Abstract
Amplified fragment length polymorphism (AFLP) analysis is a rapid and efficient method for producing DNA fingerprints and molecular characterization. Our objectives were to: estimate genetic similarities (GS), marker indices, and polymorphic information contents (PICs) for AFLP markers in almond cultivars; assess the genetic diversity of almond cultivars and wild species, using GS estimated from AFLP fingerprints and molecular characterization; and facilitate the use of markers in inter-specific introgression and cultivar improvement. The genetic diversity of 45 almond cultivars from Iran, Europe, and America, were studied assaying 19 primer combinations. In addition, several agronomic traits were evaluated, including flowering and maturity times, self-incompatibility, and kernel and fruit properties. Out of the 813 polymerase chain reaction fragments that were scored, 781 (96.23%) were polymorphic. GS ranged from 0.5 to 0.96, marker indices ranged from 51.37 to 78.79, and PICs ranged from 0.56 to 0.86. Results allowed the unique molecular identification of all assayed genotypes. However, the correlation between genetic similarity clustering as based on AFLP and clustering for agronomic traits was low. Cluster analysis based on AFLP data clearly differentiated the genotypes and wild species according to their origin and pedigree, whereas, cluster analysis based on agronomic data differentiated according the pomological characterization. Our results showed the great genetic diversity of the almond cultivars and their interest for almond breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The almond (Prunus dulcis Miller [D.A Webb] syn P. amygdalus Batsch) is one of the oldest tree nut crops, and today represents the largest production of any commercial tree nut product. Almond species grow in regions of the world that are characterized as having a subtropical Mediterranean climate, with mild wet winters and warm, dry summers (Kester and Gradziel 1996). These species originated in central Asia and represent divergent evolution under xerophytic environments (Watkins 1976). Related Prunus species are found growing wild from eastern China to mountainous areas and deserts of Western China, Kurdistan, Turkistan, Afghanistan and Iran (Browicz and Zohary 1996; Grasselly 1976; Kester and Gradziel 1996; Martínez-Gómez et al. 2007). In the year 2004, world production of almond was approximately 1,530,271 tones, of which Iran produced 80,000 tones (FAO STAT Data sources). The major cultivars grown in Iran are ‘Mamaei’, ‘Shekofeh’ and ‘Azar’, but because these are self-incompatible (Gradziel and Kester 1998), pollinizer cultivars such as ‘Rabei’, ‘Sahand’ and self-grown seedling genotypes must be included in orchards.
As Iranian varieties have been poorly characterized thus far, molecular genetic methods offer means for identifying and characterizing the relationships between almond cultivars. Similar studies have been performed on non-Iranian cultivars and wild species from Asian and Mediterranean countries, and Australian and North America states, using several types of markers including, isozymes (Arulsekar et al. 1986; Hauagge et al. 1987; Cerezo et al. 1989; Viruel et al. 1995); RFLPs (Viruel et al. 1995), RAPDs (Resta et al. 1998; Bartolozzi et al. 1998; Mir Ali and Nabulsi 2003), SSRs (Cipriani et al. 1999; Martínez-Gómez et al. 2003a; 2003b; Mnejja et al. 2004; Xu et al. 2004; Xie et al. 2006); ISSRs (Martins et al. 2003) and AFLPs (Martins et al. 2001). These studies find high levels of variation and heterozygosity. They have been used to improve the use of different genotypes in breeding programs and in the design of new crosses. Presently, the use of PCR-based molecular markers such as Amplified Fragment length polymorphisms (AFLPs) has created the opportunity for broad genetic characterization of germplasm collections. AFLPs have emerged as a powerful tool for DNA fingerprinting and molecular characterization and genetic mapping, and very high repeatability has been indicated in a wide range of plants, because of their high polymorphism and abundance (Aranzana et al. 2003; Vos et al. 1995; Zabeau 1993).
On the other hand, we also addressed breeding programs that were initiated at the Agriculture and Natural Research Center of Iran, in order to associate the high quality of Iranian cultivars with the late flowering, self-fertility and sweet kernel characters of foreign cultivars. We have identified Iranian cultivars with good production and high quality such ‘Mamaei’, ‘Sefied’, ‘Shekofeh’ and ‘Azar’. While studies of agronomic and morphological characters have previously been conducted, these studies were unable to determine genetic relationships, cultivar unique identifications, nor characterizations among varieties. For example, historically some known foreign cultivars were introduced to Iran, but their original names were lost and then they were mislabeled as ‘Shahrodi’ cultivars followed by an Arabic number. No comprehensive molecular information is available regarding almond cultivars grown in Iran. Only Vezvaei (2003) has reported the isozyme diversity of Asian almond cultivated in Iran. However, the utility of isozyme analyses has been limited, since levels of polymorphism were insufficient for genetic characterization and relatedness among closely related varieties. Thus, the present study aimed to determine the genetic characterization, diversity and relatedness among Iranian and some important foreign cultivars using AFLP molecular markers.
The specific objectives of this study were to: estimate genetic similarities (GS), polymorphism rates, and polymorphic information contents (PICs) for AFLP markers among related species and cultivated genotypes of almond; assess the genetic diversity of almond cultivars using genetic similarity of almond cultivars estimated from AFLP fingerprints and molecular characterization; and facilitate their use in inter-specific introgression and cultivar improvement.
Materials and methods
Plant material
A total of 45 genotypes including 36 cultivated genotypes and nine wild species were assayed in this study (Table 1). They included eight almond cultivars from Iran, 11 from unknown origin, five from Italy, one from Spain, three from France, one from Russia, seven from USA, and one accession of P. communis, P. orientalis, P. scoparia, P. glucea, P. bucharica, P. kuramica, P. webbii, P. tenella, and P. koteschyi. Three introduced ‘Shahrodi’ cultivars with unknown origin were also investigated. Eight interspecific almond × peach cultivars issued from several backcrosses with almond cultivars and open pollinated parent unknown. These genotypes were obtained from Agriculture and Natural Resources Research Center of Shahrekord, Iran.
Agronomic charactersitics
For two consecutive years (2004, 2005) the following characteristics were recorded in each individual using the IPGRI descriptor for Rosaceae family:
-
Flowering intensity (from 0 = null, to 5 = greatest).
-
Productivity (from 0 = null to 5 = greatest)
-
Time of maturation (Julian date).
-
Time of full bloom (Julian date).
-
Shell hardness (from 1 = very soft, to 5 = very hard).
-
Weight in shell (g).
-
Kernel weight (g).
-
Percentage of kernel (%).
-
Empty nuts (%).
-
Double kernels (%).
-
Kernels with defects (%).
-
Kernel thickness (%).
-
Kernel shrivelling (from 1 = smooth, to 3 = wrinkled)
-
Kernel flavor (from 1 = bitter, to 3 = sweet).
Total genomic DNA extraction
Total DNA was extracted from young leaves collected in early spring, following the method described by Murray and Thompson (1980), modified by Weising et al. (1995) and adapted to almond as follows: 1.5 g young leaves were ground in liquid nitrogen to fine powder and extracted with CTAB extraction buffer. The mixture was incubated at 60°C for 1 h, followed by two extractions with chloroform/isoamyl alcohol (24:1), Isopropanol was used to precipitate nucleic acids and the pellet obtained was dissolved in Tris–EDTA (TE) buffer. Co-precipitated RNA was removed by digestion with RNaseA. Remaining impurity was extracted with processed phenol and chloroform. Total DNA was precipitated using Sodium acetate and cold ethanol, washed with 10 mM ammonium acetate in 76% ethanol and dissolved in TE buffer. The purified total DNA was quantified by gel electrophoresis and verified by spectrophotometer.
PCR amplification and product electrophoresis
AFLP assays were performed as described by Zabeau (1993) and Vos et al. (1995). The 19 primers (Table 2) used were synthesized by MWG (Germany). About 6 μl of 50 ng genomic DNA was digested with MseI and PstI in 10 × buffer. MseI and PstI adapter were subsequently ligated to the digested DNA fragments. Sequencing of the adapters and adjacent restriction sites served as primer binding sites for amplifying the selected fragments. A specific population of fragments was amplified from the reaction mixture by adding nucleotides to the 3′ ends of the primers in two steps. One nucleotide was added for the first amplification step, while three nucleotides were added for the second amplification step. DNA was PCR-amplified for 26 cycles using 5 μl of template DNA and +1 primer (MseI +1 and PstI + 1). The PCR condition were as follows: an initial denaturation step at 94°C for 2 min, 26 cycles were run at 94°C for 60 s, 65°C for 60 s, 72°C for 60 s, and one last extension step of 5 min at 72°C. The second amplification step used + 3 primers (MseI + 3 and PstI + 3). The DNA template for the second amplification step was the PCR product that was produced by the first step. DNA was amplified for one cycle at 94°C for 30 s, 65°C for 30 s, and 72°C for 60 s, then for 12 cycles with a 0.7°C annealing temperature decrease per cycle, and finally for 23 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s. The PCR products produced by the second amplification step were mixed and denatured with an equal volume of loading buffer (98% formamide, 10 mM EDTA, 0.05% Xylene cyanol, 0.05% Bromophenol blue) and heated for 5–10 min at 94°C, chilled on ice then 6 μl of denatured preparation were loaded on per warmed (50°C) polyacrylamide sequencing gels (Gibco BRL, model S2) containing 7 M urea in 1 × TBE buffer. Gels were run for 1.5–2 h at 100 W until the forward running dye (Bromophenol blue) reached the end of the gel. The DNA bands were visualized by silver staining as described by Bassam and Caetano-Anolles (1993). AFLP bands ranging in length from 67 to 501 bases were scored as present (1) or absent (0).
Data analysis
In the genetic relationship study, only distinct, reproducible, well-resolved AFLP fragments in the size range of 67–501 bp were scored as present (1) or absent (0), and from band scores a binary data matrix was constructed. GS were calculated among all possible pairs of accessions using all scorable fragments of AFLP markers. A dendogram of genetic relationship was produced by clustering the data with unweighted pair group method with arithmetic average (UPGMA). The co-phenetic correlation coefficient was calculated, and the Mantel test (Mantel 1967) was performed to check the goodness of fit of cluster analysis to the similarity matrix on which it was used. Simple Matching similarity (SM) was also used for closer comparison with previously published results. All were performed using the NTSYS-pc 2.02 software package (Rohlf 1998). Bootstrap analysis (1,000 replicates) was performed to assess the relative support for different groups and the stability of the dendrogram, using the TREECON software package version 1.3 (Van de Peer and De Wachter 1994).
The information content of each AFLP marker was computed as PIC i = 1−Σp 2 i , where p i is the frequency of the ith band. The average polymorphic information content (PIC) was calculated for AFLP markers across assay units by applying the formula as above given by Powell et al. (1996). Each DNA fragment visualized within the gel was considered as a single dominant AFLP marker locus. Only polymorphic bands with strong intensity were scored, each marker was identified by the primer combination and the band number as a suffix. Markers with molecular weight lower than 100 bp were excluded from the data matrix. The discrimination power of each AFLP markers was evaluated by the polymorphism information content (PIC). Within accession diversity (H S) and total gene diversity (H T) (Nei 1973) were calculated within the species and within major groups, using the POPGENE software. The calculations were performed using all markers, both monomorphic and polymorphic. Finally, the partitioning of molecular variance within and among groups and accessions was calculated by the AMOVA technique (Excoffier et al. 1992) in ARLEQUIN software (Schneider et al. 2001). All significance tests were calculated by performing 1023 permutations.
Results
Variation for AFLP markers
The number of polymorphic bands per AFLP primer combination ranged between 18 and 60 for PstI/MseI, with an overall average of 41.11. For each of 19 primer combinations, PIC value ranged between 0.56 and 0.82, whereas, marker indices ranged between 51.37 and 78.79 (Table 3). In addition, the number of bands varied from 18 (M-GTC, P-AAC) to 65 (M-G, P-AAC) with average of 42.78 bands per assay unit. For separate assay units, PIC values ranged from 0.56 to 0.82 (Table 3). The mean PIC score for all loci was 0.68. The PIC value provides an estimate of discriminatory power of a marker by taking into account not only the number of alleles at locus but also the relative frequencies of these alleles. The distribution of PIC scores were nearly uniform (random) for the 781 polymorphic AFLP markers (Fig. 1). PIC scores were maximum (0.82) for 96.08 of the AFLP markers.
The prescreening analysis of the 45 selected accessions and 20 AFLP assay units showed that 19 primers generated strong and reproducible amplification products, all of which displayed polymorphism among the cultivars. As highly polymorphic primers were used for analysis, a relatively large number of polymorphic AFLP markers were detected by these primer combinations. Examples of amplification patterns for molecular characterization obtained by AFLP in different cultivated genotypes and related species are shown in Fig. 2.
GS estimates between replicated samples of the same cultivars of almond from 0.028 to 0.96 with an average of 0.68. In addition, genetic similarity values between cultivated genotypes and related species ranged from 0.50 to 0.96, and the mean, minimum, and maximum of similarities between almond cultivars and wild species were 0.67, 0.028, and 0.96, respectively (Fig. 3).
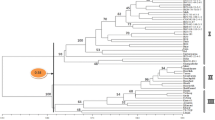
On the other hand, principle-coordinate and cluster analysis separated genotypes and wild species into three major groups. The first group consisted of cultivated genotypes of P. dulcis, the second of related wild species, and third of P. tenella as an out group in the dendrogram (Fig. 4). The first three principle coordinates accounted for 41.56% of the genetic similarity variance. Finally, the phenogram (Fig. 5) and principle-coordinate maps show the groups found with both methods.
Dendrogram obtained with the similarity Jacard coefficient pair group method with arithmetical average clustering algorithm from 781 AFLP markers for 45 almond accessions and related species of Prunus. The value on the dendrogram gives the stability of nodes estimated with a bootstrap procedure (no number indicates support less than 10%)
A total of 813 bands were revealed, of which 781 (96.23%) were polymorphic. The number of AFLP bands detected by each assay units (primer combination) depends on primer combinations, sequence and extent of variation in specific genotypes, therefore, the number of bands varied in different accessions.
AFLP polymorphism and power of discrimination
A total of 781 polymorphic fragments were generated with the 19 primer combinations used. The number of markers per primer combinations ranged from 18 to 65 with an average of 42.78. The most polymorphic primer combinations were M-CAA + P-GCA, M-CAA + P-GCC, M-CAC + P-GCC, and M-GAG + P-AGC, which produced 42, 40, 33, and 42 markers, respectively (Table 3). The fragments sizes ranged from 67 bp to 501 bp, with 81.8% of the markers between 100 bp and 400 bp. The distribution of the fragment sizes was skewed towards larger fragments. This was mainly a result of the fact that fragments smaller than 100 bp were not included.
The distribution power of each marker was estimated by the PIC (results not shown). Values of PIC ranged between 0.56 and 0.82 (the expected maximum value for a bi-allelic locus), with an average of 0.68. Hence, a large proportion of markers have a high discrimination power. In addition, the discrimination power of each primer combination was estimated by average of the discrimination power of each marker used PICav = ΣPIC i /N was calculated, where PIC i is the PIC value of the ith AFLP marker and N is the number of AFLP markers generated by a primer combinations (PC). Loci that are non-polymorphic (PIC = 0) in the germplasm of interest were excluded from this calculation. Because most loci in the study are polymorphic, this average value for a set of markers should only slightly overestimate the true PICav.
Finally, to provide an objective comparison, matrices of coephenetic values, generated from AFLP and morphological data, were compared using the Mantel test not significant and quite low correlation between the dendrograms was obtained (r = 0.33, P = 0.9741) after doing 250 random permutation with Maxcomp procedure from NTSYS program.
Partition of genetic variation and diversity within and between cultivars
Analysis of molecular variance was performed twice, separately using geographic origin and clustering as the grouping criteria. In both cases, AMOVA demonstrated highly significant variation (P < 0.001). Genetic variance was found within cultivars as well as among cultivars (Table 4): the variance within cultivars accounted for 45% of the total variance when groups were based on the clustering pattern and for 64% when groups were based on geographic origin, while the population variance contributed only 5.2 and 38.2 for origin and clustering pattern, respectively. Genetic diversity within cultivars, expressed as AMOVA mean square deviations, was positively correlated (r = 0.95; P < 0.001) with percentage of polymorphic markers detected per cultivar. The Barttle’s test for population heterosedasticity was highly significant (B.p = 3.73, P < 0.001) indicating different levels of variability within different cultivars (Table 4).
Genetic diversity, expressed as a coancestry coefficient, among the 45 cultivars ranged from 14 for the comparison of Shahrodi18 and Shahrodi16 to 186 for that of ‘Sangi26’ and P. glucea. Coancestry coefficients between Iranian and non-Iranian cultivars ranged were generally higher than coefficients between other pairs of cultivars, showing that Iranian cultivars were highly heterogeneous. Clustering (UPGMA) based on coancestry coefficients clearly separated the 45 cultivars into three clusters. Cluster one is an outgroup containing P. tenella and cluster two contained P. scoparia and P. glucea. The third cluster exclusively consisted of the related species, and P. dulcis genotypes were clearly separated from all other cultivars. Clustering of all cultivars based on Jacard similarity also clearly grouped all plants in three clusters (Fig. 5).
Gene diversity
The total gene diversity across all accessions (H T) was 0.157 and the within-accession gene diversity (H S) was 0.104 (Table 5). The gene diversity among groups of almond were investigated, thus, five main groups were grouped. Group 1 was consisted Iranian accessions, the group 2, contained Italian and Spanish cultivars, group 3 included accessions of ‘Shahrodi’ and American, group 4 consisted France and Russia cultivars and the group 5 contained related wild species of almond. Low H S relative to H T is consistent with the species being partially self-compatible in some of cultivars (Table 1). The values of H T and H S for group5 that consist of related species of almond exceed those of other groups by 33.01 and 37.78%, which indicates that group 5 contains greater genetic diversity both among and within accessions.
Although gene diversity estimates are commonly used to make comparison between species, calculations derived from AFLP data should not be compared across studies, because AFLP overestimates numbers of loci and underestimates numbers of alleles (Caicedo et al. 1999). Because of these factors and because AFLP can detect many fragments that occur at low frequency, the low diversity estimates obtained may be due to the markers technology used rather that to low genetic diversity in the species. Thus relative, but not absolute, diversity estimates from this study showed be considered reliable.
Genetic similarity and clustering of genotypes
The similarity coefficients for cultivated genotypes and related species of almond varied from a maximum of 0.96 (between ‘Nonpareil’ and ‘Shahrodi18’) to a minimum of 0.028 (between ‘Monagha’ and P. tenella), with average of 0.68, indicating the high level of genetic variation that exists in the almond gene pool. The Mantel method used for comparing the similarity matrixes produced correlation coefficients that were statistically significant for AFLP markers. The co-phenetic correlation coefficient between dendrogram and the original similarity matrix for AFLP was large and significant (r = 0.99, t = 7.22), giving a good degree of confidence in the association obtained for the accessions.
The dendrograms (Figs. 5, 6) reflect relationships among many of the cultivars in ways that, depend on their area of diffusion and/or pedigree information, analogous to reports by Woolley et al. (2000) and Martínez-Gómez et al. (2003b, 2007), which used RAPD or SSR markers, and found that cultivars originating in Europe and USA clustered in a separate group. The AFLPs discriminated most genotypes effectively and separated cultivated almond from related species. Another important aspect in cluster analysis is to determine the optimal number of cluster or number of acceptable clusters. In essence, this involves deciding where to "cut" a dendrogram to find the true or natural groups. An "acceptable cluster" is defined as "a group of two or more genotypes where the within-cluster genetic distance is lower than the overall mean genetic distance and the between cluster distances are greater than the within-cluster distance of both cluster involved (Brown-Guedira et al. 2000). Some relatively simple ways of finding the optimal number of cluster are the D 2, upper tail approach (Wishart 1987) and statistical techniques such as bootstrapping, MANOVA (Multivariate Analysis of Variance), and or discriminate analysis. We used the MANOVA method, where the optimal number of clusters or groups occurs when the F value is highest. In our study this cutting point is 0.45.
Dendrogram obtained with the dissimilarity matrix of the agronomic characteristics and unweighted pair group method with arithmetic average clustering algorithm for 45 almond accessions and related species of Prunus. The value on the cut of point on dendrogram gives the optimal clustering estimated with a MANOVA procedure
The dendrogram consists of three well-supported clusters, i.e. three groups of plants; P. dulcis cultivars (cluster I); and P. scoparia and P. glucea (cluster II) and cluster III divided into two subgroups, for which subgroup I contains P. bucharica, P. kuramica, P. webbii and P. koteschyi and subgroup II contains related species of Iranian origin, P. orientalis, P. communis and P. tenella (cluster III). The strength of relationships varied, as assessed by bootstrapping analysis. In the dendrogram, there is very strong support for clustering of cultivars that were closely related by pedigree or mislabeled. As shown in Fig. 5, the wild species of P. tenella which belongs to section Chameamygdalus (Spach), separates from other cultivated almond and related species, with a coefficient of 0.96. The wild species of P. scoparia and P. glucea which belongs to Spartioides group separates from other cultivars, with a coefficient of 0.47. The third group (cultivated and wild almond) separated from other cultivars, with a coefficient of 0.36 into two subgroups. Sub-group I is closer to almond.
The second sub-group consists of P. communis and P. orientalis. P. orientalis is not an ancestor of cultivated almonds, belongs to Euamygdalus group in which cultivated almonds have been placed, differs morphologically from the rest of the taxa in being a shrub and in having pubescent leaves (Ladizinsky 1999), but can be crossed with other almonds. It separated from other cultivated almonds with a coefficient of 0.69. The lower similarity indices and more divergent dendrogarm branch points of the wild species P. tenella, included as an outgroup, and other related species demonstrate the high genetic variability of the study material.
In the group of P. dulcis cultivars, the main cluster contained Iranian, American or European cultivar only, or mixtures of these with foreign cultivars, indicating a close relationship among cultivars. The genetic diversity of P. dulcis cultivars was high and it was possible to discriminate all almond cultivars analyzed. Foreign and Iranian cultivars were found to associate according to their origin and genotypic relatedness. This further supported the results regarding Iranian cultivars and unknown ‘Shahrodi’ cultivars for which genetic diversity, parental relations, and origins were unknown and mislabeled. Most of the Iranian cultivars grouped into the same cluster. These results have also been confirmed by morphological studies.
‘Azar’, ‘Ferragnes’ and ‘Shekofeh’, were obtained from a cross between ‘Ai’ × ‘Cristomorto’ and ‘Ai’ × ‘Nonpareil’, respectively, and they grouped into the same subcluster in the resultant dendrogram. The genetic similarity between ‘Ferragnes’ and the two Iranian Shekofeh and Azar cultivars were 0.80 and 0.79, respectively and between ‘Shekofeh’ and ‘Azar’ was 0.81, as may be expected given their common (French) parent (‘Ai’). The two cultivars of ‘Azar’ and ‘Ferragnes’ have both parental cultivars in common (‘Ai’ × ‘Cristomorto’), yet showed a fairly low genetic similarity of 0.79, presumably because of genomic differences in the parental stocks.
Discussion
AFLPs proved to be a powerful tool for fingerprinting and molecular characterization of cultivated almonds and related wild species, and they are also useful for producing genetic maps and marker-assisted selection in crop plants (Vos et al. 1995). They have virtually eliminated the DNA- marker bottleneck in almond cultivars. This bottleneck persisted for many years, and had impeded the use of markers in almond breeding and the development of a genetic map of almond. The percentage of polymorphic AFLP fragments was similar to percentage of polymorphic RFLP reported by Berry et al. (1994) and Gentzbittel et al. (1994), whereas 42.8% of AFLP fragments were polymorphic in cultivated genotypes and related species of almond.
It should be noted that AFLPs had lower PIC scores than RFLP. Berry et al. (1994) reported mean PIC of 0.49 for a selected of 57 RFLP probes (185 RFLP bands). This is significantly greater than the mean PIC we estimated for AFLPs (0.68). PIC score differences between AFLPs and RFLPs have primary causes (both markers detect DNA polymorphism caused by restriction-site mutations, insertions, or deletions). The minimum PIC scores for an AFLP marker (or any bi-allelic marker) is 0.5, where as the maximum PIC scores for an RFLP marker is 1.0. Thus for example, when an AFLP fragment is present in half and missing in half of cultivated almond and wild species, the PIC score is 0.5. Roughly 95% of the AFLP fragments in our study had maximum PIC scores. The PIC scores for a RFLP marker can be increased by testing additional restriction enzymes.
The results of this study are consistent with an earlier study of genetic diversity for RAPD and SSR markers (Shiran et al. 2007), where cultivated almond and wild species were also strongly separated into groups. These groups reflected the fundamental heterotic patterns of almond cultivars and the widespread practice of producing new genotypes by crossing cultivated almond with related species for tree improvement of almond. We found, like Woolley et al. (2000) and Martínez-Gómez et al. (2003b), a clear grouping of different almond cultivars according to geographic origin, but they found, using RAPD and SSR markers, that cultivars originating in Europe and USA clustered in a different group.
The range of the amplified bands sizes in almond and other Prunus species was also similar to those reported by Doweny and Iezzoni (2000) for black cherry and by Cipriani et al. (1999) for peach, in studies using the same primer pairs. Variation in the numbers of polymorphic AFLP markers and the total number of polymorphic bands were observed (Table 3), allowing differentiation into two groups of species, one group having high numbers of polymorphic bands (P. bucharica, P. kuramica, P. tenella, and P. webbii), the other group with lower (P. scoparia, P. glucea and other P. dulcis). Differences in amplification success for AFLP markers observed among related species and almond cultivars. Result also demonstrated the possibility of cross-species and consequently the value of markers developed in species of Prunus for molecular characterization of other cultivars species within the subgenus. Successful hybridization between P. dulcis (almond) and P. bucharica, P. kuramica, P.webbii has been also reported by different authors (Kester and Gradzeil 1996).
Prunus bucharica and P. kuramica have been described as the Prunus species most closely related to almond (Browicz and Zohary 1996; Grasselly 1976), and both species are described as ancestral species of the cultivated almond, and Kester and Gradziel (1996) proposed that P. fenzliana is a wild ancestor of almond. Unfortunately, P. fenzliana germplasm was not available for this study. Natural introgression from related Prunus species to almond has been reported, and Kester and Gradzeil (1996) suggest that sweet kernel, in addition to being a natural variant within P. dulcis, may have been transferred to P. dulcis from P. bucharica or P. kuramica. Prunus webbiii may also be the original source of self-incompatibility in European almond cultivars, including ‘Genco’, ‘Touno’, or ‘Cristomorto’, with natural introgression of this gene occurring during the centuries of almond cultivation in the Puglia region of Italy (Reina et al. 1985).
In this sense, Hesse (1975) and Scorza and Sherman (1996) described the successful hybridization between P. dulcis (almond) and P. bucharica, P. kuramica and P. webbii, as was also reported by Kester and Gradzeil (1996). The phenetic relationships among almond cultivars and related species studied here are in general agreement with previous taxonomic and genetic studies (Xie et al. 2006). The results of a genetic study of chloroplast and nuclear DNA sequence data (Bortiri et al. 2001), which included some but not all of the species studied here, supported a sister relationships between those species and P. dulcis, P. bucharica.
Both almond cultivated and wild species suffer from a limited gene pool availability for future breeding progress. Inter-specific gene transfer among these Prunus species offers a greatly expanded genetic diversity for breeders, particularly given the relative ease of initial hybridization and subsequent backcrossing (Gradziel et al. 2001). Hence, further AFLP analysis of this germplasm offers opportunities for determining more precisely genetic relationships and molecular characterization and could be an important tool for marker assisted gene transfer. DNA fingerprinting and molecular characterization using AFLP analysis could be also very useful for the selection of the most promising progeny from inter-specific crosses or back crosses, leading to greatly improved breeding efficiency. Bartolozzi et al. (1998) reported similarity coefficients among California almond cultivars/breeding line that ranged from 1.00 (between ‘Nonpareil’ and ‘Tardy Nonpareil’) to 0.484 (between ‘NeplusUltra’ and ‘Padre’), using RAPD markers.
The wild species P. communis, which is usually considered one of the ancestors of cultivated almonds (Vavilov 1930; Kovaleff and Kostina 1935; Denisov 1988), placed into cluster II. The degree of similarity between P. communis and other almond cultivars assessed in the present study is high enough that it supports the view of those scientist who concluded cultivated almond emerged by selection within the species named as P. communis. An alternate hypothesis (Evreinoff 1958) implies that P. communis arose by hybridization among P. fenzliana, P. bucharica and other species. This concept suggested that the taxon P. fenzliana Lipsky is the most likely wild ancestor of almond. Nonetheless, our AFLP marker data also indicated that P. communis is clearly related to P. dulcis cultivars.
American cultivars and one Russian cultivar grouped within the same cluster. The presence of Russian cultivar in a subgroup in which American cultivars have been placed is easily explained due to two reasons: first, ‘Primorski’ was obtained from a cross between ‘Princess2077’ × ‘Nichitsky’, and ‘Princess2077’ is a French cultivar; and second, most California almond cultivars originated from Languedoc area of southern France (Kester and Gradzeil 1996). This finding is further confirmed by the presence of S5 incompatibility allele in Primorski as well as American cultivars such as ‘Texas’ and ‘Neplus Ultra’ (Kester et al. 1994; Channuntapipat et al. 2003). The France cultivar of ‘Ferragnes’ was also classified into the same subgroup as the Iranian cultivars, indicating their close genetic relationships.
The presence of ‘Shahrodi’ cultivars in American subgroups can be explained. Varieties called ‘Shahrodi’ are mainly the same European and American varieties that were introduced from France to Iran in 1976, and their original names became lost. Probably the two ‘Shahrodi’ cultivars were ‘Tardy Nonpareil’ (with similarity of 0.91) and ‘Nonpareil’ (with a similarity of 0.94), respectively. ‘Tardy Nonpareil’ is reported to bloom 7–10 days after ‘Nonpareil’ and produce lower yield (Kester et al. 1994). ‘Tardy Nonpareil’ is usually considered to be a bud sport mutant of ‘Nonpareil’, and in the study of Bartolozzi et al. (1998) using 37 RAPD markers these two cultivars could not be separated and there were no differences between the RAPD patterns of ‘Nonpareil’ and ‘Tardy Nonpareil’. However, in the present study based on AFLP markers, these two species were clearly distinguished (with a 0.92 Jacard similarity level). This result is in agreement with the study of Martins et al. (2003) and Woolley et al. (2000) that obtained values of 93.5% (Dice) and 91% (SM similarity coefficient) between these two cultivars as based on RAPD/ISSR and RAPD, respectively.
Considering the high similarity coefficient obtained, it is clear that Tardy Nonpareil was derived from Nonpareil by a mutation in a bud sport that affected flowering time and growth habit. In the cluster, Nonpareil and Tardy Nonpareil were associated with Iranian, ‘Azar’ and ‘Shekofeh’ and ‘Shahrodi’ cultivars also ‘Texas’, ‘IXL’, ‘NeplusUltra’, ‘Kapareil’ and ‘Thompson’ from USA. This result is in accordance with the results obtained through application of ISSR and RAPD (Martins et al. 2001; 2003). Historical reports from almond development in California suggest that ‘Texas’ (‘Mission’), ‘Nonpareil’, ‘IXL’ and ‘NeplusUltra’ were the seedling selection of an early American cultivars known as ‘Languedoc’, which has been reported to have probably originated from the Languedoc region of France (Wood 1925; Kester and Gradziel 1996; Bartolozzi et al. 1998). In the present study, the similarity index showed roughly equal similarity between ‘Texas’ and ‘NeplusUltra’, ‘Nonpareil’ and ‘IXL’ (0.79, 0.78, and 0.76 respectively) and between ‘NeplusUltra’, ‘Nonpareil’ and ‘IXL’ (0.87, 0.88). The high similarity index and close proximity in dendrogram consequently support this relationships and confirming the conclusion of Bartolozzi et al. (1998) that they are siblings originating from the same ‘Languedoc’ seed.
Channnuntapipat et al. (2003) reported the presence of the S7 incompatibility allele in ‘Nonpareil’, ‘NeplusUltra’ and ‘Mission’, and this finding further supports that these cultivars are closely related. Woolley et al. (2000) reported that ‘Mission’ (‘Texas’) was too genetically distant to belong in this group for American cultivars (‘Thompson’, ‘NeplusUltra’, ‘Nonpareil’, ‘Tardy Nonpareil’ and ‘Texas’). Bartolozzi et al. (1998) produced a dendrogram based on SM similarity value ranging from 0.55 to 1.00, values roughly similar to those values we obtained for this group of cultivars (0.80–0.93) using the same similarity coefficient. Thus, all of these findings support the view of Kester et al. (1994) who believed most of the current California cultivars originated from progeny of ‘Nonpareil’ and ‘Texas’.
The European cultivars ‘Filippoceo’, ‘Facionello’, ‘Texas’, ‘Moncayo’, ‘Genco’, ‘Princess’ and ‘Ferragnes’ are clustered in the same subgroup. Somewhat surprisingly, recent self-incompatibility (S) allele analysis (Channuntapipat et al. 2003) indicates that ‘Touno’ and ‘Genco’ have the S 1 S f incompatibility/compatibility alleles in common. The S locus, which controls the gametophytic self-incompatibility response in almond, has been reported to be highly polymorphic in nature, and represents a high multiple allelic series (De Nettancourt 1997). Thus, the presence of an allele in common for ‘Touno’ and ‘Genco’ clearly suggests a high level of relatedness of these cultivars. The great diversity found among Iranian cultivars and the germplasm of Emamieh orchards plants supports the idea that Iranian cultivars are an important source of almond genes for exploitation in breeding programs. The results obtained from this study would be useful for better management, identification of cultivars, and also avoiding mislabeling of genotypes.
Our results showed great genetic diversity of the almond cultivars studied. However, the correspondence between the clustering based on AFLP and that based on agronomic traits was relatively low. Cluster analysis based on AFLP data cleary differentiated the genotypes according to their origin and pedigree information. Main groups contained genotypes of almond that originated from American, Europe, Spain and Iran, and related species. The remaining groups contained the rest of studied cultivars. In contrast, cluster analysis based on agronomic data differentiated some genotypes and wild species according to their pomological characterization although no apparent clustering by agronomic characteristics (time of maturity and flowering, self compatibility, fruit size) was observed due to the guided selection process.
The UPGMA dendrogarm obtained using morphological characters clearly separated the European and American and Iranian and other genotypes which of unknown origin. Regarding differences between genotypes and related species, out of 16 characteristics studied, all of them showed significant differences at 1% level. However, comparison of means for these traits revealed that in general such differences were not important from the breeding point of view. The most striking feature noted was the low flowering intensity and mean productivity for all the years in the case of both genotypes and related species, an indication of tardiness with which the seedling came to bearing.
Socias i company and Felipe (1988) noted that descendants resulting from self-fertilization or crossing between relatives were slow to come into bearing. For comparative purpose, it is worth mentioning the study made under similar conditions by Dicenta (1991), of over 2000 individual from 51 crosses between 12 cultivars. Although the differences were not pronounced for flowering intensity, they were larger for productivity, probably owing to the widespread phenomena of floral infertility as a result of inbreeding. In fact, our data on flower intensity are notably higher than our data of productivity (result not shown). The result obtained for fruit characteristics in both almond genotypes and wild species the fourth leafing, are very similar to those described by Dicenta (1991) in individual crosses. We believe that correlation between them could be improved if there was a more morphological marker analyzed as was reported by Martínez de Toda and Saccha (1997) or more primer combination of AFLP were used.
Working with 16 ryegrass varieties, Roland-Ruiz et al. (2001) reported correlation values of r = −0.06 between AFLP and 15 morphological characters. In comparison with ryegrass, almond genotypes appear to be environmentally more stable, as suggested by higher agreement between phenotypic and molecular analysis. Apparently, in ryegrass there is much environmental influence accounting for the morphological variability observed. Therefore, when compared with DNA fingerprinting, molecular characterization techniques, morphological traits are relatively less reliable and inefficient for precise discrimination of closely related genotypes and analysis of their GS, however, morphological traits are useful for preliminary, fast, simple, and inexpensive varietals identifications and can be used as a general approach for assessing genetic diversity among phenotipically distinguishable cultivars, although they are inefficient on account of the time and cost involved.
Both the morphological and genetic analysis allowed to separate the Iranian, France, American materials, except for some cultivars (see the dendrograms), which clustered with the same origin in the case of AFLP. The correlation between the two systems was neither significant nor very high. Our AFLP and morphology result suggest that the almond germplasm share a common genetic background differing, in genotype and morphology, from the France, American, and Spanish, varieties used for comparison in this study. The high degree of polymorphism detected and the possibility of screening a higher number of anonymous loci than morphological markers make AFLP useful for studying genetic diversity, molecular characterization within the almond germplasm. To our knowledge, this is the first report using AFLP markers to assess genetic variability on these materials. Another type of molecular markers like microsatellite, which are highly abundant in the almond genome and shows co-dominant nature, will certainly contribute to determine the relationships between Iranian, European, American, and related species of Prunus in further investigation.
References
Aranzana M, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriani G, Ryder C, Testolin R, Abbott A, King G, Arus P (2003) A set of simple sequence repeat (SSR) markers covering most of the Prunus genome. Theor Appl Genet 106:819–825
Arulsekar S, Parfitt DE, Kester DE, (1986) Comparison of Isozyme variability in peach and almond cultivars. J Hered 77:272–274
Bartolozzi F, Warburton ML, Arulsekar S, Gradziel TM (1998) Genetic characterization and relatedness among California almond cultivars and breeding line detected by randomly amplified polymorphic DNA (RAPD) analysis. J Am Sco Hort Sci 123:381–387
Bassam BJ, Caetano-Anolles G (1993) Silver staining of DNA in polyacrylamide gels. Appl Biochem Biotech 42:181–188
Berry ST, Allen RJ, Barnes SR, Caligari PDS (1994) Molecular marker analysis of Helianthus annus L. Restriction fragment length polymorphism between inbred lines of cultivated sunflower. Theor Appl Genet 89:435–441
Bortiri PE, Oh S, Jiang J, Baggett S, Granger A, Weeks C, Potter D, Parfitt DE (2001) Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL-trnF spacer DNA. Syetematic Bot 26:797–807
Browicz K, Zohary D (1996) The genus Amygdalus L. (Rosaceae): species relationships, distribution and evolution under domestication. Genet Resour Crop Evol 43:229–247
Brown-Guedira GL, Thompson JA, Nelson RL, Warburton ML (2000) Evaluation of genetic diversity of soybean introductions and North American ancestors using RAPD and SSR markers. Crop Sci 40:815–823
Caicedo AL, Gaitan E, Duque MC, Chica OT, Tohma J (1999) AFLP fingerprinting of Phaseolus lunatus L. and related wild species from South America. Crop Sci 39:1497–1507
Cerezo M, Socias I, Company R, Vargas F (1989) Identification of almond cultivars by pollen isoenzymes. J Am Soc Hort Sci 114:164–169
Channuntapipat C, Wirthensohn M, Ramesh SA, Batlle I, Arus P, Sedgley M, Collins G (2003) Identification of incompatibility genotypes in almond (Prunus dulcis Mill.) using specific primers based on the introns of the S-alleles. Plant Breed 122:164–168
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach (prunus Persica (L.) Batsch): isolation, characterization and cross-species amplification in prunus. Theor Appl Genet 100:713–722
De Nettancourt D (1997) Incompatibility in angiosperms. Monographs on theoretical and applied genetics, vol 3. Springer, Berlin
Denisov VP (1988) Almond genetics resources in the USSR and their use in production and breeding. Acta Hort 224:299–306
Dicenta F (1991) Mejora Genética del Almondro (Prunus dulcis Miller) por Cruzamientos intervarietales: Herencia de caracters y Selectión. Thesis. University Murcia, Spain, p 313
Doweny LD, Iezzoni AF (2000) Polymorphic DNA markers in black cherry are identified using sequences from sweet cherry, peach and sour cherry. J Am Soci Hort Sci 125:76–80
Evreinoff VA (1958) Contribution a letude de l’amandier. Fruits et Primeurs Afrique 28:99–104
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gentzbittel L, Zhang Y-X, Vear F, Griveau B (1994) RFLP studies of genetics relationships among inbred lines of cultivated sunflower, Helianthus annus L. evidence for distinct restorer and maintainer germplasm pools. Theo Appl Genet 89:419–425
Gradziel TM, Kester DE (1998) Breeding for self-fertility in California almond cultivars. Acta Hort 470:109–117
Gradziel TM, Martínez-Gómez P, Dicenta F, Kester DE (2001) The utilization of Prunus species for almond variety improvement. J Am Pomol Soc 55:100–108
Grasselly C (1976) Les espéces sauvages ďamandier (in French). Options Méditerr 32:28–44
Hauagge R, Kester DE, Arulsekar S, Parfitt DE, Liu L (1987) Isozyme variation among California almond cultivars. II. Cultivar characterization and origins. J Am Sco Hort Sci 112:693–698
Hesse CO (1975) Peaches. In: Janick J, Moore JN (eds) Advances in fruit breeding. Purdue Univ. Press, West Lafayette, Ind, pp 285–326
Kester DE (1994) Almond cultivar and breeding programs in California. Acta Hort 373:13–28
Kester DE, Gradziel TM (1996) Almonds In: Janick J, Moore JN (eds) Fruit breeding, Almond, vol. 1. Wiley, New York, pp 1–97
Kester DE, Gradziel TM, Micke WC (1994) Identifying pollen incompatibility groups in California almond cultivars. J Am Sco Hort Sci 119:106–109
Kovaleff NV, Kostina KF (1935) A contribution of the study of the genus prunus Focke. Leningrad
Ladizinsky G (1999) On the origin of almond. Genet Res Crop Evol 46:143–147
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martínez de Toda E, Saccha JC (1997) Ampelographical characterization of red Vitis vinifera L. cultivars preserved in Rioja. Bultten de tow, Mars-Avril, 70:220–234
Martínez-Gómez P, Arulsekar S, Potter D, Gradziel TM (2003a) An extended inter-specific gene pool available to peach and almond breeding as characterized using simple sequence repeat (SSR) markers. Euphytica 131:313–322
Martínez-Gómez P, Arulsekar S, Potter D, Gradziel TM (2003b) Relationships among peach and almond and related species as detected by SSR markers. J Am Sco Hort Sci 128:667–671
Martínez-Gómez P, Sánchez-Pérez R, Dicenta F, Howad W, Arus P, Gradziel TM (2007) Almonds. In: Kole CR (ed) Genome mapping and molecular breeding, vol. 4: Fruits & Nuts. Springer. Heidelberg, Berlin, New York, Tokio, pp 229–242
Martins M, Farinha A, Ferreira E, Cordeiro V, Monterio A, Tenreiro R, Oliveira M (2001) Molecular analysis of the genetic variability of Portuguese almond collection. Act Hort 546:449–456
Martins M, Tenreiro R, Oliveira M (2003) Genetic relatedness of Portuguese almond collection assessed by RAPD and ISSR markers. Plant Cell Rep 22:71–78
Mnejja M, Garcia-Mas J, Howad W, Badenes ML, Arús P (2004) Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol Ecol Notes 4:163–165
Mir Ali N, Nabulsi I (2003) Genetic diversity of almond (Prunus dulcis) using RAPD technique. Sci Hort 98:461–471
Murray HC, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 8:4321–4325
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci (USA) 70:3321–3323
Powell W, Morgante M, Andre C, Hanafey Mm Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Reina A, Giorgio V, Godini A (1985) Autres types autocompatibles parmi la population ďamandiers des Pouilles (in Italian). Options Méditerr 85:25–29
Resta P, Corona MG, Fanizza G, Palasciano M, Godini A (1998) Random amplified DNA polymorphism in Amygdalus communis L. and A. webbii Spach. Acta Hort 470:82–90
Rohlf FJ (1998) NYSYS-pc. Numerical taxonomy and Multivariate Analysis System, Version 2.02 Exeter Software, Setauket, NY
Roland-Ruiz L, Vaneeuwijk FA, Gilliland TJ, Dubreil P, Dillman C, Lallemand J, De loost M, Barii CP (2001) A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium Perenne L.) varieties. Theor Appl Genet 103(8):1138–1150
Schneider S, Roessli D, Excoffier L (2001) Arlequin: a software for population genetics data analysis. Version 2.000. Genetics and Biometry Lab, Dep. of Athropology, University of Geneva, Geneva
Scorza R, Sherman WB (1996) Peaches, In: Janick J, Moore JN (eds) Fruit breeding, Wiely, New York, pp 285–326
Shiran B, Ameirbakhtiar N, Kiani S, Mohamadi Sh, Tabatabaei BES, Moradi H (2007) Molecular characterization and genetic relationships among almond cultivars assessed by RAPD and SSR markers, Scientia Horticulturae 111:280–292
Socias i Company R, Felipe AJ (1988) Self-incompatibility in almond: transmission and recent advances in breeding. Acta Hort 224:307–317
Van de Peer Y, De Wachter R (1994) TREECON for Windows: as software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Vavilov NI (1930) Wild progenitors of the fruit trees of Turkistan and Caucasus and the problem of the origin of fruit trees. In: Proceeding of the IX International Horticultural Congress report and proceeding. London, pp 271–286
Vezvaei A (2003) Isozyme diversity in Iranian almond. Acta Hort 622:451–456
Viruel MA, Messeguer R, de Vicente MC, Garcia-Mas J, Puigdomenaech P, Arus P (1995) A linkage map with RFLP and isozyme markers for almond. Thero Appl Genet 91:964–971
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Watkins R (1976) Cherry, Plum, Peach, apricot and almond. In: Simmonds NW (ed) Evolution of crop plants. Longman, London, pp 342–347
Weising K, Nybon H, Wolff K, Meyer W (1995) DNA fingerprinting in plants and fungi. CRC Press, Boca Raton, USA
Wishart D (1987) CLASTAN user manual, 3rd edn. Program Library Unit, Univ. of Edinburgh, Edinburgh
Wood MN (1925) Almond varieties in the United States. USDA Bul 1282:1–42
Woolley FM, Collins GG, Sedgly M (2000) Application of DNA fingerprinting for the classification of selected almond [prunus dulcis (Miller) D. A. Webb] cultivars. Aust J Exp Agr 40:995–1001
Xie H, Sui Y, Chang FQ, Xu Y, Ma RC (2006) SSR allelic variation in almond (Prunus dulcis Mill). Theor Appl Genet 112:366–372
Xu Y, Ma RC, Xie H, Cao MQ (2004) Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 47:1091–1104
Zabeau M (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European patent Application No.0–534-858-A1
Acknowledgement
The authors are grateful to Shahrekord University for financial assistance and Dr Ali Vezvaei for helpful suggestions. Thanks to the section of Horticulture, Agriculture and Natural Resources Research Center of Shahrekord for access to trees.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sorkheh, K., Shiran, B., Gradziel, T.M. et al. Amplified fragment length polymorphism as a tool for molecular characterization of almond germplasm: genetic diversity among cultivated genotypes and related wild species of almond, and its relationships with agronomic traits. Euphytica 156, 327–344 (2007). https://doi.org/10.1007/s10681-007-9382-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9382-x