Abstract
The fungal disease cercospora leaf spot CLS (Cercospora zonata) has affected major faba bean (Vicia faba) production regions in southern Australian in the last several years. This study offers the first report of sources of resistance to CLS in faba bean and describes techniques to evaluate resistance to C. zonata in faba bean genotypes within a controlled environment. The method was rapid (43 days), repeatable (R 2 > 0.74) and demonstrated positive correlations (R 2 > 0.45–0.80) to data collected from field disease nurseries under naturally established CLS epiphytotics. All faba bean cultivars currently adopted by the Australian industry were found to be susceptible to CLS and defoliation was found to be an important component of disease expression. Genetic analysis of segregation patterns in F 2 derived F 3 families of 1322/2*Farah (resistant*susceptible) showed the mode of inheritance of resistance to C. zonata was monogenic dominant. F 3 families were shown to segregate in the ratio of 1:2:1 for homozygous resistant: heterozygous: homozygous susceptible (χ 22 = 2.78; P > 0.05) and individual plants within heterozygous F 3 families segregated in the ratio of 3:1 for resistant: susceptible responses (χ 21 = 2.93; P > 0.05). Monogenic dominant inheritance also explained the change in frequency of resistant and susceptible plants within a population of cv. Cairo following one generation of self-pollination (χ2 = 0.88, 0.3 < P < 0.5). The sources of resistance identified in this study are being used to transfer CLS resistance to adapted faba bean genotypes for future cultivar releases to the southern Australian industry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
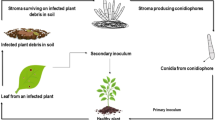
Cercospora leaf spot (Cercospora zonata) is a common fungal disease of faba bean (Vicia faba L.) in Australia but the disease has not been examined critically. The fungus is ubiquitous but influenced by seasonal conditions resulting in sporadic outbreaks. However, in recent years (2004–2009) the disease has become endemic to major faba bean production regions in southern Australia (unpublished data). The characteristics of cercospora leaf spot (CLS), predominantly appearing as red-brown to dark-grey leaf spots, are often mistaken with symptoms of ascochyta blight (Ascochyta fabae Speg.) or chocolate spot (Botrytis fabae Sard.), thus incorrect diagnosis has impacted on disease management decisions. Lesions mainly affect leaves, expanding rapidly to coalesce with adjacent lesions resulting in severe blighting of the leaf, and conidia protrude in fasciculate bundles in moist conditions from the centre (Chupp 1954; Williams 1987; Lang et al. 1993).
The Australian Faba Bean Breeding Program (AFBBP) identified a need to assess resistance to C. zonata within breeding accessions and existing cultivars but there are no known reports of phenotypic characterisation of germplasm collections of V. faba to CLS. Few countries report this disease as a major concern to faba bean production and very limited critical reviews are apparent (Woodward 1932; Yu 1947). However, many other species within the Cercospora genera are recognised as major economic concerns to production in a variety of hosts in many parts of the world (Latterell and Rossi 1983; Shane and Teng 1992; Smith and Littrell 1980; Barbetti 1991). Consequently, affected industries have often adopted stringent selection policies for approval of new cultivars, only releasing cultivars with reduced susceptibility (Windels et al. 1998; Wolf and Verreet 2002; Barbetti and Nichols 1994).
Incorporating resistance to diseases in adapted genotypes is determined by the complexity of inheritance. Studies of other CLS diseases indicate a diversity of mechanisms of inheritance. Resistance to C. beticola in sugar beet (Beta vulgaris) is under polygenic control, thus developing resistance while maintaining yield is difficult (Smith and Campbell 1996), whereas resistance to CLS diseases in soybean (Glycine max), cowpea (Vigna unguiculata) and mungbean (Vigna radiata) is monogenic expressed as either recessive, complete or incomplete dominance (Baker et al. 1999; Mishra et al. 1988; Booker and Umaharan 2008). By comparison, resistance to C. zeae-maydis in corn (Zea mays) is thought to be inherited quantitatively, associated with additive, dominant, recessive, and epistatic effects (Saghai et al. 1996; Coates and White 1998). Furthermore, a reliance on field screening under natural CLS epiphytotics can complicate quantitative assessments of resistance due to inconsistent disease distribution or mixed infections of pathogens confounding symptom expression (Booker and Umaharan 2008). Therefore, reliable and rapid screening methods for specific pathogens within controlled conditions are invaluable tools to develop disease resistance in adapted cultivars. The AFBBP currently uses such methods to screen for resistance to ascochyta blight and chocolate spot in faba bean germplasm (Kimber et al. 2004; Paull and Kimber 2006; Tivoli et al. 2006).
The objectives of this study were: to develop a method for screening for resistance to CLS in faba bean within a glasshouse and compare with results from field trials; to identify resistance to CLS in V. faba accessions within the AFBBP in controlled conditions without the confounding effect of other diseases that are frequently encountered in field trials; and to determine the inheritance of resistance to CLS in resistant genotypes, to be used by the AFBBP for future cultivar development.
Materials and methods
Inoculum production
Eight isolates, collected from different growing regions from 2000 to 2004, were chosen to represent the population of C. zonata in southern Australia and used in glasshouse experiments. Leaf tissue exhibiting CLS lesions was incubated in plastic bags containing moistened tissue paper at 3°C for 2 days. Resulting conidia were then excised from lesions with a sterilized glass needle and transferred to Potato Dextrose Agar (PDA) (in 9 cm Petri dish). Cultures were maintained at 22°C under 12 h light/dark cool-white and black-light (NUV) fluorescent light. Mass inoculum was prepared from a single culture using the following aseptic techniques. A portion of the growing culture was macerated using a scalpel, transferred into a 10 ml tube of sterile Reverse Osmosis (RO) water before being mixed using a vortex touch mixer. A 150 μl aliquot of the resulting mycelial suspension was transferred to PDA and spread with a bent-glass rod. This was repeated for each plate of inoculum required. Cultures were incubated at 22°C for 10–15 days then combined into a Warwick® commercial blender containing RO water to macerate into a mycelial suspension. Suspensions from eight isolates were combined, strained through two layers of cheesecloth, and the concentration estimated using a haemocytometer, by counting mycelial fragments. Mycelial fragments were used since attempts to induce conidia in culture were unsuccessful (unpublished data). The final inoculum concentration was adjusted to ca. 105 fragments/ml using RO water, after which 0.2% Tween-20 was added as a surfactant.
Controlled environment screening
Four glasshouse trials were conducted in 2003–2005 to evaluate 123 entries of V. faba. Four accessions of interest were selected from a naturally infected field trial conducted in 2004, while advanced breeding material and commercial cultivars commonly grown within the Australian faba bean industry were also included. Each trial was conducted as a randomised complete block design with four replicates. Variable numbers of entries were common between trials to evaluate repeatability. Plants were grown in trays (29 cm × 35 cm) containing 12 punnets per tray (MK12, 0.55L each) filled with University of California (UC) potting mix (Baker 1957). Two entries were included per tray, i.e. six punnets per entry, with one seed per punnet at 3 cm sowing depth. Plants were maintained outside (ca. means; min. 10°C, max. 22°C) for 4 weeks (ca. four-leaf nodes) then moved into two glasshouse cubicles, two replicates per cubicle. Plants were inoculated with the prepared mycelial suspension of C. zonata until run-off (ca. 2.5 ml inoculum per plant) using a Hardi® C5 spray applicator and maintained in high humidity, using overhead ultra-fine misters controlled by automated solenoids on cycled misting of 30 s every 2 h, at ca. 18°C.
Disease assessments were made on individual plants 11–21 days after inoculation (DAI), which was 5–7 days after symptoms first appeared, accounting for; lesion number and size on leaves, coalescence of lesions and defoliation, using a 1–9 scale (1 = no symptoms; 9 = severe disease) as presented in Table 1. In the 2003 experiment, disease assessments were made 11 DAI and 15 DAI. In the 2005 experiment, a preliminary assessment of entries was made 13 DAI and compared to final assessments at 15 DAI, including a separate assessment on the incidence of defoliation (%). Leaf lesions were sampled to excise conidia for examination using morphological descriptions of C. zonata (Williams 1987) and the causal pathogen was cultured to confirm Koch’s postulates.
Field disease nursery
Field disease nurseries were conducted in 2004 and 2005 at the Turretfield Research Station, 10 km north-east of Gawler, South Australia; Latitude: 34.55°S: Longitude: 138.83°E: elevation: 116 m. This site has a Mediterranean climate and annual rainfall ca. 460 mm (ca. 80% falling between April and October), with red-brown loam soils. Faba beans had historically been grown on each trial site every 3 years, consequently promoting the natural establishment of CLS. The AFBBP provided 293 entries for evaluation in 2004 and 348 entries in 2005; this included 18 entries in common for comparative analysis. Both trials were arranged as randomised block designs with two replicates, measuring 55 × 45 m, and consisted of 14 ranges of 54 plots, sown on May 21 (2004) and May 12 (2005). Plots were hand sown as two-row ‘hedge’ plots, 1.6 m long, with 25 cm row-spacing and 20 cm seed spacing within rows.
In both years CLS developed from natural in situ infection and disease assessment was made mid-winter 12 weeks after sowing (WAS), prior to confounding symptoms of Ascochyta fabae, also promoted in this trial. In 2004, the absence of ascochyta blight allowed another assessment in late spring, 22 WAS. A qualitative scale of 1–9 was used as described in Table 2.
Analysis of results
Results from screening trials were analysed using a general linear model—ANOVA, performed in Statistix® for Windows version 8.0. Regression analyses to assess repeatability and accuracy of screening techniques were conducted for faba bean entries in common between and within field and glasshouse trials, using GenStat® 11th Edition and Microsoft Excel® to fit trend-lines and calculate R 2 values.
Genetics of resistance to C. zonata in two V. faba genotypes
Two controlled environment experiments were conducted to evaluate the genetics of resistance to C. zonata in two genotypes of V. faba. The methods used for inoculum production, inoculation, sowing and maintenance of experiments were as previously described for glasshouse screening trials. Disease assessments were made using a percentage scale (%) of disease severity, which accounted for leaf area diseased (LAD%) and defoliation.
Experiment 1—Segregation patterns in F 3 progeny of S*R cross
The segregation patterns of 55 F 2 derived F 3 families from cross Farah*1322/2 were evaluated. Farah is a CLS susceptible Australian cultivar and accession 1322/2 is an Australian selection from the source population L82003 B8833, obtained from the International Center for Agricultural Research in the Dry Areas (ICARDA) that had expressed resistance to CLS in previous experiments. The trial consisted of 55 F 3 families and four entries of each parent (i.e. seeds harvested from the plants used in the hybridisation), arranged as 63 entries in a randomised complete block design with three replicates. Four entries were sown per tray, i.e. three punnets per entry, and maintained outside for 4 weeks (ca. four-leaf nodes), then moved into high humidity glasshouse cubicles and inoculated, as previously described.
Disease assessments of individual plants within the F 3 families were made 18 DAI. Based on the phenotypic responses of individual plants, F 3 families were assigned to one of three categories; homozygous resistant (<10% CLS score, as per response of resistant parent), heterozygous (including both resistant and susceptible plants) or homozygous susceptible (>10% CLS score). Family mean CLS data were plotted against the standard deviation to confirm classifications. Observed segregation patterns were tested for goodness of fit against the genotypic ratios of 1:2:1, as expected for a monogenic model controlling resistance and 1:14:1 for two genes with additive effect, using chi-squared (χ2) analysis to verify the adequacy of the genetic model.
Experiment 2—Gene frequencies in a heterogeneous population
The response patterns of two populations of cv. Cairo were examined. Cairo is a commercial cultivar that exhibited intra-population variation with both resistant and susceptible plants in the initial screening trials. One test population was derived from an open-pollinated multiplication of cv. Cairo while the second population was derived from one generation of self-pollination within a glasshouse to prevent cross-pollination by bees. The trial was a non-randomised block design with 12 replicates (trays) of each test population, each tray sown with ten seeds of a test population plus one seed each of the check entries, 1322/2 (resistant) and cv. Farah (susceptible). Plants were maintained outside (ca. means; min. 8°C, max. 16°C) for 4 weeks (ca. four-leaf nodes), then moved into high humidity glasshouse cubicles and inoculated as described previously. Disease assessment was made 18 DAI using a percentage scale (%) of CLS severity. Plants within both populations were classified as resistant or susceptible, using a threshold calculated from the mean disease score in the CLS susceptible check (cv. Farah) to accommodate for error; susceptible response threshold = (mean score cv. Farah) − (2 × standard deviation). The hypothesis of a monogenic dominant gene determining resistance was tested based on the principles of population genetics and the expected changes in genotype frequency following inbreeding. The open-pollinated population was assumed to be in Hardy–Weinberg equilibrium with genotype frequencies:
where p 2 = homozygous resistant, 2pq = heterozygous (expressed as resistant) and q 2 = homozygous susceptible. The observed frequencies for resistant and susceptible expression following one generation of inbreeding were tested for the goodness of fit against a monogenic dominant model using chi-squared (χ2) analysis to verify the adequacy of the genetic model.
Results
Controlled environment screening
Red-brown ‘specking lesions’ were evident on leaves 10–11 DAI and expanded, often coalescing with neighbouring lesions, to form large (5–25 mm in diam.) dark-red to brown lesions 15–21 DAI. Conidiophores often protruded from the centre of lesions, although small lesions (<ca. 8 mm) did not sporulate. Stem lesions (5–20 mm in length) were observed on plants in later assessments (14–21 DAI) but these were superficial and did not girdle or break stems. Intense sporulation occurred following incubation of lesions, with fascicle bundles of conidia protruding from the surface of the lesion clearly visible under binocular microscope (×10). Morphological examination of conidia excised from sampled lesions, which were then cultured on PDA, were consistent in appearance and mycelial growth habit to that of C. zonata.
There was significant variation (P < 0.05) in CLS severity among germplasm entries evaluated. Phenotypic characterisation of Australian commercial cultivars showed all were susceptible to the disease (CLS ≥4.5), with the exception of cv. Cairo (CLS = 3.0) which exhibited intra-population variation with both resistant (CLS = 1.0) and susceptible (CLS = 8.0) plants. Of 123 germplasm entries evaluated, seven resistant breeding lines were identified (mean CLS ≤3.0); Australian accessions S95007/1, 1103/3, 1107/2, 1108/2 and 1332/2 and breeding lines 1269*483/6 and AF03002 (Table 3).
Severe defoliation was found to be symptomatic of CLS infection and usually affected the lowest leaves of the canopy causing leaflets to excise at the junction with the petiole, though lesions were not always visible on affected petioles or at the terminated junction. In the 2005 experiment, significant variation (P < 0.05) in defoliation was observed amongst 48 entries assessed 15 DAI. Up to 70% defoliation was observed in susceptible entries, compared to less than 5% in resistant entries (breeding lines 1322/2 and 1269*483/6).
Field disease nursery
In 2004 and 2005, CLS established in the field trials from in situ inoculum. Small lesions (2–6 mm) first appeared on the lower leaves of seedlings (ca. three-leaf stage) 6–8 WAS although disease distribution was uneven across the site each year. High disease pressure was observed in half the 2004 trial site (replicate 1) by 22 WAS, but less disease occurred in the second replicate, thus CLS assessments were made in the first replicate only. In 2005, CLS severity was evenly distributed across 75% of the site, but diminished in bays 11–14, thus the second replicate was omitted from the analysis and mean scores calculated when multiple entries of a line or cultivar occurred. In 2004, disease assessments made at 22 WAS showed 289 entries were susceptible (scored ≥5.0) to CLS. Four entries (Accessions 1108/2, 1107/2, 1322/2 and S95007/1) exhibited a resistant reaction (CLS ≤3.0). Susceptible reactions included lesion expansion (25 mm + diam.) and blighting on leaves within the lower 70% of the plant canopy, minor lesions (<10 mm diam.) on upper leaves and stem lesions (30 mm + length) on the lower plant parts. Stem lesions were superficial and did not cause girdling or breakage. Extensive defoliation of leaflets that excised at the junction with the petiole was observed on susceptible plants and affected the lower 40% of the canopy. In 2005 disease developed faster than in 2004 and established in the trial by 12 WAS. Disease assessments showed 338 entries were susceptible to CLS (scored ≥5.0). Resistant reactions to CLS were observed in ten entries (CLS ≤3.0), four entries indentified in 2004 plus six additional entries; accessions 1517/1, 969/2 and 1777/1, and breeding lines 1269*483/6, AF04016 and AF04064.
The origin of the ten CLS resistant germplasm identified in the field and controlled environment trials are diverse, derived from germplasm collections sourced from various locations in the Mediterranean basin and South America (Table 3).
Correlation between screening techniques
Correlation analyses between CLS data in controlled environment experiments and field disease nurseries data for germplasm that was common between trials are presented in Table 4. A high correlation (R 2 = 0.74; P < 0.05) was achieved where the highest number of entries (16) were compared in controlled environment trials (2003 vs. 2004). Correlations between other controlled environment trials (R 2 = 0.65 and R 2 = 0.86) were also significant (P < 0.05) when common entries were low (nine and five entries). Where 43 entries were in common between the controlled environment and field screening trials (conducted in 2004) a significant correlation (R 2 = 0.45; P < 0.05) was achieved and identified four entries (1107/2, 1322/2, S95007/1 and 1108/2) as resistant to CLS (scored ≤3.4) in both field and controlled environment trials (Fig. 1). Positive correlations ranging from R 2 = 0.45 to R 2 = 0.62 were significant (P < 0.05) in six different analyses between the two screening techniques (Table 4). Disease data from field nurseries in the two different seasons also had positive correlations (R 2 = 0.54; P < 0.05), despite varying severity.
The relationship between mean scores for cercospora leaf spot (C. zonata) expressed in 43 faba bean (V. faba) entries in a controlled environment screening trial (assessed 21 DAI) and a field disease nursery (assessed 22 WAS) conducted in 2004. Symbols indicate resistant entries; 1322/2 (cross sign), 1108/2 (filled inverted triangle), S95007/1 (filled square) and 1107/2 (open square), and susceptible cv. Farah (plus sign)
Genetics of resistance to C. zonata in two V. faba genotypes
Experiment 1—Segregation patterns in F 2 derived F 3 families of S*R cross
Disease assessments at 18 DAI showed that all plants of the resistant parent (Acc. 1322/2) scored <5.0% and all plants of the susceptible parent (cv. Farah) scored 35–80% for CLS severity. There was considerable variation in disease reactions within and between F 3 families. Disease reactions of individual plants within F 3 families were used to characterise each family, and 11 homozygous resistant, 25 heterozygous and 19 homozygous susceptible F 3 families were identified. When mean disease data for F 3 families was plotted against their standard deviations three phenotype ‘cluster-groups’ were identified, which corresponded to the genotypic characterisation applied to each population (Fig. 2). This proportion (11:25:19) fitted the ratio of 1:2:1 expected for a monogenic model (χ 22 = 2.78, 0.2 < P < 0.3), in F 2 derived F 3 families (Table 5A). In contrast, the observed ratio did not adequately fit (P < 0.001) the principle of a two gene model with additive effect, where a ratio of 1:14:1 is expected in F 3 families (Table 5A).
Cluster groups observed when mean cercospora leaf spot severity (%) is plotted against standard deviations of means of the disease reactions in 55 F 3 families of Farah(S)*1322/2(R) in a controlled environment trial. Symbols correspond to phenotypic characterisation of families (e.g. homozygous susceptible) based on disease assessments of individual plants. Means for resistant (1322/2) and susceptible (cv. Farah) checks indicated with cross sign and open square, respectively
Disease data for individual plants (n = 220) in the 25 F 3 families characterised as heterozygous showed a bimodal distribution (Fig. 3), with the majority of plants (154) exhibiting a resistant response (≤10% CLS) and the remaining (66) plants exhibiting a susceptible response comparable to the susceptible parent (49 plants scoring 31–60% CLS). This observed ratio of 154(R):66(S) plants in F 3 heterozygous families fitted the ratio of 3:1 (χ 21 = 2.93, P > 0.05) expected for a monogenic dominant mode of inheritance (Table 5B).
Frequency distribution of cercospora leaf spot (CLS) severity (%) observed among individual plants within 25 heterozygous F 3 families identified using phenotypic characterisation in a controlled environment trial. Lines indicate the mean score (arrow) and range (error bar) of disease reactions in plants of susceptible cv. Farah
Experiment 2—Evaluating gene frequencies in a self-pollinated population
At 18 DAI the mean CLS score of the resistant check (1322/2) was 1.2% and 54.3% for the susceptible check (cv. Farah). There was considerable variation in disease reactions by individual plants within the two populations of cv. Cairo, varying from 1 to 65% CLS severity (Fig. 4). Individual plants in the self-pollinated population (n = 116) and open-pollinated population (n = 107) were characterised using a threshold of 23% CLS (Farah mean − 2*SD) to distinguish between resistant and susceptible reactions. The open-pollinated population exhibited 74 resistant (p 2 + 2pq = 0.69) and 33 susceptible (q 2 = 0.31) plants (n = 107). Therefore, q = √0.31 = 0.44 and p = 1 − 0.44 = 0.56. After one generation of self pollination, the frequency of heterozygotes at a single locus is expected to halve to:
Frequency distribution of cercospora leaf spot (CLS) scores in two populations, open-pollinated and self-pollinated, of cv. Cairo observed 18 days after inoculation with C. zonata under conducive conditions in a glasshouse experiment. The threshold to characterise resistant (≤23% CLS) and susceptible (≥23% CLS) reactions is indicated (= mean cv. Farah − 2*SD)
For a dominant gene the expected phenotype frequencies are
The expected genotype and phenotype frequencies in the self-pollinated population were calculated using the frequencies observed in the open-pollinated population:
The expected phenotype frequencies in a self-pollinated population of 116 plants were calculated as 66 resistant (0.57) and 50 susceptible (0.43) plants while the observed phenotypes were 61 resistant (0.53) and 55 susceptible (0.47) plants. The phenotype frequencies in the self-pollinated population of cv. Cairo were consistent with those expected for a monogenic dominant gene control based on the gene frequencies observed in the open-pollinated population (χ2 = 0.88, 0.3 < P < 0.5) (Table 6).
Discussion
This study has reported for the first time sources of resistance in faba bean to C. zonata. Furthermore, we describe screening techniques for phenotypic characterisation of V. faba germplasm in controlled environments to evaluate resistance to CLS using mycelial suspensions of pure C. zonata cultures. These were used effectively in inoculations and offer a viable method for pathogens that fail to sporulate in culture, such as has been reported for other Cercospora spp. (Barbetti 1985; Tessman et al. 2008). Several sources of resistant or moderately resistant lines were identified, and resistance was expressed under both field and glasshouse conditions. These include Australian accessions 1107/2, 1108/2 and 1332/2 and breeding line 1269*483/6. The first three accessions are selections that were made in Australia from germplasm obtained from ICARDA. The source populations were ILB2582, ILB2583 (both originating from Morocco) and L82003 B8833, respectively. The controlled environment screening method was rapid, 43–49 days from planting to assessment compared to 84–154 days in field trials, it was repeatable, and showed good correlations with phenotypic responses of common genotypes screened in naturally infected field trials.
The incidence of CLS has become more frequent and severe in commercial faba bean crops in southern Australia (unpublished data). This study showed all commercially available cultivars for the Australia faba bean industry are susceptible to CLS. This undoubtedly has influenced the increased prevalence of the disease and contributed to the build up of disease inoculum within intensified cropping regions in the last three decades. Broad scale adoption of highly susceptible cultivars has been reported to contribute to widespread epidemics of other Cercospora diseases, notably C. beticola of sugar beet (Windels et al. 1998) and C. zeae-maydis in maize (Latterell and Rossi 1983; Ward et al. 1999). It is important that this issue be addressed to sustain profitability of the southern Australian faba bean industry. A similar situation relevant to southern Australia has been reported recently, in which a decade of concerted breeding has resulted in over 50% of subterranean clover genotypes in the program exhibiting resistance to C. zebrina (Barbetti and Nichols 2005).
In the genotypes of V. faba tested in this study, the extent of defoliation varied in the lower canopy and this was an important component of disease susceptibility to C. zonata. Genotypic variation within germplasm, particularly with respect to the onset of premature defoliation, should be explored further as it may provide a means of quantifying partial resistance in genotypes and reduce the loss of photosynthetic leaf area by CLS. This process of quantifying partial resistance has been reported in peanut genotypes against C. arachidicola (Johnson et al. 1986; Ricker et al. 1985).
The current study provides valuable information on the genetics of resistance to CLS in faba bean. Resistance to C. zonata evaluated in accession 1322/2 and cv. Cairo was found to be governed by a single dominant gene. Phenotypic characterisation of F 2 derived F 3 families from the cross between resistant accession 1322/2 and susceptible cv. Farah exhibited segregation patterns fitting a monogenic model, where a ratio of 1:2:1 is expected. In addition, very low disease scores were identified on resistant F 3 families, corresponding to responses in the resistant parent and there was no overlap with segregating families. Segregation patterns of resistant and susceptible plants within heterozygous F 3 families indicated resistance is a dominant effect. These findings verified the method for phenotypic characterisation of F 3 populations was justified. Furthermore, a monogenic dominant model was also found to apply to resistance in cv. Cairo, demonstrated by the change in frequency of resistant and susceptible plants when comparing open-pollinated and self-pollinated populations. The sub population in cv. Cairo selected with resistance in this study has been used as an adapted source of resistance to CLS for the AFBBP.
Disease resistance inherited via monogenic dominant effects can provide breeding programs a simple pathway to incorporate resistance into adapted genotypes against new incursions, escalated disease pressure or virulent races that have compromised resistant genotypes (Baker et al. 1999). However, while single-gene resistance offers a valuable short term option for management of CLS it is important that alternative genes conferring resistance are identified to establish quantitative resistance for long term management of the disease. Eleven sources of resistance, or partial resistance, were identified in this study, including cv. Cairo. However, the relationship between these resistant sources is not known. Further studies should be undertaken to determine if there are multiple resistance genes that could be pyramided to establish durable resistance to CLS. The resistant entries identified in these experiments are being used by the AFBBP to transfer CLS resistance to adapted genotypes for release to the southern Australian faba bean industry. Industry uptake of cultivars developed by the AFBBP has historically been high thus the impact of CLS-resistant cultivars is anticipated to reduce the incidence of disease in intensive growing regions and offer growers greater confidence in disease management decisions.
References
Baker KF (1957) The UC system for producing healthy container-grown plants. University of California Press, Berkeley
Baker WA, Weaver DB, Qui J, Pace PF (1999) Genetic analysis of frogeye leaf spot resistance in PI54610 and Peking soybean. Crop Sci 39:1021–1025
Barbetti MJ (1985) Infection studies with Cercospora zebrina on pasture legumes in Western Australia. Aust J Exp Agric 25:850–855
Barbetti MJ (1991) Response of subterranean clover varieties to cercospora disease under a controlled environment and in the field. Aust J Exp Agric 31:333–336
Barbetti MJ, Nichols PGH (1994) Field resistance in Trifolium subterraneum to Uromyces trifolii-repentis and Cercospora zebrina. Aust J Exp Agric 34:1131–1136
Barbetti MJ, Nichols PGH (2005) Field performance of subterranean clover germplasm in relation to severity of Cercospora disease. Australas Plant Pathol 34:197–201
Booker HM, Umaharan P (2008) Quantitative resistance to Cercospora leaf spot disease cause by Pseudocercospora cruenta in cowpea. Euphytica 162:167–177
Chupp C (1954) A monograph of the fungus genus Cercospora. Cornell University, New York
Coates ST, White DG (1998) Inheritance of resistance to gray leaf spot in crosses involving selected resistant inbred lines of corn. Phytopathology 88:972–982
Johnson CS, Beute MK, Ricker MD (1986) Relationship between components of resistance and disease progress of early leaf spot on Virginia-type peanut. Phytopathology 76:495–499
Kimber RBE, Paull J, Davidson JA (2004) Screening methodologies to identify resistance to Ascochyta leaf blight (Ascochyta fabae) in faba bean germplasm. In: Proceedings of the 5th European conference on grain legumes, 7–11 June, Dijon France, p 326
Lang L, Yu Z, Zheng Z, Xu M, Ying H (1993) Faba bean in China: state-of-the-art review. ICARDA, Aleppo
Latterell FM, Rossi AE (1983) Gray leaf spot of corn: a disease on the move. Plant Dis 67:842–847
Mishra SP, Asthana AN, Yadav L (1988) Inheritance of Cercospora leaf spot resistance in mung bean, Vigna radiata (L.) Wilczek. Plant Breed 100:228–229
Paull JG, Kimber RBE (2006) Breeding faba bean for resistance to chocolate spot. In: Avila CM et al (eds) Proceedings of the international workshop on faba bean breeding and agronomy, 25–27 Oct. Cordoba, Spain, pp 117–120
Ricker MD, Beute MK, Campbell CL (1985) Components of resistance in peanut to Cercospora arachidicola. Plant Dis 69:1059–1064
Saghai Maroof MA, Yue YG, Xiang ZX, Stromberg EL, Rufener GK (1996) Identification of quantitative trait loci controlling resistance to gray leaf spot disease in maize. Theor Appl Genet 93:539–546
Shane WW, Teng PA (1992) Impact of Cercospora leaf spot on root weight, sugar yield, and purity of Beta vulgaris. Plant Dis 76:812–820
Smith GA, Campbell LG (1996) Association between resistance to Cercospora and yield in commercial sugarbeet hybrids. Plant Breed 115:28–32
Smith DH, Littrell RH (1980) Management of peanut foliar diseases with fungicides. Plant Dis 64:356–361
Tessman DJ, Charudattan R, Preston JF (2008) Variability in aggressiveness, cultural characteristics, cercosporin production and fatty acid profile of Cercospora piaropi, a biological agent of water hyacinth. Plant Pathol 57:957–966
Tivoli B, Baranger A, Avila CM, Banniza S, Barbetti M, Chen W, Davidson J, Lindeck K, Kharrat M, Rubiales D, Sadiki M, Sillero JC, Sweetingham M, Muehlbauer FJ (2006) Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147:223–253
Ward JM, Stromberg EL, Nowell DC, Nutter FW (1999) Grey leaf spot. A disease of global importance in maize production. Plant Dis 83:884–895
Williams MAJ (1987) CMI descriptions of pathogenic fungi and bacteria. CAB International. Martinus Nijhoff/Dr W. Junk, Dordrecht, pp 185–186
Windels CE, Lamey HA, Hilde D, Widner J, Knudsen T (1998) A Cercospora leaf spot model for sugar beet. Plant Dis 82:716–726
Wolf PFJ, Verreet JA (2002) Development and implementation of an integrated pest management system in Germany for the control of fungal leaf diseases in sugar beet: the IPM sugar beet model. Plant Dis 86:336–344
Woodward RC (1932) Cercospora fabae Fautrey, on field beans. Trans Br Mycol Soc 17:195–202
Yu TF (1947) Cercospora leaf spot of broad bean in China. Phytopathology 37:174–179
Acknowledgments
The authors sincerely thank: Kevin James, Ian Roberts and Michelle Russ for invaluable technical support; Jenny Davidson and Assoc. Prof. Eileen Scott for scientific input, and the Grains Research and Development Corporation (GRDC) for funding this study. This study was in part to fulfil requirements of a Doctorate of Philosophy (PhD) for Rohan Kimber.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kimber, R.B.E., Paull, J.G. Identification and genetics of resistance to cercospora leaf spot (Cercospora zonata) in faba bean (Vicia faba). Euphytica 177, 419–429 (2011). https://doi.org/10.1007/s10681-010-0288-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0288-7