Abstract
Thirty-eight Indian potato varieties and fifty-two advanced hybrid lines were analyzed for cytoplasm types using both plastid and mitochondrial genome specific markers. Indian genotypes thus analysed could be broadly grouped into 4 cytoplasm types i.e. T/β (69), W/α (18), W/γ (1) and A/ɛ (2). The T/β type cytoplasm, typical of common cultivated potato (ssp. tuberosum) was absent in six released varieties (Kufri Chipsona-1/-2/-3 series, Kufri Jawahar, Kufri Megha and Kufri Himalini) and fifteen out of fifty two hybrids analyzed. This information was further used to predict cytoplasm type on the basis of common shared maternal pedigree in thirty-eight other advanced hybrids, which revealed majority (25) had T/β type cytoplasm with W/α and A/ɛ cytoplasm observed in 12 and 1 genotype, respectively. T/β type cytoplasm was observed in all 28 early bulking hybrids studied along with all old genotypes. It was revealed that considerable broadening of maternal base was observed in recently developed genotypes. W/α type cytoplasm was present in most of processing (all 3 chipping varieties and 9 of 12 MP hybrids) and late blight resistant (11of 23 hybrids) genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potatoes (Solanum tuberosum L. ssp. andigena) originated in the Andean mountains of South America and are adapted to tubering in short days. In contrast, Chilean potatoes (S. tuberosum ssp. tuberosum) are adapted to tubering in long days. The potato is a highly heterozygous, autotetraploid crop that suffers inbreeding depression on selfing (Mendoza and Haynes 1974). There are no homozygous breeding lines in potato for exploiting heterotic vigour. The exploitation of a few South American clones, which were successfully introduced to Europe and USA during the 17th century, and their use in most of the potato breeding programmes worldwide resulted in a narrow genetic base of cultivated potatoes. This was further aggrevated by cytoplasmic-genetic male sterility, which restricts use of many genotypes as male parents in breeding. As a consequence, substantial gains in the yield potential of newly developed varieties have not been obtained. All these factors call for broadening of genetic base of cultivated potato.
S. tuberosum ssp. andigena has wide diversity and, therefore, has been extensively used in neo-tuberosum programmes (Glendinning 1983) and in effecting tuberosum × andigena crosses, which results in enhanced heterosis and vigour. In tuberosum × andigena crosses, yield gains are accompanied by reduced tuber size and increased male sterility (Maris 1989), while reciprocal cross is male fertile indicating that cytoplasm plays an important role in potato breeding (Grun 1990). Wild species have also been successfully used for introgression of many genes bearing desirable traits in potato. However, their use is limited due to lengthy prebreeding that often involves co-segregation of linked undesirable traits.
Considering the importance of organelle genomes (plastid and mitochondrial genomes) in potato breeding, their analysis is of immense use in studying introgression of agronomic traits. Typically, organelle genomes are haploid, non-recombinant and are usually maternally inherited. The very low mutation rates of organelle genomes as compared to the nuclear genome make them a powerful tool for such studies. Most cultivated Chilean potatoes (ssp. tuberosum) have T-type plastid DNA and β type mitochondrial genome, which is lacking in Andean potato ssp. andigena as well as wild Solanum species. Five basic cpDNA types (T/C/W/S/A) have been identified in cultivated landraces of potato (Hosaka and Hanneman 1988). These organelle genome types are not species specific but their frequencies vary in different species, with frequencies varying with latitude in the Andes within the same species. T cp-type is linked to β mt-type (thereby defined as tuberosum mt genome), while mt-types α, γ and δ were found in combination with W cp-type in wild species; and mt-type ɛ was found in A- and S-type cultivated species (Lossl et al. 1999).
In a crop like potato, which is highly heterozygous and auto-tetraploid, organelle studies are expected to provide a better picture of available diversity. This study was therefore undertaken to analyse divergence revealed by plastid as well as mitochondrial genome type in thirty-eight Indian varieties and fifty-two advanced hybrids. The present study included three markers (H1, H2, H3) based on chloroplast genome deletion (Hosaka 2003) and four cpSSR markers (Provan et al. 1999), to investigate plastid genome type. Mitochondrial genome type was investigated using PCR analysis (ALM1 + 3, ALM4 + 5 and ALM6 + 7) specific for mitochondrial genome types (Lossl et al. 1999; 2000).
Materials and methods
Plant material and isolation of DNA
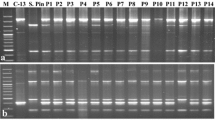
The present study included thirty-eight Indian varieties and fifty-two advanced hybrids (Table 1a, b). For DNA extraction, leaves were collected from 45 day old single plants grown in a glasshouse in the long summer days of Shimla (Latitude: 31.60 N, Longitude: 77.13 E, Altitude: 2202 msl). A modified CTAB method (Doyle and Doyle 1987) was used to isolate genomic DNA from 2 g of fresh leaves. The amount of DNA was quantified by spectrophotometer and quality was checked both by A260/280 ratio and by gel-electrophoresis.
Chloroplast and mitochondrial DNA markers
A total of ten PCR based markers consisting of three chloroplast deletions (H1, H2 and H3) (Hosaka 2003); four cpSSR (NTCP6, NTCP 8, NTCP 9 and NTCP 14) (Provan et al. 1999) and three mitochondrial PCR markers (ALM1 + ALM3; ALM4 + ALM5 and ALM6 + ALM7) (Lossl et al. 1999; 2000) were used in the present study (Table 2). H1 marker is known to amplify a 446 bp fragment in wild/andigena potato plastid genome region; however, ssp. tuberosum is known to amplify fragment of 205 bp size, thereby indicating a 241 bp deletion. These were evaluated by 1.6% agarose gel electrophoresis stained with ethidium bromide.
Similarly, in H2 and H3 markers, PCR amplification products were ethanol precipitated before digestion with HaeIII and DraI respectively. For mitochondrial genome analysis, three mitochondrial DNA specific markers mentioned above were used for PCR amplification and evaluated by 1.2% agarose gel electrophoresis.
Four cpSSR markers were analysed by semi-automated capillary-based electrophoresis. Two cpSSR forward primers, viz., NTCP6 and NTCP8 were labelled with 5′ FAM and the other two forward primers, viz., NTCP9 and NTCP14 with JOE. PCR reactions and GeneScan capillary electrophoresis based genotyping was done on the ABI Prism310 Genetic analyzer as described in our earlier studies (Chimote et al. 2004). The runs were performed at 60°C for 24 min at 15 kV with 5 s injection time. All genotypes were analyzed twice to check reproducibility of results.
Prediction analysis
Information based on organelle genome analysis in the above study and common matrilineal pedigree was used to predict cytoplasm type in 38 other advanced hybrid lines, as shown in Table 1c.
Results
Chloroplast deletion markers
Most of the varieties and advanced hybrids have T cp-type chloroplast DNA, similar to Chilean cultivated potato as revealed by H1 marker. T cp-type 241 bp chloroplast deletion typical of tuberosum type was found in 32 varieties and 37 advanced hybrids analyzed. T cp-type deletion was observed in all 17 early bulking hybrids (J series), 3 hybrids for East Indian plains (P series) and two heat tolerant hybrids (HT series). However, this deletion was absent in 6 varieties (Kufri Chipsona-1, Kufri Chipsona-2, Kufri Chipsona-3, Kufri Jawahar, Kufri Megha and Kufri Himalini), 5 out of 8 processing hybrids (MP series), 7 out of 14 late blight resistant hill hybrids (SM/HB series), 3 medium maturing hybrids for North Indian plains (MS series) and andigena clone EX/A 680-16. Exotic processing cultivar Atlantic also showed T-type cytoplasm.
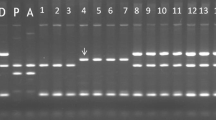
The grouping pattern of varieties and hybrids was the same with the H2 markers (Table 3). All genotypes showing T cp-type yielded two fragments of 193 and 141 bp on HaeIII digestion with H2 marker, while in non-T cp-type genotypes only an undigested 334 bp fragment was observed. In case of H3 marker, all genotypes yielded 1018 bp fragment except two advanced hybrids (MS/97-621 and EX/A680-16), which gave two fragments of 575 and 443 bp, with rejoined fragment being also observed in them.
Chloroplast microsatellite markers
Four cpSSR markers (NTCP6, NTCP8, NTCP9 and NTCP14) together amplified a total of 6 fragments, with NTCP6 and NTCP9 giving only a single fragment each. These studies classified them into three clear groupings with first major group comprising of T cp-type genotypes, second group comprising genotypes with W cp-type and last group comprising of MS/97-621 and EX/A680-16. The differences in their peak patterns are given in Table 3. Three possible allelic combinations were observed at each loci except NTCP6 (150/151), thereby clustering all genotypes into 3 groups (T, W and A cp-type).
Mitochondrial typing studies
ALM1 + ALM3 primer combination amplified atp6 region resulting in 1.2 kb band specific to α + γ mt-types in W cp-type (Table 4). ALM4 + ALM5 primer combination yielded 2.4 kb fragment specific to α type in the same genotypes with a sole exception of SM/94-43. SM/94-43 amplified 2.4 kb fragment of cob gene with ALM6 + ALM7 primer combination specific to γ mt-type.
Mitochondrial DNA specific ALM4 + ALM5 marker yielded 1.6 kb PCR product in all genotypes with T and A plastid genomes types. There are reports that β mitochondrial type coincides with T-type chloroplast (Rasmussen et al. 2000; Lossl et al. 2000). Based on these reports, all genotypes having T-type plastid genome and yielding 1.6 kb PCR product with ALM4 + ALM5 combinations were assigned β mt-type. These include 32 Indian varieties, 37 advanced hybrids and cv. Atlantic. Similarly, both A-cp-type genotypes, EX/A680-16 and MS/97-621, also yielded 1.6 kb PCR product with ALM4 + ALM5 and were assigned ɛ mt-type i.e. typical of ssp. andigena (Table 4). On this basis, we concluded that out of 90 varieties and hybrids analyzed, 69 are of β type, 17 of α type, 1of γ type and 2 of ɛ type.
Prediction analysis
Matrilineal pedigree similarity based prediction of organelle genome types using above information was used to conclude that 25 out of 38 advanced hybrid lines are T/β type; 12 are of W/α type and one is of A/ɛ type (Table 1c). All 10 early bulking hybrids (J/Jx series), 6 hybrids for East Indian plains (-P- series) and four medium maturing hybrids (MS series) showed tuberosum type cytoplasm. However, T-type deletion was absent in all 4 processing hybrids (MP series), 4 late blight resistant hybrids (SM/HB series) and three other hybrids (OP1, PS/M75, PS/M78).
Discussion
Organelle genome typing in Indian potato varieties/advanced hybrids revealed that divergence was observed only in a few recently released genotypes developed during the last 15 years primarily for specific breeding purposes like processing quality and late blight resistance, which involved wild gene introgression in their maternal pedigree.
Amplification/restriction pattern with H1, H2, H3, NTCP6 and NTCP9 matched with earlier reports (Bryan et al. 1999; Powell et al. 1999; Hosaka 2003; Sukhotu et al. 2004). However, in all the three haplotypes (T/W/A), fragments sizes were 1 bp larger with NTCP8 and 3 bp smaller with NTCP14 than reported (Hosaka, 2003; Sukhotu et al. 2004). This may be due to different electrophoresis techniques used for resolution.
In the present study the majority (73.4%) of genotypes (94 out of 128) had T-type chloroplast and β type mitochondrial genome. Present day potato cultivars have predominantly T-type organelle DNA typical of Chilean germplasm (Powell et al. 1993; Hosaka 1995; Provan et al. 1999). The predominance of T-type cytoplasm in most potato cultivars can be traced back to the 19th century Chilean introduction, Rough Purple Chili and its derived progenies i.e. Garnet Chili and Early Rose. These genotypes exhibit cytoplasmic male sterility so that they were used as female parents in a large proportion of crosses in North American and European potato breeding (Glendinning 1983).
Analysis of Indian potato varieties revealed 32 out of 38 had T-type deletion, with the only exceptions being six recently released varieties (Kufri Chipsona-1, Kufri Chipsona-2 Kufri Chipsona-3, Kufri Jawahar, Kufri Megha and Kufri Himalini). This is contrary to early reports that the Indian varieties are more like andigena type than that of tuberosum type (Swaminathan 1958; Sinha and Pushkarnath 1964). T-type marker typical of ssp. tuberosum was present in almost all improved Indian varieties, except in Kufri Jawahar and Kufri Megha and three landraces i.e., Phulwa, Gulabia, Lalmutti (Spooner et al. 2005). On nuclear SSR analysis, all Indian cultivars and landraces clustered along with Chilean landraces (ssp. tuberosum) rather than with Andean andigena accessions as expected, suggesting that Indian cultivars are not true andigena but are of neo-tuberosum type. Provan et al. (1999) reported that the extreme cytoplasm bottleneck in most European cultivars was not reflected in nuclear divergence analysis, thereby pointing towards their wide paternal base. Most of the modern Japanese cultivars had T-type cytoplasm; with a few W-type and old cultivars of A-type (Hosaka 1993). Mitochondrial genome type analysis revealed that out of 144 German potato varieties characterized, 79, 46 and 19 varieties respectively had β, α and γ mitochondrial genome type (Lossl et al. 2000).
In the present study, 34 non T-type genotypes formed 2 groups, i.e. larger W- type (31) and smaller A-type (3). EX/A680-16, MS97-621 and predicted OP1 have A/ɛ cp/mt DNA type typical of ssp. andigena type. The frequency of A-type cytoplasm was very low (2.34%) as expected because of the negative role played by andigena cytoplasmic factors (Maris 1989). These three genotypes showed a pattern typical of A-cp-type with NTCP6 (174 bp), NTCP14 (148 bp) and NTCP8 (218 bp + 251 bp fragment). Further, they also yielded 575 and 454 bp fragments with H3 marker and 289 bp fragments with NTCP9. H3 (575 and 454 bp) and NTCP9 (289 bp) markers were perfectly correlated and are observed in almost all cultivated species (except ssp. tuberosum and S. juzepczukii) of S-, A- and most C-cp types (Sukhotu et al. 2006).
None of the genotypes studied showed 127 bp band typical of S-cp-type indicating its total absence in matrilineal pedigree of Indian potato breeding. Only a few Indian varieties and hybrids are known to have either S. phureja or S. stenotomum in their pedigree (e.g. POOS16 (in MP/97-921), I1062 (in Kufri Himalini) and MEX32, etc). Kufri Jyoti’s maternal grandparent GM 2182ef7 traces back to S. phureja × S. demissum on male side. S. phureja shows cytoplasmic barrier with reciprocal differences on crossing with ssp. tuberosum (Amoah et al. 1988; Grun et al. 1977).
W/α cytoplasm type was observed in all three chipping varieties (Kufri Chipsona-1, Kufri Chipsona-2, Kufri Chipsona-3) and nine out of twelve hybrids bred specifically for processing purpose. Late blight resistant variety Kufri Himalini and hybrid SM/92-168 with good processing qualities also have W/α cytoplasm type. Interestingly most of them have Central/South American accessions like MEX-750826, F6, Muziranazara, POO16, I1062 and VB/A 64 in their matrilineal pedigree, involving wild species with good processing attributes in pedigree. There is higher variability in wild potato species than in cultivated potatoes for characters like dry matter, starch content, amylose content, mean diameter of starch granules, etc (Jansen et al. 2001). W/α genotypes identified could help in narrowing down selection for introgression of processing quality trait. However, high dry matter and processing attributes are also present in genotypes with T/β type cytoplasm i.e. in Chipping cultivar Atlantic, three processing hybrids (i.e. MP/98-31, MP/99-322 and MP/99-406) and two heat tolerant hybrids (HT-series hybrids and B-420). Atlantic has cv. Lenape cytoplasm with S. chacoense in paternal pedigree.
In case of late blight resistance breeding programme, two recent varieties (Kufri Megha and Kufri Himalini) and almost half of SM/HB-hybrids (11 of 23) developed for North Indian hills (where the duration of crop is long and late blight infection is epidemic) had W/α type cytoplasm. Wild species especially S. demissum, play an important role in incorporation of late blight resistance in potato breeding. However, most of the earlier Indian varieties (SLB series) with R gene derived late blight resistance had T/β cytoplasm as they are derived from resistant breeding material of Scottish Plant Breeding Station with S. demissum in paternal pedigree.
All of the early bulking hybrids studied had T/β type cytoplasm typical of tuberosum ssp. This can be explained by the fact that any potato breeding involving wild species/andigena results in delayed tuberization and thereby a late maturation crop which defeats the basic purpose of this breeding programme. In potato breeding programme for regular maturation crop in Indo-Gangetic plains, with yield being main objective, 3 out of 7 recent hybrids lack tuberosum typical T-type cytoplasm.
Our study highlights the importance of wild gene introgression in potato breeding for specific purposes like late blight resistance and processing. Current diversified uses of potato in processing, demands introgression of wild gene pool for improving quality characters such as cold chipping, high dry matter and starch content of tubers. Desirable genes for many quality traits can be found in Andean potato genotypes, whereas for abiotic and biotic stresses, breeding could well benefit from use of wild species.
Abbreviations
- cp-type:
-
Plastid genome type
- T:
-
tuberosum type
- A:
-
andigena type
- GM:
-
Maternal grandmother
- cpDNA:
-
Plastid DNA
- mt-type:
-
Mitochondrial genome type
- W:
-
Wild type
- cpSSR:
-
Chloroplast simple sequence repeats
- GGM:
-
Matrilineal great grandmother
- mtDNA:
-
Mitochondrial DNA
References
Amoah V, Grun P, Hill JR (1988) Cytoplasmic substitution in Solanum I. Tuber characteristics of reciprocal backross progeny. Potato Res 31:121–127
Bryan GJ, McNicoll J, Meyer RC, Ramsay G, De Jong WS (1999) Polymorphic simple sequence repeat markers in chloroplast genomes of Solaneceaous plants. Theor Appl Genet 99:859–867
Chimote VP, Chakrabarti SK, Pattanayak D, Naik PS (2004) Semi-automated simple sequence repeat analysis reveals narrow genetic base in Indian potato cultivars. Biol Plant 48:517–522
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Glendinning DR (1983) Potato introductions and breeding up to the early 20th century. New Phytol 94:479–505
Grun P (1990) The evolution of cultivated potatoes. Econ Bot 44(suppl 3):39–55
Grun P, Ochoa C, Capage D (1977) Evolution of cytoplasmic factors in tetraploid cultivated potatoes (Solanaceae) Amer J Bot 64:412–420
Hosaka K, Hanneman RE (1988) The origin of the cultivated tetraploid potato based on chloroplast DNA. Theor Appl Genet 68:55–61
Hosaka K (1993) Similar introduction and incorporation of chloroplast DNA in Japan and Europe. Jpn J Genet 90:356–363
Hosaka K (1995) Successive domestication and evolution of Andean potatoes as revealed by chloroplast DNA restriction endonuclease analysis. Theor Appl Genet 90:356–363
Hosaka K (2003) T-type chloroplast DNA in Solanum tuberosum L. ssp. tuberosum was conferred from some populations of Solanum tarijense Hawkes. Amer J Potato Res 80:21–32
Jansen G, Flamme W, Schuller K, Vandarey M (2001) Tuber and starch quality of wild and cultivated potato species and cultivars. Potato Res 44:137–146
Lossl A, Gotz M, Braun A, Wenzel G (2000) Molecular markers for potato cytoplasm: male sterility and contribution of different plastid-mitochondrial configurations to starch production. Euphytica 116:221–230
Lossl A, Adler N, Horn R, Frei U, Wenzel G (1999) Mitochondrial genome type characterization of potato: Mt α, β, γ, δ and ɛ and novel plastid-mitochondrial configurations. Theor Appl Genet 99:1–10
Maris B (1989) Analyses of an incomplete diallel cross among three ssp tuberosum varieties and seven long-day adapted ssp. andigena clones of potato (Solanum tuberosum L.). Euphytica 41:163–182
Mendoza HA, Haynes FL (1974) Genetic relationship among potato cultivars grown in United States. HortScience 9:328–330
Powell W, Baird E, Duncan N, Waugh R (1993) Chloroplast DNA variability in old and recently introduced potato cultivars. Ann Appl Bot 123:403–410
Provan J, Powell W, Dewar H, Bryan G, Machray GC, Waugh R (1999) An extreme cytoplasmic bottleneck in the modern European potato (Solanum tuberosum) is not reflected in decreased levels of nuclear diversity. Proc R Soc Ser B 266:633–639
Rasmussen JO, Lossl A, Rasmussen OS (2000) Analysis of the plastid genome and mitochondrial genome origin in plants regenerated after asymmetric Solanum ssp. protoplast fusions. Theor Appl Genet 101:336–343
Sinha SK, Pushkarnath (1964) The relationship of Indian potato varieties to Solanum tuberosum subsp. andigena. Indian Potato J 6:24–39
Spooner DM, Nunez J, Rodriguez F, Naik PS, Ghislain M (2005) Nuclear and chloroplast DNA reassessment of the origin of Indian potato varieties and its implications for the origin of the early European potato. Theor Appl Genet 110:1020–1026
Sukhotu T, Kamijima O, Hosaka K (2004) Nuclear and chloroplast DNA differentiation in Andean potatoes. Genome 47(1):46–56
Sukhotu T, Kamijima O, Hosaka K (2006) Chloroplast DNA variation in the most primitive cultivated diploid potato species Solanum stenotomum Juz. et Buk. and its putative wild ancestral species using high-resolution markers. Genet Res Crop Evol 53(1):53–63
Swaminathan MS (1958) The origin of the early European potato- evidence from Indian potato varieties. Indian J Genet Plant Breed 18:8–15
Acknowledgment
The authors are grateful to Director, Central Potato Research Institute and Head, Crop Improvement for providing necessary facilities to undertake this study. Help rendered by Meetul Kumar, Naresh and Mr. C.M.S. Bist during this study is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chimote, V.P., Chakrabarti, S.K., Pattanayak, D. et al. Molecular analysis of cytoplasm type in Indian potato varieties. Euphytica 162, 69–80 (2008). https://doi.org/10.1007/s10681-007-9563-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9563-7