Abstact
An agronomic gene pool of wheat (Triticum aestivum L.) was constructed through recurrent selection. In present research, 24 wheat SSR markers determining 25 loci on 14 different chromosomes were used to evaluate the gene pool. Thirty parents used as original materials in recurrent selection were also assessed. In total, 115 alleles were detected in gene pool with an average of 4.6, ranging from 2 to 9 alleles per locus. Statistical test showed that genetic diversities had no significant difference between the gene pool and the 30 parents. Principle coordinates analysis revealed that the individuals of the gene pool were mainly divided into three groups, which was consistent with the result of cluster analysis based on genetic distance matrix of the gene pool. Cluster analysis was carried out based on Euclidian distance calculated upon five morphological trait values and the results showed that most individuals were in a group while the others scattered. Correlation analysis of genetic distance matrix and Euclidian distance matrix showed no significant correlation between two matrices. The results suggest that the gene pool is improved after several cycles of selection, while genetic variation is still maintained. Therefore, the gene pool is suitable for further breeding program.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recurrent selection is a population improvement strategy, which can employ multiple parents in one program and accumulate favorable alleles while maintaining genetic diversity. It was applied in allogamous crop of maize at first, and subsequently extended to autogamous crops. Now, recurrent selection has been applied in many crops such as barley, rice, soybean and oat (Parlevliet and Ommeren 1988; Veillet et al. 1996; Wilcox 1998; Elizondo Barron et al. 1999). In wheat, many studies indicated that recurrent selection could be applied to improve percentage of grain protein (Loffer et al. 1983; Delzer et al. 1995), kernel weight (Busch and Kofoid 1982; Wiersma et al. 2001), grain yield (Olmeda-Arcega et al. 1995) and resistance to diseases (Jiang et al. 1994).
Evaluation of the effect of recurrent selection is commonly based on the targeted morphological traits. Morphological traits are easily to be detected, but are often influenced by the environment. In recent years, the development of molecular markers provided new approaches for evaluating genetic variation on DNA level and a variety of molecular markers had been used to assess genetic diversity in recurrent selection population. Restriction fragment length polymorphism (RFLP) markers were used to evaluate the genetic diversity of recurrent selection in oat (De Koeyer et al. 1999). Using random amplification of polymorphic DNA (RAPD) markers, Yuan et al. (2004) analyzed the genetic diversity among populations and breeding lines from recurrent selection in Brassica napus. Microsatellites (SSR), have been used to assess genetic variation in rice, wheat, barley and sorghum (McCouch et al. 1997; Röder et al. 1998; Struss and Plieske 1998; Djè et al. 2000). In maize, Huang et al. (2004) estimated the genetic diversity of recurrent selection population using SSR markers. It was more reliable to determine individual genotype by combining molecular marker and morphological traits (Franco et al. 2001). Combining molecular markers and morphological traits would precisely reveal genetic variation in breeding program of recurrent selection, but there is still no such research in wheat up to the present.
The use of male sterility reduces the labor required for hand crossing and increases crossing efficiency, facilitating application of recurrent selection in wheat. Many studies have showed that the sterility was easily to be maintained and restored and no significant side effects of cytoplasm were found in D2-type cytoplasm male sterile (CMS) line of wheat (Wu et al. 1995; Liu et al. 1999, 2002). These results suggest that D2-type CMS line is a material of potential in hybrid wheat breeding and recurrent selection program. Using D2-type CMS line, we had constructed a wheat (Triticum aestivum L.) recurrent selection population (agronomic gene pool) in which many excellent agronomic traits had high frequencies. The objectives of this study were: (1) to detect the genetic diversity of the gene pool constructed through recurrent selection; (2) to assess the agronomic value of the gene pool for wheat improvement and further application.
Materials and methods
Establishment of gene pool through recurrent selection
The D2-type CMS line msD2-CA8057 was used as male sterile material and 30 elite varieties and breeding lines (P1∼P30), possessing either good performance or special features, such as high yield, good quality, drought tolerance and disease resistance (Table 1), from six provinces of China were utilized as parents during recurrent selection process, and mass selection procedure was applied in this breeding program. In 1994, the CMS line and 30 parents were planted in field in a 2 × 3-plot matrix. Each plot contained 10 rows with 1 m in length and 0.25 m in width. The array of materials in the rows was CMS, P1, CMS, P2, CMS, P3 ... CMS, P29, CMS, P30, in order that the parents could cross randomly with CMS lines. In the next year (1995), seeds of the CMS rows were harvested and mixed together, then the mixed seeds were planted with the 30 parents as array as the year before. In flowering season of 1996, male sterile plants were marked (some of the plants in the CMS rows had been restored). Seeds from the marked plants were harvested and mixed together in harvest season, and then planted as male sterile materials in the next cycle. During the followed proceeding, male sterile individuals were further selected for multi-agronomic characters: plant structure type, plant height, spike type and resistant to powdery mildew of wheat in field. During the recurrent selection process, selection pressure was low in first several cycles, and then increased with the advance of recurrent selection cycles. The selection pressure had been arrived 8% in recent years. After several cycles of selection under high intensity, gene pool had been established. In May of 2004, 104 individuals of gene pool were randomly selected and denoted as L1∼L104. Five morphological traits of the selected individuals were surveyed. Leaves from the 104 individuals and the 30 parents were sampled for genomic DNA extraction.
Morphological traits
The five surveyed morphological traits were plant height, number of spikes of each plant, spike length, number of spikelets per spike and leaf area (leaf area = leaf length × leaf width × 0.78) (Table 2).
SSR analysis
Total genomic DNA was extracted by the CTAB method described by Saghai-Maroof et al. (1984). The sequence and character of wheat SSR primers were from Röder et al. (1998); Song et al. (2005) and website (http://wheat.pw.usda.gov/ggpages/SSRclub/Sourdille/). 150 pairs of wheat SSR primers covering whole genome of wheat were screened, and finally 24 pairs of primers determined stable, legible, and polymorphic bands were chosen in present research (Table 3). Polymerase chain reactions (PCR) were performed in a volume of 20 μl using a thermocycler (Whatman Biometra T1 Thermocycler, Göttingen, Germany). The reaction mixture contained 10 mM Tris-HCl (pH 8.0), 10 mM KCl, 2 mM MgSO4, 8 mM (NH4)2SO4, 0.05% NP-40, 150 μM dNTPs, 0.2 μM primers, 50 ng template DNA and 1 U Taq DNA polymerase. PCR reactions were programmed for an initial step for 3 min at 94°C, followed by 39 cycles of 45 s at 94°C, 45 s for annealing at primer-specific temperature and 45 s for extension at 72°C, then a final step at 72°C for 5 min.
The amplification products were separated on 4% denaturing polyacrylamide gel for about 40 min with constant power of 75 W and visualized by silver staining method.
Data analysis
The presence or absence of each single fragment was coded by 1 or 0, respectively and scored for a binary data matrix. The number of alleles in the 30 parents and the gene pool was counted respectively. Frequency of each allele was calculated and compared between the 30 parents and the gene pool. Genetic diversity was calculated according to the formula of Nei (1973):
where P ij is the frequency of the jth allele for ith locus summed across all alleles for the locus. The Student’s t-test was used to estimate the significance of difference in genetic diversity between the 30 parents and the gene pool.
Nei’s genetic distances were computed for each pair of lines in the 30 parents and for each pair of individuals in the gene pool using NTSYS-pc (Version 2.1) software (Rohlf 2000). The significance of difference in genetic distance was estimated by Mann-Whitney U test between the 30 parents and the gene pool.
Principal coordinates analysis (PCoA) was performed based on the SSR dataset to examine the internal genetic structure of the gene pool. An UPGMA cluster analysis was undertaken based on the genetic distance matrix and a dendrogram was constructed. To test the goodness of fit of the clustering to the genetic distance data, the cophenetic correlation coefficient was calculated. All the trait values were standardized and Euclidian distances between individuals were calculated based on the standardized trait values. Another UPGMA cluster analysis was performed based on Euclidian distance matrix producing a second dendrogram and the cophenetic correlation coefficient was computed. Correlation coefficient of genetic distance matrix derived from SSR data and Euclidian distance matrix was calculated to detail the correlation of molecular markers and morphological traits.
Results
SSR analysis
The 24 SSR markers detected 25 loci on 14 different chromosomes. In total, 117 alleles were detected in the 30 parents while 115 alleles were detected in the gene pool. The number of alleles per locus was on average 4.7 in the 30 parents and 4.6 in the gene pool, with a range from two (Xbarc228, Xbarc130, Xbarc320 and Xbarc175) to nine (Xbarc319). Considering different genomes, it was shown that the largest number of alleles per locus occurred in the B genome (6.3) compared with A (5.4) and D (3.5) genomes (Table 4). For different homologous chromosome groups (without group 1 data), the largest allele number was detected in groups 2 and 7 in the 30 parents (group 2 was largest in the gene pool), whereas the least number was observed in group 6.
Frequency of each allele in the 30 parents and the gene pool was compared. The results revealed that there were 44 alleles whose frequency variations were large than 10 percent. These alleles were in 18 loci, accounting for 72 percent of total loci. The allele frequency variation indicated the genetic constitution of the gene pool had been different from that of the 30 parents.
Genetic diversity
The mean value of genetic diversity of the total loci in the gene pool (0.54) was slightly smaller than that in the 30 parents (0.56), but the range of variation in the gene pool (0.07–0.82) was larger than that in the 30 parents (0.18–0.8) (Table 3). In the gene pool, the mean values of genetic diversity for the A, B and D genomes were 0.49, 0.73 and 0.52, while they were 0.52, 0.73 and 0.53 in the 30 parents (Table 4). The largest value occurred in the B genome, which was consistent with previous studies (Huang et al. 2002; Khlestkina et al. 2004). For different homologous chromosome groups (without group 1 data), it was shown that the largest genetic diversity value was found in group 7 (0.68) followed by group 2 (0.66) and the least was group 3 (0.45) in the gene pool; whereas in the 30 parents the genetic diversity value in group 2 was largest (0.7) followed by group 7 (0.65) and the least was group 3 (0.48) (Table 4). It could be observed that genetic diversity was basically accordant in the 30 parents and the gene pool. Although the genetic diversity of the gene pool was smaller than that of the 30 parents on most genomes and homologous chromosome groups (Table 4), statistical test revealed that it had no significant difference between genetic diversities of the gene pool and that of the 30 parents.
Genetic distance
Genetic distance of the 30 parents was 0.91 on average with a range from 0.08 to 2.12, by comparison, that of the gene pool averaged 0.64 and ranged from 0.05 to 1.65 (Table 5). The distributions of genetic distance in the 30 parents and the gene pool were quite different (Fig. 1). Evaluation of difference of mean value showed that there was a significant difference in genetic distance between the gene pool and the 30 parents.
Principle coordinates analysis (PCoA) and cluster analysis
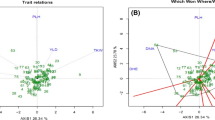
PCoA result based on SSR data of the gene pool showed that most of the 104 individuals clustered in three distinct groups, but there were still high variations between and within groups (Fig. 2).
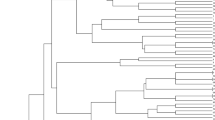
Dendrogram based on the genetic distance matrix of the gene pool was shown in Fig. 3. The cophenetic correlation coefficient was 0.66, indicating that cluster result had a good fit to the genetic distance matrix. The gene pool could be divided into three main groups at 0.57 point: A (24 individuals), B (26 individuals) and C groups (22 individuals), which was consistent with the result of PCoA analysis (Figs. 2, 3). Mean value of genetic distance within the three main groups was calculated, respectively, and analysis of variance was performed to evaluate difference of the mean values among the three groups. The results showed that the three clusters had significant difference one another (Table 6).
The dendrogram derived from cluster analysis of morphological traits was shown in Fig. 4. The result of clustering had a good fit to the Euclidian distance matrix with cophenetic correlation coefficient 0.71. Clustering result showed that most individuals were in a group while the others scattered. The result indicated that phenotypes of individuals in the gene pool became relatively uniform under high selection pressure. Correlation coefficient of genetic distance matrix and Euclidian distance matrix was 0.038, showing no significant correlation between the two matrices.
Discussion
Different number of alleles has been detected in wheat using microsatellite markers. Huang et al. (2002) reported an average allele number of 18.1 in 998 gene bank accessions of hexaploid wheat originated from 68 countries of five continents. Khlestkina et al. (2004) found an average allele number of 6.6 in 54 Siberian old and modern common spring wheat varieties. Roussel et al. (2005) reported an average allele number of 16.4 in 480 wheat varieties originating from 15 European geographical areas and released from 1840 to 2000. The average number of allele obtained in this study was 4.7 in 30 parents. The value was lower than most previous studies, but it was comparable with Zhang’s results, which detected 5.5 alleles per locus in Chinese 43 wheat varieties (Zhang et al. 2002).
After cycles of recurrent selection, the genetic variation in the gene pool didn’t statistically decrease, indicating genetic diversity was maintained in the gene pool. It was consistent with the result from Brassica napus (Yuan et al. 2004).
Genetic distance within the gene pool was significantly different to that within the 30 parents (Table 5, Fig. 1). This result didn’t mean that the variation was severely decreased under high selection intensity. In our research, most loci in 30 parents are homozygous, whereas a majority of loci in gene pool are heterozygous after recurrent selection. Hybridization and recombination may have caused the significant reduction of genetic distance. In maize, Huang et al. (2004) reported that genetic distance was reduced but not significantly after two cycles of recurrent selection, in which base population evaluated was heterozygous.
PCoA and clustering analysis based on SSR data showed that a majority of individuals in gene pool clustered in three distinct groups: A, B, and C groups, and the analysis of variance in genetic distance among the three groups indicated that significant difference was found between each two of them. It may reflect that favorable genes or alleles assembled in most individuals after eliminating disadvantageous genetic background in the gene pool; on the other hand, it is possibly related to the distributive difference of different favorable genes or alleles, especially quantity trait loci (QTLs) in population.
We detected no correlation between the SSR data and morphological data in present study, which was consistent with the result obtained by Marić et al. (2004). But Hamza et al. (2004) reported a relatively high correlation between morphological traits and SSR markers in Tunisian winter barley. Therefore, correlation between molecular markers and morphological traits should be further studied.
In conclusion, the microsatellite analysis in this study revealed that the genetic diversity was maintained in gene pool after recurrent selection, while the gene pool had become convergent whether on DNA level or on morphological traits. The effect of recurrent selection is evident and the gene pool is suitable for further wheat breeding program.
References
Busch RH, Kofoid K (1982) Recurrent selection for kernel weight in spring wheat. Crop Sci 22:568–572
De Koeyer DL, Phillips RL, Stuthman DD (1999) Changes in genetic diversity during seven cycles of recurrent selection for grain yield in oat, Avena sativa L. Plant Breed 118:37–43
Delzer BW, Busch RH, Hareland GA (1995) Recurrent selection for grain protein in hard red spring wheat. Crop Sci 33:924–928
Djè Y, Heuertz M, Lefèbvre C, Vekemans X (2000) Assessment of genetic diversity within and among germplasm accessions in cultivated sorghum using microsatellite markers. Theor Appl Genet 100:918–925
Elizondo Barron J, Pasini RJ, Davis DW, Stuthman DD, Graham PH (1999) Response to selection for seed yield and nitrogen (N2) fixation in common bean (Phaseolus vulgaris L.). Field Crops Res 62:119–128
Franco J, Crossa J, Ribaut JM, Betran J, Warburton ML, Khairallah M (2001) A method for combining molecular markers and phenotypic attributes for classifying plant genotypes. Theor Appl Genet 103:944–952
Hamza S, Hamida WB, Rebaï A, Harrabi M (2004) SSR-based genetic diversity assessment among Tunisian winter barley and relationship with morphological traits. Euphytica 135:107–118
Huang SH, Teng WT, Wang YJ, Dai JR (2004) Genetic diversity analysis of maize recurrent selection populations by SSR marker. Acta Gene Sin 31:73–80
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Jiang G, Wu Z, Huang D (1994) Effects of recurrent selection for resistance to scab (Gibberella zeae) in wheat. Euphytica 72:107–113
Khlestkina EK, Röder MS, Efremova TT, Börner A, Shumny VK (2004) The genetic diversity of old and mordern Siberian varieties of common spring wheat as determined by microsatellite markers. Plant Breed 123:122–127
Liu CG, Wu YW, Hou N, Liu GQ, Zhang CL, Zhang Y (2002) Genetic analysis on restorer genes of D2-type CMS lines of common wheat. Acta Gene Sin 29:531–536
Liu CG, Wu YW, Hou N (1999) Studies of the genetic characters of four isonucleus-alloplasmic lines of common wheat. Acta Gene Sin 26:657–665
Loffer CM, Busch RH, Wiersma JV (1983) Recurrent selection for grain protein percentage in hard red spring wheat. Crop Sci 23:1097–1101
Marić S, Bolarić S, Martinčić J, Pejić I, Kozumplik V (2004) Genetic diversity of hexaploid wheat cultivars estimated by RAPD markers, morphological traits and coefficients of parentage. Plant Breed 123:366–369
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol 35:89–99
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Olmeda-Arcega OB, Elias EM, Cantrell RG (1995) Recurrent selection for grain yield in durum wheat. Crop Sci 35:714–719
Parlevliet JE, van Ommeren A (1988) Recurrent selection for grain yield in early generations of two barley populations. Euphytica 38:175–184
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Publications, New York, USA
Roussel V, Leisova L, Exbrayat F, Stehno Z, Balfourier F (2005) SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theor Appl Genet 111:162–170
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Struss D, Plieske J (1998) The use of microsatellite markers for detection of genetic diversity in barley populations. Theor Appl Genet 97:308–315
Veillet S, Filippi MC, Gallais A (1996) Combined genetic analysis of partial blast resistance in an upland rice population and recurrent selection for line and hybrid values. Theor Appl Genet 92:644–653
Wiersma JJ, Busch RH, Fulcher GG, Hareland GA (2001) Recurrent selection for kernel weight in spring wheat. Crop Sci 41:999–1005
Wilcox JR (1998) Increasing seed protein in soybean with eight cycles of recurrent selection. Crop Sci 38:1536–1540
Wu YW, Zhang CL, Liu CG (1995) Breeding of wheat male sterile line with Aegilops crassa (6 x) cytoplasm and research of its characters. Chin Sci Bull 40:243–247
Yuan M, Zhou Y, Liu D (2004) Genetic diversity among populations and breeding lines from recurrent selection in Brassica napus as revealed by RAPD markers. Plant Breed 123:9–12
Zhang X, Li C, Wang L, Wang H, You G, Dong Y (2002) An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties. I. Information from large-scale planted varieties and cornerstone breeding parents in Chinese wheat improvement and production. Theor Appl Genet 106:112–117
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, J., Liu, L., Hou, N. et al. Genetic diversity of wheat gene pool of recurrent selection assessed by microsatellite markers and morphological traits. Euphytica 155, 249–258 (2007). https://doi.org/10.1007/s10681-006-9326-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-006-9326-x