Abstract
Strain NF87–2 is an aerobic, non-motile and rod-shaped Gram-negative bacterium. It was isolated from the rhizosphere of green pepper. In the present study, sequence analyses of the 16S rRNA and copA genes revealed that strain NF87–2 belongs to the species Lysobacter capsici. Strain NF87–2 could produce chitinase, cellulase, protease and siderophore. The strain showed a broad spectrum of antifungal activities against phytopathogens, including Alternaria brassicae, Rhizoctonia solani, Sclerotinia sclerotiorum, Botrytis cinerea, Colletotrichum gloeosporioides and Fusarium oxysporum. The secondary metabolites secreted by strain NF87–2 could inhibit the growth of both bacteria and fungi, but the mixture of peptides and proteins extracts from a suspension of strain NF87–2 could only inhibit the mycelia growth of fungi. Our results also have shown that strain NF87–2 could control pepper damping off caused by R. solani effectively in a greenhouse setting. Our findings provide a new source for a biocontrol agent and shed light on the mechanism of the antagonistic activity of L. capsici.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil-borne plant pathogens can cause significant reductions in both yield and quality in vegetable crops. Management of plant pathogens with pesticides has resulted in environmental pollution and pathogen resistance (Fernando et al. 2005). It was reported that soils suppressive of various soil-borne plant pathogens such as Gaeumanomyces graminis var. tritici, Fusarium oxysporum, Fusarium solani, Phytophthora cinnamomi, Rhizoctonia solani and Sclerotinia sclerotiorum, can limit disease development and provide a favorable environment (Cook and Baker 1983). The suppressive activity is due to the presence of antagonistic microbes. For example, plant growth-promoting rhizobacteria (PGPR), such as Pseudomonas, Bacillus and Azospirillum species, play a vital role in disease control, crop protection and growth promotion (Fernando et al. 2005).
The genus Lysobacter was first described as PGPR bacteria by Christensen and Cook (1978). Members of this genus are strongly proteolytic and characteristically lyse a variety of microorganisms such as Gram-positive (including Actinomycetes), Gram-negative bacteria, fungi, and green algae, as well as nematodes (Qian et al. 2009). Many strains of Lysobacter can produce exoenzymes and antibiotics that are of ecological and biotechnological interest (Postma et al. 2010). These strains can be potentially used as biocontrol agents (Hayward et al. 2010).
Lysobacter enhances the disease resistance of plants via various mechanisms, such as production of chitinases, lipases, proteases and β-1,3-glucanases (Ko et al. 2009; Palumbo et al. 2003), plant colonization (Islam et al. 2005; Ji et al. 2008), and induction of systemic resistance (Kilic-Ekici and Yuen 2004). In addition, Lysobacter is a source of bioactive natural products (Nett and Konig 2007; Xie et al. 2012; de Bruijn et al. 2015). For example, dihydromaltophilin, a heat stable anti-fungal factor (HSAF), has been shown to have antifungal activity and was isolated from L. enzymogenes OH11, a bacterium used in the biological control of fungal diseases in plants (Lou et al. 2011). 4-Hydroxyphenylacetic acid, which is used for control of Phytophthora blight, was isolated from L. antibioticus HS124 (Ko et al. 2009). L. enzymogenes 3.1 T8 was found to produce a polyketide synthase that can be used against Pythium aphanidermatum (Folman et al. 2004). Several Lysobacter species produce potent antibacterial metabolites, including cyclic peptide lysobactin and cyclic lipodepsipeptide WAP-8294A2 (Wang et al. 2013).
Strain NF87–2 was isolated from the rhizosphere of green pepper and was described as an antagonist to many plant pathogens, including Alternaria brassicae, Botrytis cinerea, Colletotrichum gloeosporioides, F. oxysporum, R. solani and S. sclerotiorum (Liu 2016). In the current study, we further characterized the strain NF87–2 by identifying its active compounds and determining its effects on soil-borne pathogens in vitro. Our results provide insight into the antifungal mechanism of strain NF87–2 against soil-borne plant pathogens.
Materials and methods
Microorganisms, media and culture conditions
Bacterial and fungal strains used in this study are listed in Table 1. The bacterial strains were routinely cultured at 28 °C on Luria-Bertani agar (LBA) and preserved in 20% glycerol at −80 °C. The plant-pathogenic fungi were cultured on potato dextrose agar (PDA) at 28 °C for 2–7 days and stored at 4 °C.
Morphological, biochemical, and physiological features
Gram staining, flagella staining, and motility assays were performed according to standard protocols (Fang 1998). The morphology of individual cells was observed using a transmission electron microscope (TEM) (Tecnai G2 F30, FEI, Thermo Fisher Scientific, Waltham, MA). Activities of chitinase, cellulase, protease and siderophore were detected as described previously (Liu et al. 2012). Briefly, nutrient broth (NB) medium was used for the chitinase assay. NF87–2 cells were spotted onto the plates and incubated at 28 °C for 3 days to observe the clear zones around the colony. Carboxymethyl cellulose (CMC) agar medium was used for cellulase assay. NF87–2 cells were spotted onto the plates and incubated at 28 °C for 3 days, and Gram’s iodine solution was poured onto the CMC plates to observe the clear zones around the colony. Skim milk agar was used for the protease assay. NF87–2 cells were spotted onto the surface of the medium and incubated at 28 °C for 3 days. Protease production was determined by formation of a clear zone around the colony due to breakdown of milk protein. Chrome Azurol S (CAS) agar containing 0.01 mmol L−1 FeCl3·6H2O was used for the siderophore assay. NF87–2 cells were spotted onto the surface of the medium and incubated at 28 °C for 3 days. Siderophore activity was represented by a yellow zone around the colony.
NF87–2 cells were prepared as follows: strain NF87–2 was cultured in LB liquid medium at 28 °C on a shaker (150 r/min) for 36 h. Then the culture was centrifuged at 10,000×g at 10 °C for 10 min, after which the cell pellet was re-suspended in 0.05 M phosphate buffer (pH 7.2) to the desired concentration of 2 × 108 colony-forming units per mL (CFU/mL). Next, a 5-μL suspension of strain NF87–2 was inoculated onto each plate and incubated.
16S rRNA sequence and phylogenetic analysis
Genomic DNA extraction of strain NF87–2 was performed using the cetyl trimethylammonium bromide (CTAB) protocol (Ausubel 1988). The 16S rRNA was amplified by polymerase chain reaction (PCR) from genomic DNA by using universal primers 27f and 1492r (Lane 1991). PCR was performed as previously described (Sambrook et al. 1989). Direct sequencing of the PCR products was performed by Invitrogen (Shanghai, China). The 16S rRNA sequence was generated from three independent sequencing reactions, and the Seqman program of the Lasergene software package (DNASTAR Inc., Madison, WI) was used to construct the final sequence of the 16S rRNA gene. Multiple sequence alignment with 16S rRNA gene sequences from the GenBank database (www.ncbi.nlm.nih.gov) was performed using the Lasergene software package. Phylogenetic analysis with a bootstrap analysis of 1000 samplings was conducted using MEGA version 4.0 software (Tamura et al. 2007).
Cloning of copA gene from Lysobacter sp. NF87–2 and phylogenetic analysis
Lysobacter is able to resist copper ions and its resistance is probably due to the presence of conserved genes coding for copper oxidase (copA) and copper exporting PIB-type ATPases (ctpA) (Puopolo et al. 2014b). The gene encoding CopA in Lysobacter sp. NF87–2 was cloned using previously reported primers. PCR was performed as previously described (Puopolo et al. 2014b). The amplified PCR product was used for direct sequencing by Invitrogen (Shanghai, China), and alignment of nucleotide sequences was carried out using the BLAST function of GenBank (www.ncbi.nlm.nih.gov).
Antibacterial and antifungal activities of Lysobacter sp. NF87–2
A 5-μL suspension of strain NF87–2 sample as described above was inoculated onto each LBA plate and incubated at 28 °C for 72 h. Distilled water was used as the control. The bacterial strains Bacillus megaterium 329 and Xanthomonas oryzae pv. oryzicola (Xooc) strain RS11 (Table 1) were used as test strains. Suspensions of the two test strains were prepared in the same way as strain NF87–2.The antibacterial effect of NF87–2 was evaluated using the plate bioassay protocol on LBA plates as described previously (Gu et al. 2009). Each treatment was replicated three times. Each experiment was repeated three times independently. The diameters of the inhibitory zones of Lysobacter sp. NF87–2 were measured when the test strains covered the entire control plate.
The fungal pathogens (Table 1) were grown on PDA plates at 28 °C for 72 h. A 0.5-cm-diameter mycelial disk was placed at one side of a new PDA plate (9-cm-diameter), and a 5-μL suspension of strain NF87–2 was inoculated on the other side of the PDA plate, equidistant between the plate edge and center. Distilled water was used as the control. Each treatment was replicated three times. Each experiment was repeated three times independently. The diameters of the pathogen colonies and the inhibition bands of Lysobacter sp. NF87–2 against the fungal pathogens were measured when the mycelia covered the entire control plate.
The inhibition rate was calculated using the following formula: inhibition rate = (diameter of colonies of control – diameter of colonies of treated group)/(diameter of colonies of control) × 100%.
Extraction and evaluation of active compound of strain NF87–2
Strain NF87–2 was grown on LB liquid medium at 28 °C on a shaker (150 rpm) for 72 h. Secondary metabolites were extracted from a 100-mL fermented liquid of strain NF87–2 following a previously described method (Ko et al. 2009) and dissolved in 1 mL water/methanol (vol/vol = 1:2). The mixture of peptides and proteins (MPP) was extracted from 100 mL fermented liquid of strain NF87–2 by the ammonium sulphate method (Liu et al. 2010) and dissolved in 10 mL of 25 mM phosphate buffer (pH 7.0).
Secondary metabolites and MPP were evaluated using the plate bioassay protocol on LBA and PDA plates as described previously (Liu et al. 2010). Briefly, four 0.5-cm-diameter holes were punched in each LBA or PDA plate equidistant from the plate edge and center using a stainless-steel rod. Then 5 μL, 10 μL or 20 μL of suspension was added to each hole. As a control, 20 μL of sterile water/methanol (for secondary metabolites) or phosphate buffer (for MPP) was applied in the left hole. Each treatment was replicated three times. Each experiment was repeated three times. The diameters of the inhibitory zones or bands were measured to evaluate antimicrobial activities of secondary metabolites or MPP secreted by strain NF87–2 when the test strains covered the entire control hole.
Biocontrol effect of strain NF87–2 against R. solani under greenhouse conditions
Pepper damping off caused by R. solani is a common soil-borne disease throughout pepper-growing areas. The pepper cultivar “sujiao 5” was used as the host plant to test the biocontrol effect of strain NF87–2 against R. solani. The sujiao 5 seeds were provided by the Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences. Fifty seeds were planted in each plastic tray (35 mm × 25 mm) and supplied with a vermiculite/nutrition soil mixture at a 1:1 ratio. The plants were grown in a greenhouse at the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, with 12-h light/dark cycle at 28 °C and 75% relative humidity.
The fungal pathogen R. solani was grown on PDA plates at 28 °C for 3 days. A 0.5-cm-diameter mycelial disk was then transferred to a new PDA plate (9-cm diameter) and grown for 3 days when mycelia covered the entire PDA plate. The suspension of strain NF87–2 with the concentration of 2 × 108 CFU/mL was prepared as described above. The bacterial strain B. subtilis PTS-394, an antagonistic bacterium against many fungal pathogens (Liu et al. 2012), was used for comparison. B. subtilis PTS-394 was cultured under the same condition as the strain NF87–2.
The biocontrol experiment involved five different treatment (1) treatment with R. solani only (used as control): the soil was inoculated with the fungal pathogen R. solani collected from 12 plates (heavy inoculation), six plates (medium inoculation) or three plates (light inoculation); The plates were prepared as described in above paragraph; (2) treatment with R. solani and water-diluted suspension of strain NF87–2: the soil was inoculated with different amounts of R. solani (above (1)) and then treated with 20 mL of a100-fold diluted suspension of strain NF87–2; (3) treatment with R. solani and original solution of NF87–2: the soil was inoculated with different amounts of R. solani (above (1)) and then treated with 20 mL of the original suspension of strain NF87–2; (4) treatment with R. solani and water-diluted suspension of strain PTS-394: the soil was inoculated with different amounts of R. solani (above (1)) and then treated with 20 mL of a 100-fold diluted suspension of strain PTS-394; and (5) treatment with R. solani and original solution of PTS-394: the soil was inoculated with different amounts of R. solani (above (1)) and then treated with 20 mL of the original suspension of strain PTS-394. Soil supplied with the same amount of sterile water was used as a blank treatment. Each treatment was replicated three times. The experiment was repeated three times. All the treatments were kept at constant humidity and temperature for 3 weeks, and then the rate of seedling emergence was measured and the biocontrol efficiency was calculated as follows:
Control efficiency = (the rate of seedling emergence of treatment – the rate of seedling emergence of control)/(the rate of seedling emergence of control) × 100%.
Statistical analysis
Data were processed by analysis of variance (ANOVA) using the SAS GLM procedure (SAS Institute, Inc., Cary, NC). P ≤ 0.05 was considered statistically significant, and significant means were further compared by Fisher’s protected least significant difference (PLSD).
Results
Phenotypic characteristics of strain NF87–2
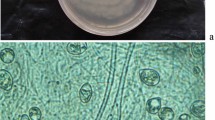
Culturing of strain NF87–2 on LBA plates at 28 °C for 2 days yielded colonies that were yellow in color, circular, and glossy in appearance. Strain NF87–2 was observed to be a non-motile, rod-shaped, Gram-negative bacterium (Fig. 1). Strain NF87–2 was able to grow at 5–35 °C, and the optimal growth temperature was 28–30 °C. Spot assays showed that strain NF87–2 produced chitinase, cellulase, protease and siderophore (Fig. 2).
Assay of chitinase (a), cellulase (b), protease (c) and siderophore (d) secretion by strain NF87–2. Nutrient broth (NB) medium was used for the chitinase assay (a). NF87–2 cells were spotted onto the plates and incubated at 28 °C for 3 days to observe the clear zones around the colony. Carboxymethyl cellulose (CMC) agar medium was used for the cellulase assay (b). NF87–2 cells were spotted onto the plates and incubated at 28 °C for 3 days. Gram’s iodine solution was poured onto the CMC plates to observe the clear zones around the colony. Skim milk agar was used for the protease assay (c). NF87–2 cells were spotted onto the surface of the medium and incubated at 28 °C for 3 days. Protease production was determined by formation of a clear zone around the colony due to breakdown of milk protein. Chrome Azurol S (CAS) agar medium containing 0.01 mmol L−1 FeCl3·6H2O was used for the siderophore assay (d). NF87–2 cells were spotted onto the surface of the medium and incubated at 28 °C for 3 days. A, B and C, enzyme activity is shown as clear zones around colonies on chitin agar, CMC medium and milk medium, respectively. D, siderophore activity is shown as a yellow zone around the colony
Phylogenetic analyses of the 16S rRNA and copA genes
The nearly full-length 16S rRNA gene sequence (1457 bp) of strain NF87–2 was determined and deposited into the GenBank database (Accession No. KJ147476). This sequence shared high identities with those of the Lysobacter species, further confirming that strain NF87–2 belongs to the genus Lysobacter. A phylogenetic tree derived from these 16S rRNA sequences was constructed to illustrate the position of strain NF87–2 and 16 other Lysobacter species. The phylogenetic tree revealed that strain NF87–2 is very closely related to the species L. capsici (Fig. 3).
The copA gene is one of the genetic determinants involved in bacterial resistance to copper (Lejon et al. 2007). Several copA genes belonging to Lysobacter species were collected from GenBank and used for phylogenetic analyses. The results further indicate that strain NF87–2 is a member of L. capisici (Fig. 4).
Antibacterial and antifungal activities of strain NF87–2
The antibacterial activities of strain NF87–2 were determined on LBA plates. The results showed that strain NF87–2 possessed antibacterial activities against the Gram-negative bacterium Xooc strain RS11 and the Gram-positive bacterium B. megaterium 329 (Fig. 5a, b), with clear inhibitory surrounding the colonies of strain NF87–2. The diameters of the inhibitory zones against B. megaterium 329 and Xooc strain RS11 were 19.2 ± 1.6 mm and 20.5 ± 1.4 mm, respectively. In addition, strain NF87–2 strongly inhibited the radial growth of some phytopathogenic fungi on PDA agar (Fig. 5c–h), with clear inhibitory bands surrounding the colonies of strain NF87–2. Compared with those on the control plates, the inhibitory bands against A. brassicae, B. cinerea, C. gloeosporioides, F. oxysporum, R. solani and S. sclerotiorum were 38.1 ± 0.6 mm, 24.2 ± 1.3 mm, 32.4 ± 0.8 mm, 30.5 ± 0.5 mm, 34.3 ± 1.1 mm and 23.7 ± 1.1 mm, with the inhibition rate 42.33 ± 0.67%, 26.89 ± 1.44%, 36.00 ± 0.89%, 33.89 ± 0.56%, 38.11 ± 1.22% and 26.33 ± 1.22%, respectively.
Antibacterial (a–b) and antifungal (c–h) activities of strain NF87–2. For analysis of antibacterial activity, 5 μL suspension of strain NF87–2 was spotted on the center of LBA plates, incubated at 28 °C for 3 days and then sprayed with B. megaterium 329 (a) or X. oryzae pv. oryzicola (Xooc) strain RS11 (b). The over-sprayed plates were further incubated for 24 h at 28 °C. For analysis of antifungal activity, strain NF87–2 was co-cultured with the following fungi on PDA plates (spotted on opposite sides of the plate):A. brassicae (c); B. cinerea (d); C. gloeosporioides (e); F. oxysporum (f); R. solani (g) and S. sclerotiorum (h). The plates were incubated at 28 °C for 2–5 days
Antimicrobial activities of active compounds from the culture suspension of strain NF87–2
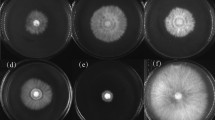
We first examined the antimicrobial activities of the secondary metabolites secreted by strain NF87–2 toward B. megaterium 329, Xooc strain RS11 and several fungal pathogens. As shown in Fig. 6, the inhibitory against B. megaterium 329 (Fig. 6a) and Xooc strain RS11(Fig. 6b) increased significantly as the volume of suspension increased from 5 to 20 μL. Similar results were observed for the tested fungal pathogens. The inhibitory bands for R. solani (Fig. 6c), A. brassicae (Fig. 6d), and S. sclerotiorum (Fig. 6e) were 6.0 ± 0.3 mm, 4.5 ± 0.4 mm, and 11.0 ± 0.6 mm, respectively, when treated with 20-μL suspensions of secondary metabolites of strain NF87–2.
Antibacterial (a–b) and antifungal (c–e) activities of secondary metabolites secreted by strain NF87–2. For analysis of antibacterial activity, different dosages (5 μL, 10 μL and 20 μL) of secondary metabolites secreted by strain NF87–2 were poured into 0.5-cm-diameter holes on LBA plates. 20 μL water/methanolwas used as control (CK). The plates were sprayed with B. megaterium 329 (a) and X. oryzae pv. oryzicola (Xooc) strain RS11 (b) and then incubated at 28 °C for 3 days. For analysis of antifungal activity, different dosages (5 μL, 10 μL and 20 μL) of secondary metabolites secreted by strain NF87–2 were poured into 0.5-cm-diameter holes on PDA plates. 20 μL water/methanol was used as control (CK). Different fungi, including R. solani (c), A. brassicae (d), and S. sclerotiorum (e), were spotted at the center of the plate (equidistant from each hole) and incubated at 28 °C for 3 days. The diameters of the inhibitory zones or bands were measured to evaluate antimicrobial activities of secondary metabolites secreted by strain NF87–2
Next, we examined the antimicrobial activities of MPP extracted by the ammonium sulphate method. The results showed that MPP had no obvious inhibitory effect against B. cinerea, R. solani, B. megaterium 329, or Xooc strain RS11, but an apparent antifungal effect toward A. brassicae, C. gloeosporioides, F. oxysporum, and S. sclerotiorum (Fig. 7). However, the inhibition by MPP was much weaker than that by the strain NF87–2 itself, suggesting that secondary metabolites and some unknown natural products secreted by strain NF87–2 may also contribute to the antimicrobial activity in addition to MPP. Among the tested fungal pathogens, F. oxysporum and S. sclerotiorum showed the highest sensitivity to MPP. The inhibitory bands were 7.5 ± 1.2 mm and 9.2 ± 1.6 mm, respectively, after treatment with 20-μL suspensions of MPP of strain NF87–2.
Antifungal activities of the mixture of peptides and proteins (MPP) secreted by strain NF87–2. Different dosages of MPP (5 μL, 10 μL and 20 μL) secreted by strain NF87–2 were poured into 0.5-cm-diameter holes on PDA plates. 20 μL phosphate buffer was used as control (CK). Different fungi, including A. brassicae (a), C. gloeosporioides (b); F. oxysporum (c), and S. sclerotiorum (d), were spotted at the center of the plate (equidistant from each hole) and then incubated at 28 °C for 3 days. The diameters of the inhibitory bands were measured to evaluate antimicrobial activities of MPP secreted by strain NF87–2
Biocontrol efficacy of strain NF87–2 against R. solani in greenhouse settings
Greenhouse experiments revealed that inoculation of the soil with R. solani greatly reduced the rate of seedling emergence of the control (Table 2). Addition of suspension of NF87–2 significantly recovered the rate of seedling emergence, especially in the medium-inoculated soil, in which the rate of seedling emergence increased from 52.95 ± 1.31% to 80.24 ± 1.50%, with a biocontrol efficiency of 51.54 ± 1.44%. The biocontrol efficacy of the original suspension of NF87–2 was higher than that of the water-diluted liquid, which was 35.03 ± 1.26% in the medium-inoculated soil. No significant difference in the seedling emergence rate was observed between the groups treated with original suspension and diluted liquid of NF87–2 in the light-inoculated soil. The biocontrol efficiency ranged from 21.69 ± 1.25% to 15.50 ± 1.58%. With application of the same treatment, the rate of seedling emergence in the heavy-inoculated soil was much lower than those in the light-inoculated soil and medium-inoculated soil.
B. subtilis PTS-394 was also a good biocontrol strain. In this study the biocontrol efficiency treatment with PTS-394 suspension reached 53.22 ± 2.03% and 23.09 ± 1.87% in the medium inoculated soil and heavy inoculated soil, respectively. When treated with diluted liquid of PTS-394, the biocontrol efficiency reduced from 39.91 ± 2.33% to 6.35 ± 1.93%. (Table 2).
Discussion
Some strains of Lysobacter spp., such as 3.1 T8, HS124, AZ78, and OH11, have been reported to be effective at controlling both soil-borne and foliar diseases (Folman et al. 2004; Ko et al. 2009; Puopolo et al. 2014a; Qian et al. 2009). In the present work, the antagonistic effects of Lysobacter strain NF87–2 against some bacteria and plant-pathogenic fungi were studied. In addition, repeated experiments indicated that the antagonistic capability of the NF87–2 strain was stable and persistent. These data suggest that strain NF87–2 has potential as a biocontrol agent.
It has been reported that resistance to copper is a trait shared by Lysobacter species and associated with the presence of the copper oxidase gene (copA) (Puopolo et al. 2014b). Phylogenetic analysis indicated that the copA sequence of L. capsici AZ78 clustered with gene homologs from Azotobacter vinelandi strain CA6, Pseudomonas fluorescens strain PfO-1, and P. stutzeri strain CCUG 29243, bacterial species that are phylogenetically distant from the genus Lysobacter (Puopolo et al. 2016). In the present study, genes coding for CopA were collected in Lysobacter spp. and some other bacterial strains for phylogenetic analysis. We found that gene copA was commonly expressed in many Lysobacter species. Phylogenetic analysis of copA sequences further confirms the sequence analysis of 16S rRNA gene and strongly suggests that Lysobacter sp. NF87–2 maybe a member of L. capsici.
It is well known that certain Lysobacter strains can produce antibacterial and antifungal products. In the present study, we demonstrated that secondary metabolites and MPP play a key role in the biocontrol activity of strain NF87–2 against fungal pathogens. Secondary metabolites secreted by strain NF87–2 could significantly inhibit the mycelial growth of R. solani, A. brassicae, and S. sclerotiorum, while MPP extracted from the suspension of strain NF87–2 could inhibit the mycelial growth of A. brassicae, C. gloeosporioides, F. oxysporum, and S. sclerotiorum. The results indicate that different phytopathogenic fungi have different sensitivities to active compounds of strain NF87–2. We also showed that secondary metabolites secreted by strain NF87–2 could inhibit the growth of B. megaterium and Xooc, whereas MPP could not inhibit the growth of bacteria. This suggests that different mechanisms may be employed by strain NF87–2 against bacterial and fungal pathogens. The bacteria tested in the study were only sensitive to secondary metabolites, whereas the tested fungi were sensitive to both secondary metabolites and MPP.
It is currently unknown which types of antimicrobial compounds are released by the strain NF87–2. Previous studies showed that Lysobacter species could produce secondary metabolites with antimicrobial activities, such as antifungal HSAF and antibacterial WAP-8294A2 produced by L. enzymogenes C3 and L. capsici 55, lysobactin produced by L. gummosus 3.2.11, and phenazine with antifungal activity produced by L. antibioticus ATCC29479 (de Bruijn et al. 2015; Wang et al. 2013). These reports provide a good basis for us to explore whether NF87–2 could produce these characterized and potentially new secondary metabolites via genome mining in future.
Most L. capsici strains have been recently recognized as biocontrol agents for plant-pathogenic fungi, oomycetes, and nematodes. For example, L. capsici YS1215 can produce lactic acid and lytic enzymes to control the root-knot nematode (Lee et al. 2014; Lee et al. 2015). Four strains of L. capsici showed in vitro activity against 9 fungi and 4 oomycetes pathogens, except for X. campestris (Gómez et al. 2015). Interestingly, antibacterial compounds released by L. capsici AZ78 are toxic to Gram-positive bacteria only (Puopolo et al. 2016). In this study, we found that the strain NF87–2 has two advantages compared to most previously characterized L. capasici strains: (1) it has both antibacterial and antifungal activities in vitro, and (2) it can inhibit the growth of both Gram-positive and Gram-negative bacteria. These results suggest that the antibacterial substances secreted by NF87–2 may be different from those secreted by other L. capsici strains as reported previously.
Biocontrol bacteria with in vitro antagonistic activity may not effectively control plant diseases in the field. For example, strains of L. capsici showed strong in vitro activity against R. solani. However, no significant and consistent suppression of R. solani damping-off disease was observed when the strains were introduced into soil (Gómez et al. 2015). In this study, the strain NF87–2 could inhibit the mycelial growth of many pathogenic fungi including R. solani in vitro and control well the damping off of pepper in the greenhouse. The results suggest that strain NF87–2 is a promising biocontrol bacterium. Additional studies about the biological characteristics of strain NF87–2, such as colonization, survival ability and product formulation, are needed to confirm the biocontrol effect of strain NF87–2 in the field.
Conclusions
In summary, our study identified strain NF87–2 as a new strain of L. capsici that possesses significant antagonistic activities against plant-pathogenic fungi and bacteria, including both Gram-positive and Gram-negative bacteria. Secondary metabolites and mixture of peptides and proteins from the strain NF87–2 were identified as active compounds against bacteria and plant-pathogenic fungi. The antibacterial substances secreted by NF87–2 appeared to be different from other L. capsici strains reported previously. These findings could help expand the biocontrol spectrum of L. capasici strains currently used. Our study could also provide a new source for a biocontrol agent and a solid basis for understanding the antagonistic mechanism of strain NF87–2.
References
Ausubel, F. M. (1988). Current protocols in molecular biology. New York: Greene Pub, Associates.
Christensen, P., & Cook, F. D. (1978). Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. International Journal of Systematic and Evolutionary Microbiology, 28, 367–393.
Cook, R. J., & Baker, K. F. (1983). The nature and practice of biological control of plant pathogens. St. Paul: APS Press.
de Bruijn, I., Cheng, X., de Jager, V., Gómez, E. R., Watrous, J., Patel, N., Postma, J., Dorrestein, P. C., Kobayashi, D., & Raaijmakers, J. M. (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics, 16, 991–1006.
Fang, Z. D. (1998). Methods in plant pathology. Beijing: Chinese Agriculture Press.
Fernando, W., Nakkeeran, S., & Zhang, Y. (2005). Biosynthesis of Antibiotics by PGPR and its Relation in Biocontrol of Plant Diseases, pp 67–109. PGPR: Biocontrol and Biofertilization, Springer Netherlands.
Folman, L. B., Klein, M. J. E. M. D., Postma, J., & Veen, J. A. V. (2004). Production of antifungal compounds by Lysobacter enzymogenes isolate 3.1T8 under different conditions in relation to its efficacy as a biocontrol agent of Pythium aphanidermatum in cucumber. Biological Control, 31, 145–154.
Gómez, E. R., Postma, J., Raaijmakers, J. M., & de Bruijn, I. (2015). Diversity and activity of Lysobacter species from disease suppressive soils. Frontiers in Microbiology, 6, 1243.
Gu, G., Smith, L., Wang, N., Wang, H., & Lu, S. E. (2009). Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochemical and Biophysical Research Communications, 380, 328–332.
Hayward, A. C., Fegan, N., Fegan, M., & Stirling, G. R. (2010). Stenotrophomonas and Lysobacter: Ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. Journal of Applied Microbiology, 108, 756–770.
Islam, M. T., Hashidoko, Y., Deora, A., Ito, T., & Tahara, S. (2005). Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne Peronosporomycetes. Applied and Environmental Mmicrobiology, 71, 3786–3796.
Ji, G. H., Wei, L. F., He, Y. Q., Wu, Y. P., & Bai, X. H. (2008). Biological control of rice bacterial blight by Lysobacter antibioticus strain 13-1. Biological Control, 45, 288–296.
Kilic-Ekici, O., & Yuen, G. Y. (2004). Comparison of strains of Lysobacter enzymogenes and PGPR for induction of resistance against Bipolaris sorokiniana in tall fescue. Biological Control, 30, 446–455.
Ko, H. S., Jin, R. D., Krishnan, H. B., Lee, S. B., & Kim, K. Y. (2009). Biocontrol ability of Lysobacter antibioticus HS124 against Phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Current Microbiology, 59, 608–615.
Lane, D. J. (1991). In E. Stackebrandt & M. Goodfellow (Eds.), 16S/23S rRNA sequencing in nucleic acid techniques in Bacterial Systematics (pp. 115–175). New York: John Wiley & Sons.
Lee, Y.S., Naning, K.W., Nguyen, X.H., Kim, S. B, Moon, J.H., & Kim, K.Y. (2014). Ovicidal activity of lactic acid produced by Lysobacter capsici YS1215 on eggs of root-knot nematode, Meloidogyne incognita. Journal of Microbiology and Biotechnology, 24, 1510–1515.
Lee, Y. S., Nguyen, X. H., Naning, K. W., Park, Y. S., & Kim, K. Y. (2015). Role of lytic enzymes secreted by Lysobacter capsici YS1215 in the control of root-knot nematode of tomato plants. Indian Journal of Microbiology, 55, 74–80.
Lejon, D. P., Nowak, V., Bouko, S., Pascault, N., Mougel, C., Martins, J. M., & Ranjard, L. (2007). Fingerprinting and diversity of bacterial copA genes in response to soil types, soil organic status and copper contamination. FEMS Microbiology Ecology, 61, 424–437.
Liu, Y.Z. (2016). Application of Lysobacter capsici NF87-2, in: Patent, C. (Ed.).
Liu, B., Huang, L., Buchenauer, H., & Kang, Z. (2010). Isolation and partial characterization of an antifungal protein from the endophytic Bacillus subtilis strain EDR4. Pesticide Biochemistry and Physiology, 98, 305–311.
Liu, Y. Z., Chen, Z. Y., Liang, X. J., & Zhu, J. H. (2012). Screening, evaluation and identification of antagonistic bacteria against Fusarium oxysporum f. sp. lycopersici and Ralstonia solanacearum. Chinese Journal of Biological Control, 28, 101–108.
Lou, L., Qian, G., Xie, Y., Hang, J., Chen, H., Zaleta-Rivera, K., Li, Y., Shen, Y., Dussault, P. H., Liu, F., & Du, L. (2011). Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. Journal of the American Chemical Society, 133, 643–645.
Nett, M., & Konig, G. M. (2007). The chemistry of gliding bacteria. Natural Product Reports, 24, 1245–1261.
Palumbo, J. D., Sullivan, R. F., & Kobayashi, D. Y. (2003). Molecular characterization and expression in Escherichia coli of three beta-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. Journal of Bacteriology, 185, 4362–4370.
Postma, J., Nijhuis, E. H., & Yassin, A. F. (2010). Genotypic and phenotypic variation among Lysobacter capsici strains isolated from Rhizoctonia suppressive soils. Systematic and Applied Microbiology, 33, 232–235.
Puopolo, G., Cimmino, A., Palmieri, M. C, Giovannini, O., Evidente, A., & Pertot, I. (2014a). Lysobacter capsici AZ78 produces cyclo(L-pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangiaof Phytophthora infestans and Plasmopara viticola. Journal of Applied Microbiology, 117, 1168–1180.
Puopolo, G., Giovannini, O., & Pertot, I. (2014b). Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiological Research, 169, 633–642.
Puopolo, G., Tomada, S., Sonego, P., Moretto, M., Engelen, K., Perazzolli, M., & Pertot, I. (2016). The Lysobacter capsici AZ78 genome has a gene pool enabling it to interact successfully with phytopathogenic microorganisms and environmental factors. Frontiers in Microbiology, 7, 96.
Qian, G. L., Hu, B. S., Jiang, Y. H., & Liu, F. Q. (2009). Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agricultural Sciences in China, 8, 68–75.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor laboratory Press.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Wang, Y., Qian, G., Li, Y., Wang, Y., Wang, Y., Wright, S., Li, Y., Shen, Y., Liu, F., & Du, L. (2013). Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One, 8, e66633.
Xie, Y., Wright, S., Shen, Y., & Du, L. (2012). Bioactive natural products from Lysobacter. Natural Product Reports, 29, 1277–1287.
Funding
This study was funded by the National Key R&D Program of China (grants 2017YFD0200400) and the Science and Technology Project of Jiangsu Province (BE2018359). This research was also partially funded by Suzhou Science and Technology Projects (No. SNG2018095).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Rights and permissions
About this article
Cite this article
Liu, Y., Qiao, J., Liu, Y. et al. Characterization of Lysobacter capsici strain NF87–2 and its biocontrol activities against phytopathogens. Eur J Plant Pathol 155, 859–869 (2019). https://doi.org/10.1007/s10658-019-01817-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01817-9