide gels, re-amplified and sequenced (Table 5). The determined sequences are available from the National Center for Biotechnology Information (accession numbers given in Table 5). Bands 1 and 2 showed the same sequences with the size of 170 bp. Since each band was sequenced from both directions using the EcoRI and HpaII/MspI primers used for the selective amplification, it was found that Band 2 was amplified by a mismatch of one base in the region corresponding to the selective nucleotides of the HpaII/MspI primer. Similarly, Bands 4 and 5 had the same sequences with the size of 334 bp, the latter band having been amplified by a mismatch. BLAST-search indicated that Bands 1, 6 and 7 showed no or less than 40% homology with any known sequences (Table 5). Band 3 (205 bp) shared 97% homology with a part of S. tuberosum cv. Desiree chloroplast DNA. Band 4 shared 97% homology with a part of Nicotiana tabacum mitochondrial DNA. Band 8 shared 87% homology with a part of non-coding region of Vitis vinifera mitochondrial DNA, but not with any Solanaceous mitochondrial DNA. Instead, it shared 42% homology with a part of S. lycoperisicum chromosome 2 or 38% homology with a part of S. demissum chromosome 5.
Discussion
Using 126 primer pairs, pollen DNA generated a total of 23,527 AFLP bands from the S. tuberosum × S. demissum (TD) hybrid and 23,525 bands from the reciprocal one (DT). With unknown reason the second set primers amplified 1.1 times more number of bands than the first set primers, although both primer sets had the same GC content with only difference of one nucleotide substitution from A to T in the first position of three selective nucleotides. Even comparing DNA samples digested with methylation-sensitive isoscizomers HpaII and MspI, TD and DT hybrids showed almost identical AFLP banding patterns (Fig. 1) and the same methylation rates (8.88% in TD and 8.85% in DT, respectively). Yet, we found at least six differences between TD and DT. As expected, one difference possibly occurred in chloroplast DNA (Band 3) and another one in mitochondrial DNA (Band 4). In addition, we found Band 1, which occurred by a DNA sequence level difference or a difference of DNA methylation that did not cause differential sensitivity between the two enzymes, and Bands 6–8, which occurred as DNA methylation level differences. To our knowledge, this is the first report detecting reciprocal differences in DNA sequence or DNA methylation other than those in cytoplasmic DNA.
Band 1 was detected in all DT hybrids and the S. demissum parent. BC1 plants from a cross between DT × T shared Band 1 in common (data not shown). These indicated that the Band 1 was transmitted maternally, implying chloroplast or mitochondrial DNA origin. Potato chloroplast DNA was completely sequenced (Chung et al. 2006). Mitochondrial DNA in the Solanaceous model plant tobacco (Nicotiana tabaccum) has also been completely sequenced (Sugiyama et al. 2005). Nevertheless, Band 1 shared no homology with any known sequences. A further research is absolutely necessary to clarify whether the Band 1 originated from S. demissum-specific region of chloroplast or mitochondrial DNA, or from nuclear DNA of S. demissum that was maternally transmitted with some unknown reason.
Bands 3 and 4 were apparently originated in chloroplast and mitochondrial DNA, respectively, because of the high sequence homology and specificity to maternal parents. It has been known that the parental S. demisssum and S. tuberosum genotypes have different chloroplast DNA (W and S types, respectively) and mitochondrial DNA (α and ε types, respectively), which might be associated with Bands 3 and 4. Previously, we found that the cytoplasmic difference was a major contributory factor to the reciprocal difference in crossability between TD and DT (Sanetomo et al., unpublished). However, it remains unknown whether Bands 3 or 4 is related to contributory factors to crossability difference.
In plant, apparent non-Mendelian inheritance or re-modeling of parental methylation patterns has been observed to occur in certain situations, like in several plant inter-specific hybrids, allopolyploids and introgression lines (Madlung et al. 2002; Liu and Wendel 2003; Levy and Feldman 2004; Liu et al. 2004; Salmon et al. 2005; Lukens et al. 2006; Marfil et al. 2006). Unlike such epigenetic methylation, the presence or absence of Bands 6–8 were consistent within all of 16 plants of TD or all 9 plants of DT. Thus, these methylated DNA regions should have some functional meanings. Imprinted genes or parent-of-origin dependent genes are regulated by differential DNA methylation between parental alleles (Kiyosue et al. 1999; Ingouff et al. 2005; Kinoshita 2007; Köhler et al. 2010). Imprinting was extensively studied for endosperm genes in maize (Gutiérrez-Marcos et al. 2004) and Arabidopsis (Gehring et al. 2009; Hsieh et al. 2009). By comparison of mRNA and protein profiling screened for cold germination and desiccation tolerance, Kollipara et al. (2002) identified 336 of 32,496 and 656 of 32,940 cDNA fragments, or 117 of 2,641 and 205 of 1,876 detected proteins to be differentially expressed between reciprocal maize hybrids. They hypothesized that these differentially expressed genes were either directly or indirectly associated with the imprinting phenomenon. Although the functional roles of Bands 6–8 remain unknown, these might be related to one of regulating factors, especially for the differential crossability or endosperm development. Particularly, Band 8 would be interesting because its difference between TD and DT was specifically found in pollen.
The methylation rates of pollen and leaf DNA were almost similar to each other (8.85–8.88% and 8.83–8.87%, respectively). Since HpaII and MspI do not show differential sensitivity to DNA methylation of full-methylated external cytosine and hemi-methylated internal cytosine, the present DNA methylation rates were apparently underestimated. Yet, we can suggest that the sporophyte and the male gametophyte DNA are methylated to the same degree at least in a genome-wide view. On the other hand, hypomethylation in endosperm is well documented in Arabidopsis (Adams et al. 2000; Vinkenoog et al. 2000; Gehring et al. 2009; Hsieh et al. 2009). In maize, the endosperm exhibited 13% reduction in total cytosine methylation level compared with leaf and embryo (Lauria et al. 2004), and in sorghum, endosperm exhibited 11% reduced methylation rate than leaf in inbred and hybrid lines (Zhang et al. 2007). In addition, Zhang et al. (2007) revealed higher expression of the endosperm-specific hypomethylated six genes by RT-PCR analysis. Thus, hypomethylation of the endosperm is generally recognized to induce expression of many genes, which plays an important role for seed development or seed formation (Lund et al. 1995a, b; Gutiérrez-Marcos et al. 2004). By genome-wide high-throughput bisulfate sequencing, Hsieh et al. (2009) demonstrated that virtually the entire endosperm genome was hypomethylated in Arabidopsis. Further, they found endosperm hypomethylation was accompanied by hypermethylation of embryo transposable elements (TEs). Likewise in the Arabidopsis pollen, Slotkin et al. (2009) found that sperm-derived DNA was hypermethylated and vegetative nucleus-derived DNA was hypomethylated. TEs, triggered by hypomethylation and reactivated in endosperm or vegetative cell in pollen, might generate short interfering (si)RNAs that move to embryo or sperm cell and enhance silencing of TEs in the embryo or sperm cell to secure the progeny (Hsieh et al. 2009; Slotkin et al. 2009; Mosher and Melnyk 2010). Therefore, what we observed in this study for pollen DNA might be the total or average methylation rate of vegetative and sperm cell DNA, resulting in the almost similar methylation rate to that of leaf DNA.
Although the comparison was made using bulked DNA samples, 0.81–0.82% of bands were differently methylated between leaf and pollen (Table 2). By the whole genome transcriptome analysis of the sperm cell in Arabidopsis, Borges et al. (2008) suggested that distinct mechanisms might be involved in regulating the epigenetic state of the paternal genome, and identified numerous candidate genes involved in sperm cell development and fertilization pathways. Differentially methylated DNA fragments observed in this study might be associated with such tissue-specific expression as known in various tissues in various plant species (Adams et al. 2003; Ingouff et al. 2005; Wang et al. 2006; Chaudhary et al. 2009).
References
Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127:2493–2502
Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100:4649–4654
Bagnara D, Daynard TB (1983) Reciprocal differences in kernel growth among four maize inbreds. Maydica 27:357–363
Baroux C, Spillane C, Grossniklaus U (2002) Genomic imprinting during seed development. Adv Genet 46:165–214
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces, and chromatin. Cell 24:451–454
Black W (1943) Inheritance of resistance to two strains of blight (Phytophthora infestans de Bary) in potatoes. Trans R Soc Edinburgh 61:137–147
Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148:1168–1181
Burke JM, Voss TJ, Arnold ML (1998) Genetic interactions and natural selection in Louisiana Iris hybrids. Evolution 52:1304–1310
Campbell DR, Waser NM (2001) Genotype-by-environment interaction and the fitness of plant hybrids in the wild. Evolution 55:669–676
Cervera MT, Ruiz-García L, Martínez-Zapater JM (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol Genet Genomics 268:543–552
Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, Wendel JF (2009) Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploidy cotton (Gossypium). Genetics 182:503–517
Chung HJ, Jung JD, Park HW, Kim JH, Cha HW, Min SR, Jeong WJ, Liu J (2006) The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep 25:1369–1379
Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B (2000) Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12:1551–1568
Dionne LA (1961) Cytoplasmic sterility in derivatives of Solanum demissum. Am Potato J 38:117–120
Ehrlich KC (1993) Characterization of DBPm, a plant protein that binds to DNA containing 5-methylcytosine. Biochim Biophys Acta 1172:108–116
Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324:1447–1451
Gierl A, Lütticke S, Saedler H (1988) TnpA product encoded by the transposable element En-1 of Zea mays is a DNA binding protein. EMBO J 20:4045–4053
Gonzalo M, Vyn TJ, Holl JB, McIntyre LM (2007) Mapping reciprocal effects and interactions with plant density stress in Zea mays L. Heredity 99:14–30
Groszmann A, Sprague GF (1948) Comparative growth rates in a reciprocal maize cross: 1. The kernel and its component parts. J Am Soc Agron 40:88–98
Gutiérrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O’Sullivan DM, Wormald M, Perez P, Dickinson HG (2004) Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16:1288–1301
Hosaka K (1986) Who is the mother of the potato?—restriction endonuclease analysis of chloroplast DNA of cultivated potatoes. Theor Appl Genet 72:606–618
Hosaka K, Hanneman RE Jr (1998) Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 2. Localization of an S locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica 103:265–271
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454
Ingouff M, Haseloff J, Berger F (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42:663–674
Irikura Y (1968) Studies on interspecific crosses of tuber-bearing Solanums. 1. Overcoming cross-incompatibility between Solanum tuberosum and other Solanum species by mean of induced polyploids and haploids. Hokkaido Agric Exp Stn Shuho 92:21–37
Kimball S, Campbell DR, Lessin C (2008) Differential performance of reciprocal hybrids in multiple environments. J Ecol 96:1306–1318
Kinoshita T (2007) Reproduction barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet Syst 82:177–186
Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada J, Goldberg RB, Fischer RL (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96:4186–4191
Köhler C, Scheid OM, Erilova A (2010) The impact of the triploid block on the origin and evolution of polyploidy plants. Trends Genet 26:142–148
Kollipara KP, Saab IN, Wych RD, Lauer MJ, Singletary GW (2002) Expression profiling of reciprocal maize hybrids divergent for cold germination and desiccation tolerance. Plant Physiol 129:974–992
Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A, Lund G (2004) Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16:510–522
Levy AA, Feldman M (2004) Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol J Linn Soc 82:607–613
Liu B, Wendel JF (2003) Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol 29:365–379
Liu ZL, Wang YM, Shen Y, Guo WL, Hao S, Liu B (2004) Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia. Plant Mol Biol 54:571–582
Lössl A, Adler N, Horn R, Frei U, Wenzel G (1999) Chondriome-type characterization of potato: mt α, β, γ, δ, ε and novel plastid-mitochondrial configurations in somatic hybrids. Theor Appl Genet 99:1–10
Lössl A, Götz M, Braun A, Wenzel G (2000) Molecular markers for cytoplasm in potato: male sterility and contribution of different plastid-mitochondrial configurations to starch production. Euphytica 116:221–230
Lu Y, Rong T, Cao M (2008) Analysis of DNA methylation in different maize tissues. J Genet Genomics 35:41–48
Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 140:336–348
Lund G, Ciceri P, Viotti A (1995a) Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J 8:571–581
Lund G, Messing J, Viotti A (1995b) Endosperm-specific demethylation and activation of specific alleles of alpha-tubulin genes of Zea mays L. Mol Gen Genet 246:716–722
Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L (2002) Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol 129:733–746
Marfil CF, Masuelli RW, Davison J, Comai L (2006) Genomic instability in Solanum tuberosum × Solanum kurtzianum interspecific hybrids. Genome 49:104–113
Martienssen RA, Colot V (2001) DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293:1070–1074
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Mosher RA, Melnyk CW (2010) siRNAs and DNA methylation: seedy epigenetics. Trends Plant Sci 15:204–210
Plaisted RL, Hoopes RW (1989) The past record and future prospects for the use of exotic potato germplasm. Am Potato J 66:603–627
Reyna-López GE, Simpson J, Ruiz-Herrera J (1997) Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet 253:703–710
Rhode JM, Cruzan MB (2005) Contributions of heterosis and epistasis to hybrid fitness. Amer Nat 166:E124–E139
Roach DA, Wulff RD (1987) Maternal effects in plants. Annu Rev Ecol Syst 18:209–235
Ross H (1986) Potato breeding-problems and perspectives. Verlag Paul Parey, Berlin
Rudorf W (1950) Methods and results of breeding resistant strains of potatoes. Am Potato J 27:332–339
Rutherford S, Henikoff S (2003) Quantitative epigenetics. Nat Genet 33:6–8
Salmon A, Ainouche ML, Wendel JF (2005) Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol Ecol 14:1163–1175
Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472
Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272:603–615
Tiffin P, Olson MS, Moyle LC (2001) Asymmetrical crossing barriers in angiosperms. Proc R Soc Lond Ser B Biol Sci 268:861–867
Vinkenoog R, Spielman M, Adams S, Fischer RL, Dickinson HG, Scott RJ (2000) Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12:2271–2282
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23:4407–4414
Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, Chen ZJ (2006) Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172:507–517
Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261:439–446
Zhang MS, Yan HY, Zhao N, Lin XY, Pang JS, Xu KZ, Liu LX, Liu B (2007) Endosperm-specific hypomethylation, and meiotic inheritance and variation of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) inter-strain hybrids. Theor Appl Genet 115:195–207
Acknowledgments
We thank the US Potato Genebank (NRSP-6), Sturgeon Bay, Wisconsin, for providing seeds of S. demissum. This study was carried out at the Food Resources Education and Research Center, Kobe University, and supported partly by Grant-in-Aid for Scientific Research (C) (18580005), MEXT, and Calbee Potato, Inc.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sanetomo, R., Hosaka, K. Reciprocal differences in DNA sequence and methylation status of the pollen DNA between F1 hybrids of Solanum tuberosum × S. demissum . Euphytica 182, 219–229 (2011). https://doi.org/10.1007/s10681-011-0444-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-011-0444-8