Abstract
Flooding is one of the major hazards of rice production for the rainfed lowland rice ecosystem, and tolerant cultivars are urgently needed to help protect farmers from submergence damage. A quick and efficient strategy was implemented to introgress SUB1, a major QTL for submergence tolerance, into a rainfed lowland mega variety BR11 of Bangladesh by only two backcrosses and one selfing generation. In marker-assisted backcrossing (MABC), one tightly-linked simple sequence repeat (SSR) and two gene-based markers, four flanking SSR and 116 background SSR markers were used for foreground, recombinant and background selection, respectively, in backcrosses between a SUB1 donor IR40931-33-1-3-2 and BR11. BR11-Sub1, identified in a BC2F2 plant, possessed BR11 type SSR alleles on all fragments analyzed except the SUB1 QTL. The introgression size in BR11-Sub1 was 800 Kb indicating approximately 99.8% identity to BR11. BR11-Sub1 along with other introgression lines showed submergence tolerance similar to the tolerant parent. Yield, yield-component parameters and grain physico-chemical properties showed successful recovery of the BR11 traits in BR11-Sub1, with yield potential ranging from 5.2 to 5.6 t/ha, not significantly different from the recurrent parent mega variety BR11. Producing a large number (~1000) of backcross F1 plants was considered essential to achieve recombination on both sides of the gene, limiting linkage drag with only two backcrosses. A large number of background markers ensured proper recovery of the recurrent parent genome in the BC2F2 generation. The study demonstrates a rapid and highly precise strategy to introgress a major QTL by BC2F2 generation into a modern rice variety using an unadapted donor. The variety can replace BR11 on more than 2 million of ha in Bangladesh and provide major increases in rice production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Submergence caused by flash-flooding is one of the important hazards to the agriculture of tropical Asia. In Bangladesh, of 10.6 million ha of rice production, more than two million ha is affected by flash flooding. Flash floods regularly affect rainfed lowland rice (RLR) ecosystems where floodwater remains for around 2 weeks in many parts of the country. Traditional varieties adapted to the submergence-prone environments are low yielding due to their low tillering ability, long droopy leaves, susceptibility to lodging and poor grain quality. Improved varieties are needed that combine high yield with submergence tolerance. Because of the sensitivity of rice and the prevalence of the stress, submergence tolerance has been an important breeding objective for decades in rainfed lowland areas of Asia (Mackill 1986). Improved submergence tolerant cultivars have been under development for more than three decades (HilleRisLambers and Vergara 1982; Mackill 1986; Mohanty and Chaudhary 1986). However, improved cultivars have generally not been adopted by the farmers in submergence prone areas. One reason is that these tolerant varieties lack many of the desirable characteristics of the popular and widely grown varieties (‘mega varieties’), such as high yield and good grain quality (Septiningsih et al. 2009).
There are several rice mega-varieties grown in Bangladesh, of which BR11 is the most important, covering 40% of the RLR area of Bangladesh (BRRI 2007). Unfortunately these mega-varieties are susceptible to major yield losses due to submergence. Mackill (2006) proposed that adoption of a completely new variety could take considerable time, whereas the chances of rapid adoption of popular varieties converted through marker assisted backcrossing (MABC) were relatively higher.
MABC is a precise and effective method to introgress a single locus controlling a trait of interest while retaining the essential characteristics of the recurrent parent (Collard and Mackill 2008; Hospital 2001; Hospital and Charcosset 1997). MABC has three main advantages over conventional backcrossing. Firstly, DNA markers can be used for simple and efficient selection of the target locus (‘foreground selection’). Secondly, the size of the donor chromosome segment containing the target locus can be minimized (‘recombinant selection’). Thirdly, the recovery of the recurrent parent can be accelerated by selecting backcross lines with a higher proportion of recurrent parent genome (‘background selection’). This approach has been used with great success for ‘enhancing’ rice varieties for traits such as bacterial blight resistance gene XA21; (Chen et al. 2000), the waxy locus for grain quality (Zhou et al. 2003) and submergence tolerance SUB1 (Neeraja et al. 2007; Septiningsih et al. 2009).
At the International Rice Research Institute (IRRI), a project was undertaken to transfer SUB1, a major QTL on chromosome 9 explaining almost 70% of the phenotypic variance (Xu and Mackill 1996; Xu et al. 2006), into at least six widely-grown rice cultivars in Asia. For the first variety Swarna, a mega-variety grown in India, Swarna-Sub1 seeds were sent to India and Bangladesh for testing in 2005 (Neeraja et al. 2007) and the variety was officially released in 2009. However, in Bangladesh, the variety BR11 is more widely grown under RLR conditions. The conversion of BR11 into a submergence tolerant type while maintaining all its other characteristics unchanged, would assure increased production from the submergence prone areas of the country.
The main objective of the present study was to convert the mega rice variety BR11 into a submergence tolerant variety by incorporating SUB1 using a MABC approach. We fully exploited recombinant and background selection in order to gain high precision for reducing the size of the donor chromosome segment, to minimize linkage drag, and to rapidly recover the recurrent parent genome as quickly as possible. The SUB1 lines developed through this strategy were tested for their submergence tolerance and other important traits. The present investigation was also aimed to compare the BR11-Sub1 lines to the recurrent parent with respect to grain yield, yield-contributing attributes and grain physico-chemical properties.
Materials and methods
Plant materials and crossing scheme
IR40931-33-1-3-2, one of the FR13A-derived submergence-tolerant breeding lines (Mackill et al. 1993), was used as the donor of SUB1. IR40931-33-1-3-2 was a submergence tolerant indica advance breeding line of rice with moderate yield of 3–4 t/ha and improved plant type. The SUB1 gene in this line was inherited from the landrace FR13A, a widely-used submergence tolerance donor with poor agronomic properties. The recipient variety was BR11, a widely grown cultivar in Bangladesh. This variety was derived from the cross IRRISail/IR5, and was originally designated BR52-87-1-HR88. The variety was released by BRRI in 1980 for the Transplanted Aman (RLR) season. The yield potential of this variety is 6.5 ton/ha under optimum management.
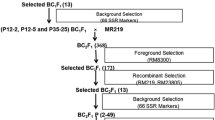
For the MABC scheme (Fig. 1), BR11 was crossed with IR40931-33-1-3-2 to obtain F1 seeds. F1s were backcrossed to BR11 to obtain a large number of BC1F1 seeds. A large amount of backcross seeds was produced for MABC (Collard et al. 2008b).
Development of the submergence tolerant BR11-Sub1 with details of markers used for foreground, recombinant and background selection. The numbers of plants selected in each generation is indicated in parentheses; P311 means BC2F1 population produced from plant number 311 of BC1F1 generation. SRP Single recombinant at proximal end, SRD Single recombinant at distal end, DR Double recombinant
Molecular marker analysis
DNA was extracted from young leaves of 2-week-old plants using a modified protocol as described by Zheng et al. (1995). PCR was performed in 10 μl reactions containing 25 ng of DNA template, 1 μl 10× PCR buffer (containing 200 mM Tris-HCl pH 8.3, 500 mM KCl, 15 mM MgCl2), 1 μl of 1 mM dNTP, 0.50 μl each of 5 μM forward and reverse primers and 0.25 μl of Taq DNA polymerase (4 U/μl) using an MJ Research single or dual 96-well thermal cycler or G-Storm thermal cycler. After initial denaturation for 5 min at 94°C, each cycle comprised 1 min denaturation at 94°C, 1 min annealing at 55°C, and 2 min extension at 72°C with a final extension for 5 min at 72°C at the end of 35 cycles. The PCR products were mixed with bromophenol blue gel loading dye and were analyzed by electrophoresis on 8% polyacrylamide gel using mini vertical polyacrylamide gels for high throughput manual genotyping (CBS Scientific Co. Inc., CA, USA). The gels were stained in 0.5 mg/ml ethidium bromide and photos were taken using Alpha Imager 1220 (Alpha Innotech, CA, USA). Microsatellite or simple sequence repeat (SSR) markers were used for selection (IRGSP 2005; McCouch et al. 2002; Temnykh et al. 2001).
For foreground selection, gene-based STS marker Sub1C173 specific to SUB1C and cleaved amplified polymorphic site (CAPS) marker GnS2 specific to SUB1A of the SUB1 QTL were used for the confirmation of SUB1 (Neeraja et al. 2007; Septiningsih et al. 2009). Eleven tightly-linked and gene-based foreground markers (IRGSP 2005; Neeraja et al. 2007; Septiningsih et al. 2009; Xu et al. 2006) were surveyed over the two parents BR11 and IR40931-33-1-3-2. Five primers were polymorphic between two parents (See Table 1 in Supplementary Material). The maximum physical distance of tightly-linked markers with the SUB1 QTL was 0.3 Mb. Out of five polymorphic markers, one primer RM8300 (Neeraja et al. 2007) was used as tightly-linked marker initially in foreground selection because of its clear codominant nature and capability of producing easily-scorable bands.
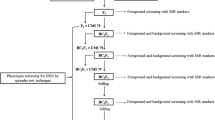
For flanking markers used for recombinant selection, 0–8 Mb region on the distal region of chromosome 9 around the SUB1 QTL was targeted. A total of 28 flanking markers (IRGSP 2005; Neeraja et al. 2007; Xu et al. 2004, 2006) were surveyed for identifying polymorphism between recipient and donor parents. Out of 28 flanking markers, 15 markers were obtained as polymorphic (See Table 2 in Supplementary Material). The position of the SUB1 QTL was at 6.2–6.3 Mb or 4.4–6.8 cM on chromosome 9 (Xu et al. 2006). Out of 15 polymorphic flanking markers, two flanking markers, SC34 (RM23679) and SC16 (RM23805), were used at the proximal end of SUB1 QTL (6.3 Mb/4.4–6.8 cM) and SC26 (RM23915) and SC30 (RM23958) were used at the distal end. The closest flanking markers on either side were 2.3 (proximal end) and 3.2 (distal end) cM from the SUB1 QTL (Fig. 2). The position of the centromere in chromosome 9 was at 2.8 Mb (TIGR data base, http://rice.plantbiology.msu.edu/) which was again at the proximal end of the SUB1 QTL, and the flanking markers (at the proximal end) were taken from both sides of the centromere as the likelihood of crossovers were less in the centromeric region (Chen et al. 2002). The flanking markers were also selected based on their distance from SUB1, nature of codominance and capability of producing scorable bands. The flanking markers that revealed fixed (homozygous) alleles of the recipient parent at non-target loci in one generation were not screened in the next backcross generation.
Microsatellite markers unlinked to SUB1 covering all the chromosomes, that were polymorphic between the two parents, were used for background selection to recover the recipient genome. Out of 524 SSR primers surveyed, 77 markers including the four flanking markers of the target QTL were used for background selection initially. The maximum number of background markers used was 10 for chromosome 3 and the minimum number was 5 for chromosomes 4, 5, 7, 8, 10 and 12. The average number of background markers over all the 12 chromosomes was 6 (See Table 3 in Supplementary Material). The average distance between adjacent markers ranged from 13 cM (chromosome 9) to 29 cM (chromosome 8). The microsatellite markers that revealed fixed (homozygous) alleles at non-target loci in one generation were not screened in the next backcross generation. The segregants with fixed donor alleles were discarded from the selection.
For the selected plants from the BC2F2 generation, additional microsatellite markers were used to check the fixation of the recipient genome. The total number of additional background markers used over all the 12 chromosomes was 43. The maximum number of background markers used was 9 in chromosome 5 and the minimum number of background markers was 1 in chromosome 12 (See Table 4 in Supplementary Material). The maximum distance of additional background markers with the closest initial background marker was 28.4 cM which was obtained for RM227 of chromosome 3. The marker RM227 should have been used as initial background marker because the distance of this marker with the closest initial background marker was 29 cM. Otherwise, for all other additional background markers, the distance with the closest initial background marker was less than 13 cM. However, the average distance of the additional background markers with the closest initial background marker was 5 cM.
Data analysis
The molecular weights of the different alleles were measured using Alpha Ease Fc 5.0 software. The marker data was analyzed using the software Graphical Genotyper (GGT 2.0) (Van Berloo 2008). The homozygous recipient allele, homozygous dominant allele and heterozygous allele were scored as ‘A’, ‘B’ and ‘H’. The percent markers homozygous for recipient parent (%A) and the percent recipient alleles including heterozygous plants (%R) were calculated.
Screening of PILs for submergence tolerance
Submergence screening was performed in the greenhouse at IRRI, Los Baños, Philippines, following standard protocols (Xu et al. 2000). Seeds of the six PILs including BR11-Sub1 of the BC2F2 generation with parents and susceptible check IR42 and resistant check FR13A were germinated in rows in 20 cm × 15 cm × 10 cm trays with three replications. Fourteen-day-old seedlings were submerged for 21 days. The duration of submergence was longer as the prevailing temperature during the period of the experiment was comparatively low. The survival percentage and elongation ratio of plants were taken 21 days after de-submergence. Submergence tolerance score was given just after de-submergence and 7 days after recovery following SES of IRRI (IRRI 2002) for confirmation of the presence of the SUB1 locus.
Eighteen genotypes, including eight PILs, four submergence tolerant enhanced mega-varieties (BR11-Sub1, Swarna-Sub1, IR64-Sub1, Samba Mahsuri-Sub1), two mega varieties (BR11, Swarna), two tolerant checks (IR40931-33-1-3-2, FR13A), and two susceptible checks (BR5, IR42), were tested under submergence stress with two replications. 14-day old seedlings were transplanted in the plant physiology submergence tank on 8 July 2008. Twenty-one seedlings were transplanted in three rows at spacing of 20 × 20 cm using a single seedling per hill. At 7 days after transplanting (DAT), the crop was submerged completely maintaining 70 cm water depth for 14 days. During the submergence period, the water of the tank was made turbid twice daily and monitored through light meter (L1-250). Water was drained out completely 14 days after submergence. The survival and recovery data were taken 5 days and 30 days after de-submergence following SES of IRRI (IRRI 2002).
Phenotyping of Precision Introgression Lines (PILs)
Phenotypic evaluations were conducted at the experimental farm of the Bangladesh Rice Research Institute (BRRI), Gazipur during the irrigated rice season in 2007-08, with six PILs (lines with a small introgressed fragment containing the target gene/QTL, Collard et al. 2008a) and two parents. Forty-five day old seedlings were transplanted using a single seedling per hill. The unit plot size was 5.4 m × 10 rows. RCB design with three replications was followed with the spacing between plants in a row 15 cm and the rows 25 cm apart. The full plot was harvested for taking grain yield which was adjusted to 14% moisture level. Missing hills were adjusted in calculating grain yield. Grain yield (ton/ha) along with plant height (cm), days to maturity, number of panicles per plant, number of grains per panicle, and 1000-grain weight (g) were recorded and used in the analysis done by MSTAT-C software of Michigan State University of USA.
Grain physical parameters were taken for the six PILs. Data of 10 grains from 10 random panicles were recorded on grain length (mm), grain width (mm), grain thickness (mm), kernel length (mm), kernel width (mm) and kernel thickness (mm). Grain and kernel length, width and thickness were measured by a digital slide calliper, model no. SD-089. Dehulled and unpolished brown rice was designated as kernel. The length and shape of the kernel were classified following standard evaluation system (SES) of IRRI (IRRI 2002).
The seeds were dehulled and milled for the analysis of grain chemical properties. The grain chemical properties analyzed were head-rice recovery percentage, chalkiness, waxyness, amylose content (%), gelatinization temperature and gel consistency. The analyses were done in collaboration with the researchers at the Grain Quality, Nutrition, and Postharvest Center, IRRI. Gelatinization temperature with the range of 70–74°C was classified as intermediate.
Results
F1 and backcross seed production
Forty-four F1 plants were produced which were confirmed using two polymorphic SSR markers RM488 and RM541 and one sequence tagged site (STS) marker Sub1C173. A total of 1797 BC1F1 seeds were produced from crosses with 44 F1 plants and 1421 plants survived in the main field. Selection was carried out dividing all the BC1F1 plants into three populations for convenience in managing the fieldwork.
A total of 3635 BC2F1 seeds were produced from crosses with 14 plants of all the three populations. The maximum number of BC2F1 seeds was 676 obtained from plant number 337 of population 2 of BC1F1 generation and the minimum number of BC2F1 seeds was 31 obtained from plant number 290 of population 2. The low number of seeds produced from plant number 290 of population 2 indicated the practical difficulties of producing large seed numbers for the BC2F1 generation. Out of 3635, the total number of BC2F1 seeds used was 763 obtained from the best three plants (P1-95, P2-311 and P3-06) of the three BC1F1 populations for the execution of MABC in this study.
Foreground selection
Out of 1421 plants, 649 plants were heterozygous for the marker RM8300 tightly linked to SUB1, 761 plants were found with the locus fixed for the recipient allele (Score A) and only 11 plants were found with the locus fixed for donor allele (resistant allele) (Score B). The eleven plants with score B were produced from accidental self-pollination. The 649 plants with the ‘H’ score for the tightly-linked marker were subjected to recombinant selection with flanking markers.
In the BC2F1 generation, foreground selection was carried out over 763 plants where 375 plants showed ‘H’ score and 386 plants showed ‘A’ score. BC2F2 seeds were produced by self-pollinating the best plant (number 311-391 of population 2) of the BC2F1 generation, where foreground markers RM8300, Sub1C173 and GnS2 were heterozygous. Out of 1221 plants, 627 were heterozygous for the tightly-linked marker (Score H), 289 plants were fixed for recipient alleles (susceptible allele) (Score A) and 305 plants were fixed for donor alleles (resistant allele) (Score B). The results fitted the expected 1:2:1 ratio for this generation (χ2 = 1.31. P > 0.05).
Recombinant selection
Recombinant selection was carried out on 649 plants that were heterozygous for the marker used for foreground selection in the BC1F1 generation. Thirteen single recombinants were obtained which minimized the size of the donor segment at the distal end while the other eight single recombinants minimized introgression size at the proximal end.
In BC2F1 generation, recombinant selection was carried out using heterozygous flanking markers specific to the three BC2F1 populations produced from the three best plants of the BC1F1 generation. Double recombinants were not obtained from two BC2F1 populations produced from plant number 6 of population 3 (P3-06) and P1-95 as there were no recombination events between the SUB1 QTL and the flanking markers at the proximal end. A total of 216 positive plants of P2-311 population were surveyed with the heterozygous flanking markers RM23915 and RM23958 during foreground selection. Eight segregants showed recipient alleles for both or one of those two markers. Out of these eight plants, five (68, 123, 271, 304 and 391) had recipient alleles for both flanking markers while the other three (292, 325 and 336) obtained the recipient alleles for only one marker RM23958.
Selection using gene-based markers
Before carrying out background selection, the selected BC1F1 plants were verified with the gene-based markers Sub1C173 for SUB1C and GnS2 for SUB1A to confirm that the SUB1 gene was indeed present in the lines. In case of plant number 145, the tightly-linked marker RM8300 was heterozygous in the foreground selection. All tolerant types including the check IR40931 showed the tolerant allele of SUB1, but the marker allele of plant 145 was cut generating two alleles having lower size which was also obtained for the susceptible allele of BR11. Plant number 145 showed homozygous recipient allele for the SUB1C marker Sub1C173. The results indicated that plant number 145 neither contained SUB1C nor SUB1A. This shows the importance of using gene-based primers since there will be no recombination at all between the markers and the gene of interest.
Background selection
Initially, background selection was carried out using 73 markers in BC1F1. The percent markers homozygous for the recipient parent ranged from 32.5 to 70.1% in those 22 plants (Table 1). In plant number 6 of population 3 of BC1F1 generation, 54 out of 77 markers (70.1%) were homozygous for the recipient parent type, and the percentage of recipient alleles was 85.1%. So this plant was selected as the best plant of the population. In this plant, chromosome 1, 5, 11 and 12 were completely of the recipient type, whereas the remaining ranged from 50% (chromosome 8) to 90% (chromosome 4 & 10). The second best plant P2-311 had 83.1% recipient alleles and the third P1-95 had 80.5% (Table 1).
In BC2F1, double recombinants were not obtained in the P3-06 population. However, double recombinants were obtained for population P2-311 and background selection was carried out over 8 double recombinants using 23 heterozygous background markers. The maximum percentage of recipient alleles was 97.3% obtained in plant number 311-391. The minimum percentage of recipient alleles was 89.7%, which was obtained in plant number 311-123 (Table 2). The presence of SUB1 QTL was confirmed in the selected plants by using gene-based markers Sub1C173 and GnS2 as before.
Importantly, the best plant 311-391 was a double recombinant for all the four flanking markers. While plant 311-391 had four heterozygous markers remaining, they were located in only two positions of the genome on chromosomes 1 and 8. Here, the two markers RM212 and RM486 used in BC1F1 generation were located within 5 cM distance, so these two markers were considered as one SSR-background-locus. Based on the ease of producing scorable bands, RM212 was selected for obtaining homozygosity and recovering recipient genome of BR11 in BC2F2 generation. The best plant 311-391 had 100% recipient alleles for all the chromosomes without considering foreground markers except chromosome 1 and 8.
In the BC2F2 generation, background selection was carried out over 305 segregants that were homozygous for the SUB1 QTL using the 4 markers that were heterozygous in the BC2F1 for the population P311-391. Ten plants possessed homozygous recipient alleles for all the four background markers. This seemed unusually high, because out of 1221 BC2F2 plants originally genotyped for SUB1, only one plant would be expected to be homozygous for four background and the target locus markers. The phenotypes of the selected plants were carefully observed and some variation was noticed for flowering and tillering pattern, grain length, grain width and grain thickness from visual observation. An additional 43 markers over 10 selected BC2F2 segregants showed that all background markers had fixed recipient alleles of BR11 except three markers: RM486 on chromosome 1, RM249 on chromosome 5 and RM264 on chromosome 8 (Fig. 2). RM486 showed heterozygous alleles for the plants 391-202 and 391-472. RM249 showed heterozygous alleles for the plants 391-25, 391-370, 391-500, 391-865 and 391-925 and homozygous alleles of the donor parent for the plants 391-217 and 391-1192. RM264 showed heterozygous alleles for the plants 391-25 and 391-865. Only, the plant number 391-148 had all the alleles fixed for BR11 for all the initial and additional background markers. The genotypic constitution of plant number 391-148 was unique out of 1221 BC2F2 plants and this plant was considered as BR11-Sub1 (Fig. 2, referred to as the fixed line IR 05F290 or ‘IR85260-391-148’ in the IRRI pedigree designation). The other nine BC2F2 plants were also selected as precision introgression lines (PILs) (Collard et al. 2008a) as the lines were different from BR11 for only one or two markers.
The 10 selected BC2F2 PILs were tested with the GnS2, the gene-based CAPS marker specific to SUB1A. The tested 10 PILs showed the same banding pattern of resistant allele of the donor of SUB1 IR40931-33-1-3-2. Hence, it was possible to confirm that all the selected plants contained SUB1.
Introgression size and position of the SUB1 QTL
The introgression size of the SUB1 QTL was measured by using seven very closely spaced primers located around the SUB1 QTL of chromosome 9 in the SUB1 lines including BR11-Sub1. The seven very closely spaced primers were RM23835, RM23852, RM23865, RM23869, RM23911, RM23916 and RM23917 and their positions were 5.5, 5.9, 6.2, 6.3, 7.1, 7.2 and 7.3 Mb. All the markers had recipient alleles of BR11 in all the lines, which indicated that the maximum size of the SUB1 introgression was 0.8 Mb (800 Kb). The result also indicated that the position of the SUB1 gene was below 6.3 Mb in BR11-Sub1.
Submergence screening
The PILs including BR11-Sub1 showed submergence tolerance just after de-submergence and after recovery against submergence stress of 21 days. The submergence tolerance scores of these lines were similar to those of the tolerant donor IR40931-33-1-3-2 and tolerant check FR13A (Table 3). The result confirmed the introgression of SUB1 QTL in these lines. On the other hand, the control BR11 was susceptible to submergence stress like the susceptible check IR42. It was observed that BR11 and IR42 did not recover and showed poor survival, but all the PILs including resistant checks showed very good recovery and survival capability.
All the tested Sub1 breeding lines including BR11-Sub1 performed well regarding the survival ability (90–100%) which was similar to tolerant check FR13A and tolerant parent IR40931-33-1-3-2 during the RLR season of 2008 in Bangladesh (Table 4). The results confirmed the presence of SUB1 in the tested lines. On the other hand, two mega varieties of the RLR ecosystem, BR11 and Swarna, were susceptible to submergence stress like the susceptible checks IR42 and BR5. Among the other submergence tolerant Sub1 mega varieties, Swarna-Sub1 and IR64-Sub1 performed well but Samba Mahsuri-Sub1 did not perform well. However, in addition to BR11-Sub1, the three other PILs IR85260-391-217, IR85260-391-25-Gaz 2, and IR85260-391-202-Gaz 1 also showed very good to excellent recovery against the submergence stress of 2 weeks during RLR season, 2008 at BRRI, Gazipur.
Phenotyping of precision introgression lines
Out of 10 PILs, six were selected based on their visual acceptability during seed increase in the RLR season 2007-08 conducted at BRRI farm, Gazipur, Bangladesh (data not shown). Phenotyping of six submergence tolerant PILs was done to compare yield and yield-contributing characters, grain physical parameters and chemical properties of the submergence tolerant lines with the recurrent parent BR11. All the characters showed significant variation among the eight genotypes except filled grains/panicle. However, there were no significant differences between BR11 and BR11-Sub1 for yield and all the yield-contributing characters (Table 5). Based on kernel length (5.51–6.6 mm) and the ratio of kernel length and kernel width (2.1–3.0), the brown rice shape of BR11 and all the PILs viz. BR11-Sub1, IR85260-391-202, IR85260-391-217, IR85260-391-370, IR85260-391-500 was classified as medium (both length and shape) (IRRI 2002) (Table 6). The grain chemical properties of BR11-Sub1 compared to BR11 showed no change also (Table 7).
Discussion
The present study clearly demonstrated the rapid and precise enhancement of the mega variety BR11, which is grown on around 40% RLR areas in Bangladesh, for submergence tolerance by only two backcrosses and one selfing generation. We called the enhanced mega-variety ‘BR11-Sub1’. The specific aim of reducing the size of a donor segment using closely spaced flanking markers (i.e. recombinant selection) was achieved, limiting the size of the donor introgression to less than 1 Mb in the BC2F2 generation. Considering the size of the rice genome in Nipponbare as 389 Mb, BR11-Sub1 was 99.8% similar to BR11. The conversion of Swarna-Sub1 resulted in an introgression size of 2.3–3.4 Mb in the BC3F2 and 6.5 Mb in the BC2F2 (Neeraja et al. 2007). In the present study, production of a large number of BCnF1 seeds facilitated limiting the introgression size (Collard and Mackill 2008).
Takeuchi et al. (2006) reported the introgression size of the heading date QTLs in three isogenic lines ranged from 170 to 625 Kb in the background of the recurrent parent which was possible due to performing recombinant selection using very closely spaced flanking markers. The introgression size of the waxy locus controlling cooking and eating quality from Minghui 63, a restorer line into Zhenshan97 through MAS was less than 6.1 cM in length (Zhou et al. 2003). Chen et al. (2000) reported that the individual selected in BC3F2 generation carried a fragment of less than 3.8 cM from the donor line in the Xa21 region on chromosome 11 and about 98.8% of the genetic background of the recurrent parent was recovered.
NILs produced using conventional backcrossing may not be genetically “clean” and possess many donor introgressions on non-carrier and carrier chromosomes and large donor chromosomal segments on the carrier chromosome. The extent of donor segments within NILs remains unknown. As BR11-Sub1 and other BR11-Sub1 lines developed through MABC possessed minimal donor segments flanking SUB1 and precise regions of donor segments in other genomic positions was known, these lines were termed as Precision Introgression Lines (PILs) as proposed by Collard et al. (2008a).
There appeared to be some differences among the tested BR11-derived Sub1 lines due to variable amount of introgression from the donor parent. The excellent consistent performance of the PIL IR85260-391-217 for submergence tolerance might be due to fixed donor allele of RM249 on chromosome 5 which occurred by chance. This entry showed quick recovery in all the experiments. Otherwise, there was little morphological variation among the PILs, which supported the genotyping data, because they were different from one another for only one SSR locus. The similar grain chemical properties of BR11 and BR11-Sub1 also indicated successful recovery of the BR11 genome in BR11-Sub1.
The target of recombinant selection in BC1F1 generation was to obtain single recombinants at one side of the SUB1 QTL. These were segregants in which the size of the donor segment was minimized at one side of the QTL. Four flanking markers from the SUB1 region of chromosome 9 were used in BC1F1 generation. For the development of Swarna-Sub1, RM316 (1.8 cM) was used as flanking marker at proximal end and RM219 (11.7 cM) at distal end (Neeraja et al. 2007). According to Hospital (2001) flanking markers for recombinant selection should be chosen as closely linked as possible to the introgressed gene to minimize linkage drag. He suggested computing the minimal population sizes required to obtain double recombinants for such closely linked markers and to optimize the population size in the context of a multi-generation backcross program which was first proposed by Young and Tanksley (1989). Tighter flanking markers were used for the development of BR11-Sub1 compared to Swarna-Sub1. The distance between the two closest flanking markers (RM23805 and RM23915) at both side of the QTL was 7.9 cM. Interestingly, Neeraja et al. (2007) also found fewer recombinants at the proximal end of SUB1 QTL in the BC2F1 generation. Hospital (2001) indicated that marker data points for recombinant selection could be reduced by using additional backcross generations. This approach was not adopted here because the main objective of the present study was to reduce the timeframe of the backcrossing scheme.
Foreground selection was confirmed by use of markers from the SUB1 genes in all the plants. Even when a marker is tightly linked with the target QTL, there may be a crossing over between them, which might produce false-positive results in foreground selection. As the number of selected plants was reduced considerably after recombinant selection, this was also economical to use these CAPS gene-based markers after recombinant selection that are more costly to use. The importance of gene-based marker in confirming the presence of the target gene was demonstrated.
Hospital et al. (1992) and Visscher et al. (1996) reported that, as a general rule, two to four markers per 100 cM could be efficiently used to accelerate the recovery of the recurrent parent genome in the early generations such as BC1F1 or BC2F1. Takeuchi et al. (2006) used 116 RFLP markers covering 12 chromosomes in background selection in rice. Neeraja et al. (2007) used 56 SSR markers as initial background markers for the development of Swarna-Sub1. Background selection was confined to the individuals which had the target gene as well as which minimized the size of the donor segment containing the QTL in the BC1F1 generation. However, these types of segregants were very low in number, which limited performing background selection with respect to recovery of the recurrent parent genome. This highlights the need to increase the population size in order to recover the recurrent parent genetic background by only two backcrosses in the BC2F2 generation. If the number of background markers remaining in the best plant of BC2F1 generation is more than four, another backcrossing is usually needed to recover the recurrent parent genome by BC3F2 generation using around 1000 selfed progenies, although an additional selfing generation can also be used.
As the average distance of additional background markers with the closest initial background markers was 5 cM, the use of these additional markers turned out to be unnecessary. It is also known that the crossing-over interference is generally considered within at least 15 cM and the number of crossovers per chromosome per meiosis rarely exceeds four (Kearsey and Pooni 1996). However, it was possible to find genetic differences among the 10 PILs using 43 additional background markers. Out of those 43 markers, 3 were segregating among the finally selected 10 BC2F2 plants. Some phenotypic variations were also observed among those plants with respect to flowering, tillering and importantly for grain length, breadth and thickness. The additional background markers were thus found to be important in detecting very small introgressions from the donor parent in the BC2F2 generation. Since recombination events accumulate over time, the number of donor chromosome segments spread throughout the genome increases as their length decreases. Hence, more markers are required to detect them at more advanced backcross generations (e.g. BC3) (Collard et al. 2008b; Hospital et al. 1997). Neeraja et al. (2007) used 32 additional background markers in the finally selected BC2F2 and BC3F2 version of Swarna-Sub1 for ensuring the proper recovery of recurrent parent genome. However using additional background markers for identifying Swarna-Sub1 was unnecessary because no additional donor segments were identified.
In conclusion, the MABC approach described in the present study was successfully adopted to introgress SUB1 into BR11 and could be successfully utilized to introgress SUB1 in the other important RLR varieties with a minimum introgression segment and within a short time frame. By limiting the size of the introgression, the chance of introducing donor genes that might change the essential characteristics of this popular variety were reduced. This will become increasingly important as other desirable genes are introduced into BR11-Sub1. For the purpose of introducing SUB1 into other RLR varieties, BR11-Sub1 or other PILs generated in this study could be used instead of IR40931, because BR11 has considerably better agronomic and quality characteristics. Population sizes as large as the one in this study may not be required because the donor is an adapted, high-yielding variety, and introgression of deleterious alleles would not be so likely. It is expected that the newly developed Sub1 lines will be able to increase rice production in the submergence prone areas of Asia.
References
BRRI (2007) BRRI annual internal review 2004–2005. Agricultural Economics Division, Bangladesh Rice Research Institute, Gazipur-1701, Bangladesh
Chen S, Lin XH, Xu CG, Zhang QF (2000) Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci 40:239–244
Chen MS, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang FC, Kim H, Frisch D, Yu YS, Sun SH, Higingbottom S, Phimphilai J, Phimphilai D, Thurmond S, Gaudette B, Li P, Liu JD, Hatfield J, Main D, Farrar K, Henderson C, Barnett L, Costa R, Williams B, Walser S, Atkins M, Hall C, Budiman MA, Tomkins JP, Luo MZ, Bancroft I, Salse J, Regad F, Mohapatra T, Singh NK, Tyagi AK, Soderlund C, Dean RA, Wing RA (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14:537–545
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the 21st century. Phil Trans Royal Soc B Rev 363:557–572
Collard BCY, Cruz CMV, McNally KL, Virk PS, Mackill DJ (2008a) Rice molecular breeding laboratories in the genomics era: current status and future considerations. Int J Plant Genomics 2008: Article ID 524847, 25 pp
Collard BCY, Iftekharuddaula KM, Thomson MJ, Pamplona AM, Mackill DJ (2008b) An electronic manual on marker assisted backcrossing in rice: theory and applications, 1st edn. GCP Wiki, International Rice Research Institute, Manila, Philippines. http://mcclintock.generationcp.org/index.php?option=com_content&task=view&id=92&Itemid=114
HilleRisLambers D, Vergara BS (1982) Summary results of an international collaboration on screening methods for flood tolerance. In: Proceedings of the 1981 international deepwater rice workshop. Int. Rice Res. Inst., Los Baños, Philippines, pp 347–353
Hospital F (2001) Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics 158:1363–1379
Hospital F, Charcosset A (1997) Marker-assisted introgression of quantitative trait loci. Genetics 147:1469–1485
Hospital F, Chevalet C, Mulsant P (1992) Using markers in gene introgression breeding programs. Genetics 132:1199–1210
Hospital F, Moreau L, Lacoudre F, Charcosset A, Gallais A (1997) More on the efficiency of marker-assisted selection. Theor Appl Genet 95:1181–1189
IRGSP (2005) The map-based sequence of the rice genome. Nature 436:793–800
IRRI (2002) Standard evaluation system for rice. Int. Rice Res. Inst, Los Baños, Philippines
Kearsey MJ, Pooni HS (1996) The genetical analysis of quantitative traits. Stanley Thornes (Publishers) Ltd., Wiltshire
Mackill DJ (1986) Varietal improvement for rainfed lowland rice in South and Southeast Asia: results of a survey. Progress in rainfed lowland rice. Int. Rice Res. Inst, Los Baños, Philippines, pp 115–144
Mackill DJ (2006) Breeding for resistance to abiotic stresses in rice: the value of quantitative trait loci. In: Lamkey KR, Lee M (eds) Plant breeding: The Arnel R Hallauer international symposium. Blackwell Publishes, Ames, IA, pp 201–212
Mackill DJ, Amante MM, Vergara BS, Sarkarung S (1993) Improved semidwarf rice lines with tolerance to submergence of seedlings. Crop Sci 33:749–753
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Zing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Mohanty HK, Chaudhary RC (1986) Breeding for submergence tolerance in rice in India. Progress in rainfed lowland rice. Int. Rice Res. Inst, Los Baños, Philippines, pp 191–200
Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ (2009) Development of submergence tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot 103:151–160
Takeuchi Y, Ebitani T, Yamamoto T, Sato H, Ohta H, Hirabayashi H, Kato H, Ando I, Nemoto H, Imbe T, Yano M (2006) Development of isogenic lines of rice cultivar Koshihikari with early and late heading by marker-assisted selection. Breed Sci 56:405–413
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Van Berloo R (2008) GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered 99:232–236
Visscher PM, Haley CS, Thompson R (1996) Marker-assisted introgression in backcross breeding programs. Genetics 144:1923–1932
Xu K, Mackill DJ (1996) A major locus for submergence tolerance mapped on rice chromosome 9. Mol Breed 2:219–224
Xu K, Xu X, Ronald PC, Mackill DJ (2000) A high-resolution linkage map in the vicinity of the rice submergence tolerance locus Sub1. Mol Gen Genet 263:681–689
Xu KN, Deb R, Mackill DJ (2004) A microsatellite marker and a codominant PCR-based marker for marker-assisted selection of submergence tolerance in rice. Crop Sci 44:248–253
Xu K, Xia X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene response factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Young ND, Tanksley SD (1989) RFLP analysis of the size of chromosomal segments retained around the tm-2 locus of tomato during backcross breeding. Theor Appl Genet 77:353–359
Zheng K, Subudhi PK, Domingo J, Magpantay G, Huang N (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newsl 12:255–258
Zhou PH, Tan YF, He YQ, Xu CG, Zhang Q (2003) Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor Appl Genet 106:326–331
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iftekharuddaula, K.M., Newaz, M.A., Salam, M.A. et al. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178, 83–97 (2011). https://doi.org/10.1007/s10681-010-0272-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0272-2