Abstract
In fungi, the expression of genes encoding proteins related to parasitism is regulated by several factors, including pH. This study reports the structural and functional characterization of the pacCl gene, which encodes the transcription factor PacC of C. lindemuthianum. The pacCl gene showed reduced expression in acidic pH, and its transcription was activated by elevated extracellular pH. The importance of this gene was demonstrated by the development of a pacC1 disruption mutant line of C. lindemuthianum. The mutant line was able to penetrate the host tissue through differentiation of primary hyphae. However, it was not able to cause maceration on the infected plant tissue. The results suggest that PacCl is a regulator of gene activation, and its expression is required for fungal growth in alkaline conditions, as well as for the transcription of genes necessary for the passage from the biotrophic to the necrotrophic phase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Colletotrichum represents one of the most important groups of phytopathogenic fungi; it is widely distributed worldwide and occurs mainly in tropical and sub—tropical regions of Latin America. Many of these species have been used as models to study differentiation and plant–pathogen interaction (Bailey and Jeger 1992; Perfect et al. 1999).
The species Colletotrichum lindemuthianum (Sacc. & Magnus) is a phytopathogen with a hemibiotrophic life cycle and is responsible for anthracnose of common bean (Phaseolus vulgaris L.), a widespread disease in Brazil, especially in the south and southeast and in mountainous areas such as the southern region of Minas Gerais (Ansari et al. 2004; Damasceno e Silva et al. 2007; Bonett et al. 2008). The use of phytopathogen-resistant cultivars is the main strategy to contain this disease in Brazil (Damasceno e Silva et al. 2007). However, due to the large genetic variability of C. lindemuthianum, the breakdown of resistance prevents the use of cultivars for long periods of time, thus hindering the establishment of a permanent solution to the disease (Rodriguez-Guera et al. 2003; Ansari et al. 2004; Mahuku and Riascos 2004; Barcellos et al. 2011). The large genetic and pathogenic variability found in Colletotrichum species has been extensively studied, and it can be partially explained by recombinant events and mutations caused by the presence of transposons (Casela and Frederiksen 1994; Santos et al. 2012), and by the presence of anastomoses among conidia (Roca et al. 2003; Castro-Prado et al. 2007; Rosada et al. 2010; Franco et al. 2011).
Along with trying to understand the genetic variability in C. lindemuthianum, many researchers have focused on the molecular and regulatory mechanisms involved in the pathogen–host interaction, from the initial contact until disease establishment. In this context, the ambient pH is the most important factor because to survive and multiply, micro-organisms must have mechanisms to adapt to different ambient conditions (Peñalva et al. 2008). In particular, in fungi, the ambient pH plays an important role in determining the transcriptional levels of many genes, influencing growth, physiology, and differentiation processes (Lamb et al. 2001). Moreover, in plant–pathogen interactions, the ambient pH can be dynamically changed by the fungi. While some fungi change the extracellular pH through the production of organic acids, such as oxalic, citric, and glucuronic acids, other fungi, as in the case of Colletotrichum, alkalize the medium by producing and releasing ammonium (Caracuel et al. 2003a; Kramer-Haimovich et al. 2006).
Considering the ambient pH, a regulatory system controlling gene expression was identified and characterized for the first time in the filamentous fungus Aspergillus nidulans (Caddick et al. 1986). Since then, this mechanism has been studied and characterized in a wide variety of filamentous fungi and yeasts, including Saccharomyces cerevisiae (Su and Mitchell 1993), Yarrowia lipolytica (Blanchin-Roland et al. 1994), Penicillium chrysogenum (Suárez and Peñalva 1996), Candida albicans (Brown and Gow 1999), Sclerotinia sclerotiorum (Rollins and Dickman 2001), Fusarium oxysporum (Caracuel et al. 2003a), Ustilago maydis (Elva et al. 2005), Colletotrichum acutatum (You et al. 2007), Wangiella (Exophiala) dermatitidis (Wang and Szaniszlo 2009), Clonostachys rosea (Zou et al. 2010), and Fusarium graminearum (Merhej et al. 2011).
The general function of this regulatory system is well characterized in A. nidulans and requires the expression and integrity of seven dedicated genes, designated pacC, palA, palB, palC, palF, palH, and palI. In this gene group, pacC plays a central role, as it encodes a transcriptional regulator directly involved with the activation and repression of genes controlled by the ambient pH. (Denison 2000; Peñalva and Arst 2002, 2004; Arst and Peñalva 2003; Peñalva et al. 2008). In this work, we characterized the nucleotide sequence that encodes the protein PacC from C. lindemuthianum (denoted PacCl) and functionally demonstrated its essential role as a pathogenic determinant in the interaction with its host.
Methods
Organisms and culture conditions
The experiments were conducted with the isolates UPS9, LV 49 (pathotype 81), and LF (pathotype 89 A2 2–3) of the fungus C. lindemuthianum. These isolates were kept at −80 °C in 25 % glycerol, using a concentrated suspension of conidia (approximately 108 conidia/ml). When necessary, the isolates were activated in potato dextrose agar (PDA) culture medium, pH 6.8, or malt-agar medium (Parisot et al. 2002), for growth at a temperature of 22 °C for 7 days. The susceptible bean (P. vulgaris L.) cultivar La Victoire was used in the pathogenicity experiments, and the resistant cultivar Jalo was used together with La Victoire to measure the apoplastic pH in bean leaves.
Isolation and structural characterization of the pacCl gene
For the isolation of the pacCl gene, the methods described by Benton and Davis (1977) were used for DNA hybridization to single plaques, from the genomic library of C. lindemuthianum, constructed in the Lambda EMBL3 vector using the Lambda EMBL3/BamHI Vector Kit (Stratagene®). For the screening, the probe was a plasmid containing the cDNA corresponding to the pacCl gene, obtained from a cDNA library constructed by the LLPM Laboratory of the Institute of Plant Biology, University of Paris XI, France. The primer pairs pacCl1/pacCl9 and pacCl10/pacCl11 (Table 1) were designed based on the cDNA sequences of the pacCl gene and used for complete sequencing by PCR and inverse PCR (Ochman et al. 1988), respectively. The developed clones were propagated in ultra-competent E. coli DH5α (Inoue et al. 1990). From the deduced amino acid sequence, the programs WoLF PSORT (Horton et al. 2007) and ngLOC (King and Guda 2007) were used for prediction of subcellular localization. For the phylogenetic inferences, the neighbour-joining reconstruction method in MEGA 6.0 software (Tamura et al. 2007) was applied, using homologous sequences deposited in GenBank. The sequences of 31 organisms, including filamentous fungi and yeasts, were used for the analysis. GenBank accession numbers: A. nidulans (CAA87390); A. niger (Q00203); A. fumigatus (XP_754424); A. oryzae (BAB20756); A. parasiticus (Q96UW0); A. giganteus (Q5XL24); P. chrysogenum (Q01864); S. cerevisiae (NP_011836); C. albicans (Q9UW14); S. Sclerotiorum (Q9P413); A. chrysogenum (Q96X49); F. oxysporum (Q870A3); F. verticillioides (Q873X0); N. crassa (Q7RVQ8); C. glabrata (Q6FV94); D. hansenii (Q6BSZ4); K. Lactis (XP-453982); C. neoformans (XP_572292); M. grisea (Q52B93); Y. lipolytica (CAA67927); C. acutatum (ABL96218); T. harzianum (ABK60115); B. fuckeliana (AAV54519); C. dubliniensis (Q873Y3); G. fujikuroi (Q8J1U9); T. rubrum (Q9C1A4); U. maydis (CAG34353); C. gloeosporioides (ACD56154); P. brasiliensis (AAN06812); C. lindemuthianum (JX679084). C. graminicola and C. higginsianum: Colletotrichum Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/).
pacCl gene expression analysis
pacCl gene expression was analyzed by northern blot. Total RNA was extracted from mycelia that had grown for 5 days in buffered liquid medium with an initial pH ranging from 3 to 9. Samples containing 20 μg of RNA were separated by electrophoresis in agarose gel containing formaldehyde and transferred to nylon N+ membranes (Amersham France SA, Les Ulis, France) by capillarity (Sambrook et al. 1989). Hybridizations were performed as previously described (Dufresne et al. 1998). Fragments of the pacCl gene obtained by PCR with the primers Opac1/Opac2 (Table 1) were used as probes. To measure the transcript of the constitutively expressed clgpd gene, the PCR product obtained with the primers Ogpd1/Ogpd2 was labelled and used as probe. The probes were 32P-labelled using the Ready-To-Go DNA labeling kit (Pharmacia Biotech S.A., Saint Quentin-en-Yvelines, France).
qRT-PCR was performed to study the transcription of the pacCl during the course of infection of bean plants. Total RNA was extracted with TRIzol reagent (TRI Reagent, Sigma®). The cDNA synthesis was performed from 1.0 μg total RNA using the Im Prom-IITM Reverse Transcription System kit (Promega). The gpd gene from C. lindemuthianum was used as an endogenous control. The expression analysis was performed according to the standard curve based method for relative real time PCR data processing, proposed by Larionov et al. (2005). The experiments were repeated two times using preparations of total RNA obtained from independent biological samples. The sequences of the oligonucleotides used in this study are listed in Table 1.
Construction of pacCI gene-inactivation vector
Amplification products obtained by the PCR of genomic DNA and the RT-PCR of total RNA from C. lindemuthianum with the primers Opac1/Opac3 (Table 1) were cloned into the TOPO® TA vector (Invitrogen®), resulting in the plasmids pPacgen and pPacrt, respectively. A DNA fragment from pPacrt cleavaged with EcoRI was subcloned into the pBluescript II KS + vector (Stratagene®), producing pBSKpac. This plasmid was used in the construction of the inactivation vector, through the mutagenesis system GPSTM-M (New England Biolabs®). The plasmid pGPS3. Hygro-Amp (kindly provided by Marc-Henri Lebrun, CNRS Bayer Cropscience) was used as a donor vector. For the transposition reaction, 40 ng of donor vector pGPS3. Hygro-Amp and 60 ng of the recipient pBSKpac were mixed with the transposase TnsABC. After the transposition reaction, the restriction enzyme PI-SceI was used for the destruction of the donor vector. The transposition reaction products were used to transform ultra-competent E. coli DH5α (Invitrogen®) cells in kanamycin selective medium. Clones of interest were selected by PCR with the primer pairs Opac1/N and Opac1/S (Table 1) and confirmed using the primer pair Opac1/Opac2 (Table 1), according to the conditions suggested by the manufacturer of the GPSTM-M kit. From this selection, the plasmid pBKSpac::hph2 was developed and used to disrupt the pacCl gene in C. lindemuthianum.
Development of the pacCl mutant
The pBKSpac::hph2 vector was transformed into C. lindemuthianum line USP9. The mycelium, grown in GPYECH liquid media (Ansari et al. 2004) for 48 h, was washed and placed in 100-ml Erlenmeyer flasks, containing 20 ml of 10 mg/ml glucanex in 10 mM phosphate buffer containing 0.8 M KCl as osmotic stabilizer. The obtained protoplasts were washed 3x in STC solution (1 M sorbitol, 50 mM CaCl2 in 10 mM Tris–HCl buffer). At the end of the washes, the protoplasts were re-suspended in STC at a concentration of 108 protoplasts/ml. Next, 200 ml of protoplast suspension, 5 μg of pBKSpac::hph2, and 50 μl of 60 % polyethylene glycol (PEG) 6000 in STC were added to a tube, and the tube was kept on ice. After 20 min, 500 ml of PEG 6000 was added, and the solution was incubated at room temperature for 20 min. At the end of this period, the protoplasts were plated on PDA medium containing 0.56 M sucrose as osmotic stabilizer. After 48 h of protoplast regeneration, a new layer of PDA medium containing 100 μg/ml hygromycin was added to the Petri dishes. The hygromycin-resistant transformants were transferred to new culture medium containing 50 μg/ml hygromycin and subsequently purified by monosporic isolation.
Molecular characterization of the functional deletion of the pacCl locus
The possible pacCl mutants were detected by PCR using the primer set Opac1/Opac2 (Table 1).
DNA hybridization was used to confirm pacCl interruption. First, DNA hybridization was performed with the wild-type isolate to determine the number of pacCl copies in the genome of C. lindemuthianum. For this purpose, 5 μg of total DNA was digested with the restriction enzymes XbaI, EcoRV, and HindIII (Promega®). The probe used was a 1.8-kb DNA fragment corresponding to the pacCl gene, obtained with the primer set pacCl1/pacCl9. To characterize the pacCl − mutant, total DNA from wild-type and mutant isolates were cleaved with HindIII. In this case, two probes were used: the aforementioned 1.8-kb DNA fragment and the empty pBluescript II KS + vector (Stratagene®). To label and detect the DNA fragments, the Gene Images AlkPhos Direct Labeling and Detection System (Amersham Biosciences®) was used. Total DNA from isolates was extracted by the method of Specht et al. (1982).
Phenotypic characterization of transformants in alkaline pH
To verify the influence of the pacCl gene on the growth of C. lindemuthianum, the conidium lines USP9 and Mutpac2 and the transformants Tr1 and Tr3 were inoculated in buffered solid media at pH 3, 6, and 9. In addition, growth curves were performed by inoculating 7-mm mycelial discs from the lines UPS9 and Mutpac2 in the center of Petri dishes containing culture media buffered at different pH values. The radial growth was evaluated at 2-day intervals.
Pathogenicity test
The common bean plant (P. vulgaris cv. La Victoire) was used as the susceptible cultivar in the pathogenicity tests. The seeds were germinated, and the bean plantlets were maintained in growth chambers for 7 days. The surface of the excised cotyledonary leaves were inoculated with a suspension of 107 conidia/ml. These leaves were kept in Petri dishes containing filter-paper discs soaked in water, then incubated at 19 °C in a growth chamber with 8-h dark and 16-h light periods (166 μE/s/m). Six leaves were used from each transformant line tested. The symptoms were observed 5–7 days after inoculation. The isolates showing a reduction or loss of pathogenicity were subsequently inoculated on plants kept in a greenhouse, to confirm their phenotypes. For microscopic observation of infection kinetics, excised bean hypocotyls were inoculated with 10 μl of the same conidial suspension and maintained under the same light, humidity, and temperature conditions.
Observation of infection process by fluorescence microscopy
The hypocotyl epidermal tissues were marked with a stylus, cleared twice for 10 min in 1 M KOH at 70 °C, and mounted in 67 mM K2HPO4 pH 9.0, containing 0.1 % aniline blue. The observations were obtained by epifluorescence microscopy in an Axioskop microscope (using filters BP365, FT395, and LP397 with UV light at 340 to 380 nm wavelength) (Zeiss, Oberkochen, Germany), and digital images were recorded with the aid of a video camera (Sony, Kolen, Germany) coupled to the computer.
Influence of the pacCl gene on the production and secretion of lipase
Tests were performed on solid medium containing 1.5 % agar or in liquid medium (10 g peptone, 5 g NaCl, 0.1 g CaCl2 · 2H2O, 10 ml Tween 20, 100 ml H2O). The dye bromocresol purple (25 mg/l) was used only on solid medium. Plaques were inoculated in the centre with 7-mm discs taken from the edge of the colony, and then the plaques were grown in M3S medium and maintained at 21 °C. After 7 days, the plaques were placed in a refrigerator to allow halos to form, which contained precipitated Ca+2 salts. Enzymatic activity was estimated using an enzymatic index (I), which expresses the ratio between the medium diameter of the halo and the medium diameter of the colony.
Liquid culture media were used to measure the amount of lipase produced by the different lines of C. lindemuthianum. Bottles containing 50 ml of culture medium were inoculated with 1 ml of a conidial suspension containing 107 conidia/ml and shaken at 150 rpm at 21 °C. After 8 days, the culture supernatant was removed, and the amount of lipase was estimated using the lipase dosage kit from Bioclin Laboratories.
Apoplastic pH measurement in the leaves of the susceptible common bean cultivar La Victoire and the resistant cultivar Jalo
Ten leaves were used for each experiment. These leaves were completely immersed in 100 ml of Milli-Q water in a beaker. The beaker was placed under a vacuum for 15 min. The vacuum was broken slowly to allow infiltration of the apoplast. Then, the excess surface water was removed from the leaves. These leaves were placed in syringe into Falcon tubes. The liquid obtained after 5 min of centrifugation at 4000 × g was collected, and the pH was measured with a pH meter.
Results
Structural characterization of pacCl gene
Sequence analysis identified a protein containing 582 amino acids and a stop codon. This ORF was flanked by a 204-bp 5’ UTR and by a 403-bp 3’ UTR. The start codon (ATG) was present in a 5’-CACCATGTC-3’ sequence, similar to that found in other filamentous fungi (Gurr et al. 1987). This sequence included the important A nucleotide of the Kozak consensus sequence (CAMMATGNC) (Kozak 1984) at position −3 relative to the ATG. The comparison between the nucleotide sequence of the pacCl gene and the cDNA library sequence identified three putative introns. The first intron was 60 bp long and located at position 172–232. The second intron was 58 bp long and located at 446–504. The third intron was 74 bp long and located at 582–656. The cis element 5’-GT…AG-3’ was found in all of the identified introns (Ballance 1986). The C. lindemuthianum pacCl gene nucleotide sequence was deposited in GenBank with accession number JX679084.
The alignment of the obtained protein sequence with sequences available in databases showed that its amino acid sequence had high identity and similarity values with the pH-response transcriptional regulator PacC from filamentous fungi. The PacC1 subcellular location prediction by WoLF PSORT and ngLOC showed nuclear localization in both programs with indices 26.5 and 23.51 %, respectively. Using the PacC/Him101 protein sequences in the NCBI database, we performed an alignment and constructed a phylogenetic tree. To construct the phylogenetic tree, sequences from 32 organisms were used, including filamentous fungi and yeast belonging to the Basidiomycota and Ascomycota phyla (data not shown). Interestingly, most clades grouped taxonomically related organisms at the class level. A monophyletic group was formed by the Eurotiomycetes class, including all species of the genus Aspergillus and the species P. chrysogenum, T. rubrum, and P. brasiliensis, with the group being supported by a bootstrap value of 99. The species S. sclerotiorum and B. fuckeliana were grouped in the Leotiomycetes class, with a consistency value of 100. The monophyly of the Sordariomycetes class, which included the three Colletotrichum species examined here (which were also grouped into a single clade, supported by a bootstrap value of 100), the genus Fusarium, and the species M. grisea, N. crassa, T. harzianum, A. chrysogenum, and G. fujikuroi (the species C. lindemuthianum is highlighted in the tree). This clade was supported by a consistency value of 99. Finally, more basal branches included the Saccharomycetes class, represented here by three species of the genus Candida and species D. hansenii, K. lactis, and S. cerevisiae.
pacCl gene expression is pH-dependent
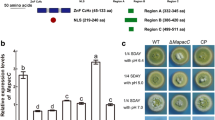
To evaluate the influence of ambient pH on pacCl gene expression, its transcription was analyzed by northern blot. Total RNA was extracted from mycelia grown for 5 days in buffered liquid medium, with an initial pH between 3 and 9. Similarly to other works (Dufresne et al. 1998; Parisot et al. 2002), the constitutively transcribed gpd gene was used as the normalization control. The results are expressed as the ratio of pacCl to gpd band intensity (Fig. 1a). The densitometry values of the amplified products were obtained using ImageJ v1.37 software (http://rsb.info.nih.gov/ij). pacCl transcript was detected at all pH values tested, but the quantitative determination (Fig. 1a) clearly shows that the expression level is pH-dependent, increasing at more alkaline pH values.
Analysis of pacCl gene expression. (a) Effects of ambient pH on pacCl expression in the wild-type isolate UPS9. Isolate UPS9 was cultured for 5 days in media with buffered pH values varying from 3 to 9. (b) pacCl expression during the infection kinetics. gpd - transcript of the constitutively expressed gene
qRT-PCR was used to analyze pacCl expression in C. lindemuthianum during the course of infection of bean plants. The pacCl gene was transcribed in the cells of C. lindemuthianum during all the infection process, but the highest levels were found at the end of the necrotrophic phase. (Fig. 1b).
pacCl gene inactivation
The pacCl gene in C. lindemuthianum was interrupted by homologous recombination. The plasmid pBKSpac::hph2 (Electronic Supplementary Material A1), containing the hygromycin resistance cassette interrupting the cDNA sequence of pacCl, was transformed into the wild-type isolate UPS9.
Mutants of the pacCl gene were detected by PCR using the primer set Opac1/Opac2 (Table 1), as the interrupted pacCl sequence was inserted between these two oligonucleotides in the pBKSpac::hph2 vector. After PCR amplification using the transformant DNA and the Opac1/Opac2 primer set, the expected 1,221-bp fragment from DNA genomic was not amplified in one of the four lines tested (Electronic Supplementary Material A1). It was probable that in this transformant, denominated Mutpac2, the pacCl gene was interrupted. PCR experiments with the primer set Ohph3/Ohph4 (Table 1), which amplified a 712-bp DNA fragment of hph, were performed to verify the transformation event. In all transformant lines, the hph gene was inserted in the genome. The transformants Tr1, Tr3, and Tr4, were uncharacterised hygromycin resistant transformants having heterologous integration. In these lines, pacCl was likely not interrupted. The Tr2 line had a rearrangement in the pacCl locus because the amplified DNA fragment was smaller (861 bp) than expected when using the Opac1/Opac2 oligonucleotides (Electronic Supplementary Material A1).
To confirm pacCl interruption in Mutpac2, DNA hybridization was performed. First, this technique was used to determine pacCl copy number in the genome of the wild-type C. lindemuthianum. The result obtained from total DNA digested with the restriction enzymes ApaI, EcoRV, and HindIII, which did not cleave pacCl, allowed the visualization of a single positive band, confirming that the pacCl gene, similar to pacC/HIM101 of other filamentous fungi and yeasts, was present in only one copy in the genome (Electronic Supplementary Material A1). Then, to analyze and confirm the pattern of integration of the pBKSpac::hph2 vector in Mutpac2, DNA hybridization was again used. As shown in Electronic Supplementary Material A1, with the wild-type DNA and the pacCl probe, a single band was observed because the probe used was a fragment of the gene of interest, which exists as a single copy in the genome. In the case of Mutpac2, two positive bands were observed. When the pBluescriptSK+ cloning vector was used as a probe, no positive bands were observed in the mutant or in the wild-type line. This could be explained by the strategy used in the transformation and by the type of inactivation vector used. Because site-directed mutagenesis was used, we expected that homologous recombination would take place; in other words, a recombination between the pacCl gene contained in the inactivating vector and the wild-type gene contained in the C. lindemuthianum genome. Moreover, because the inactivation vector was designed to carry the pacCl gene interrupted by a central region containing a transposon and the E. coli hph gene, we also expected homologous recombination with gene exchange to occur, leaving no traces of the vector in the fungus genome. Finally, we expected that the hybridization with the 1.8-kb pacCl probe would show two bands in the mutant line because the new construct contained a HindIII restriction site. Fig. 2a and d show that these objectives were successfully achieved.
Effect of ambient pH on the growth of transformants obtained with pBKSpac::hph2 transformation. (a) Conidia from the wild-type isolate (UPS9), the two transformants Tr1, Tr3, and Mutpac2 (mutant with loss of pacCl function) were inoculated in media with pH values of 3, 6, and 9, three days after conidium inoculation and six days after. (b) Growth curve of the wild-type isolate UPS9 and of the null mutant Mutpac2 at different pH values in buffered media
PacCl is necessary for the vegetative growth of C. lindemuthianum in alkaline pH
Six days after inoculation, all tested lines had similar growth patterns in culture medium at pH 6 (Fig. 2a). The radial growth at pH 3 was much smaller compared to neutral pH, demonstrating that C. lindemuthianum grows preferably at a pH close to neutrality compared to more acidic values. The lines UPS9, Tr1, and Tr3 were capable of growing at pH 9, with a radial growth larger than at pH 3. Clearly, high pH values inhibit C. lindemuthianum growth. Interestingly, the growth of the Mutpac2 mutant was strongly inhibited at pH 9 (Fig. 2a).
The growth of the lines UPS9 and Mutpac2 was estimated measuring the radial growth every 2 days after inoculation in buffered media at different pH values (Fig. 2b). The wild-type line grew at pH values from 3 to 9. pH values of 5, 6, and 7 provided for the best growth of UPS9, followed by pH 8 and 9, at which the growth rates were the same. More acidic environments (pH 3 and 4) did not favour growth, which was more noticeable at pH 3, as observed above. The isolate UPS9 is able to change the pH of the culture medium, even if the medium is buffered.
In contrast, the growth of the Mutpac2 line suffered greater inhibition in more alkaline conditions. This line did not show any growth until day 5 after its inoculation in the culture medium. (Fig. 2b). This result clearly demonstrates that PacCl is necessary for the normal growth of C. lindemuthianum in alkaline pH, and it supports the hypothesis that in the transformant Mutpac2, the pacCl gene was interrupted, causing its loss of function.
The pacCl gene is essential for the pathogenicity of C. lindemuthianum
Infection tests in excised leaves and bean plants were performed to determine the effect of pacCl loss of function on C. lindemuthianum pathogenicity.
When inoculated in excised leaves from the susceptible cultivar La Victoire, the C. lindemuthianum wild-type isolate UPS9 induced symptoms typical of anthracnose, with the formation of dark lesions in the primary veins 5 days after inoculation and subsequently extending to the secondary veins, resulting in complete maceration of the tissue 7 days after inoculation (Fig. 3a). The line Mutpac2, which carried an interrupted pacCl gene, did not cause anthracnose symptoms (Fig. 3a), even after longer incubation times. Rare, small, light-brown lesions could be observed on the leaf principal veins; however, the area of the lesions did not expand, showing a nonpathogenic phenotype.
Effect of pacCl gene inactivation on the pathogenicity of C. lindemuthianum inoculated on the susceptible cultivar La Victoire. (a) Excised cotyledonary leaves were inoculated with conidia from the wild-type isolate UPS9, the transformants Tr1 and Tr3, and the mutant line Mutpac2. (b) Plants from the anthracnose-susceptible cultivar La Victoire were pulverized with a conidial suspension, which was obtained from the same lines as in part A. Photographs were taken 9 days after inoculation
Plants of the cultivar La Victoire were inoculated to confirm the phenotype shown by the mutant line. Symptoms appeared 4–5 days after inoculation, beginning with typical brown lesions on leaves, petioles, and hypocotyls. The lesions expanded with time, resulting in tissue maceration, followed by brownish exudates and eventually the death of these plants (Fig. 3b). The inoculation of conidia from this mutant line resulted in light lesions in the primary veins 6 days after inoculation. In contrast to what was observed in excised leaves, some lesions extended to the secondary veins, but no liquid exudates were observed from these regions. Small and rare lesions were also observed in the secondary veins, petioles, and hypocotyls. Because these lesions did not cause plant tissue maceration, the plants remained alive even 9 days after conidium inoculation.
To determine the Mutpac2 phenotype during the process of infection, the infection kinetics of this line were followed microscopically. Two days after conidium inoculation, the Mutpac2 mutant differentiated melanized appressoria, as did the wild-type line (Fig. 4a). Primary and secondary hyphae were observed in wild-type UPS9 7 days after inoculation, coinciding with the macroscopic visualization of plant tissue maceration (Fig. 4b). Mutpac2 differentiated conidia were functional and capable of epithelial tissue penetration, as primary hyphae were seen 7 days after inoculation of conidia (Fig. 4b). Contrary to the wild-type line, Mutpac2 did not differentiate secondary hyphae. In no field of observation were hyphae characteristic of the second phase of nutrition of this fungus observed, which suggests that PacCl is required for the transition from the biotrophic to necrotrophic phase in C. lindemuthianum. Because an organism’s pathogenicity is characterized by its capacity to efficiently colonize the host and complete its life cycle, we propose that PacCl is involved in C. lindemuthianum pathogenicity.
Fluorescence microscopy analysis of the effect of pacCl inactivation on the infection cycle of C. lindemuthianum on the susceptible cultivar La Victoire. Excised hypocotyls were inoculated with conidia from wild-type isolate UPS9 and from Mutpac2. (a) Two days after inoculation, conidia from both lines developed melanized appressoria. (b) The wild-type line infected the vegetative tissue, developing primary and secondary hyphae and thereby completing its infection cycle. The Mutpac2 infected the vegetative tissue developing only primary hyphae (a): appressorium; (c): conidium; (PH): primary hyphae; (SH): secondary hyphae; (cd): conidiophore. Scale bar represents 25 μm
The apoplastic pH from the leaves of the susceptible bean cultivar La Victoire was approximately 6.35. At this pH, the PacCl protein might be processed or modified in a way that converts it to its active form and confers the ability to activate the transcription of its own gene. Therefore, PacCl was functional in the C. lindemuthianum UPS9 cells, while its hyphae grew in the interior of the La Victoire host tissue. Interestingly, the apoplastic pH of the resistant cultivar Jalo was approximately 5.6. Felle et al. (2004) concluded that changes in the apoplastic pH are important indicators of the plant–pathogen interaction, such that the different stages of infection of Blumeria graminis f. sp. hordei correlate with the different types of defence response of the host plant.
To evaluate the influence of pacCl inactivation on the expression of the lipase-encoding gene, lipase activity was detected in the culture media in the wild-type isolate UPS9 as well as the mutant Mutpac2. The lipase enzymatic activity index was lower in the wild-type, indicating that the pacCl gene might influence the production capacity of this enzyme (Fig. 5a). To confirm this result, both lines were cultured in liquid medium and the amount of lipase in the supernatant determined. Lipase activity was detected in the wild-type culture, but not in the Mutpac2 culture supernatant (Fig. 5b).
Lipase activity in solid and liquid culture media from the isolates UPS9 and Mutpac2. (a) Index of lipase activity in solid medium. (b) Lipase units (UI) in the supernatant of liquid cultures. One international lipase unit is defined as the enzymatic activity capable of releasing 1 μmol of -SH from the substrate dimercaprol tributyrate per minute per milliliter
Discussion
In this work, a 2,549-bp sequence of the C. lindemuthianum genome was isolated, containing a 1,749-bp ORF. The identified sequence was translated in silico into a putative polypeptide of 582 amino acids, which showed high similarity to the proteins from the PacC/Rim101 family of transcription factors identified in A. nidulans (Tilburn et al. 1995), A. niger (MacCabe et al. 1996), F. oxysporum (Caracuel et al. 2003a), S. scleotiorum (Rollins and Dickman 2001), Y. lipolytica (Lambert et al. 1997), and C. albicans (Ramon et al. 1999). The comparative analysis between the pacCl nucleotide sequence and the cDNA sequence identified three introns. These pacCl sequence characteristics are similar to those obtained in other pacC genes. In F. oxysporum (Caracuel et al. 2003a), T. harzianum (Moreno-Mateos et al. 2007), C. acutatum (You et al. 2007), and C. gloeosporioides (GenBank EU671075), for example, the putative ORFs have nucleotide sequences varying from 1,770 bp (C. acutatum) to 2,107 bp (T. harzianum). Moreover, the pacC sequences of all these fungi show three introns, with sizes varying from 50 to 127 bp.
Similarly to the PacC/Rim101 protein from many fungi, the polypeptide analysis of PacCl identified the major elements that characterize this protein family. In the amino-terminal region, there were three zinc finger motifs, which are common in proteins that interact with DNA, such as transcription factors. In most cases, the zinc finger motif presents the C(X 2–4)C(X12)H(X3–5)H consensus sequence, i.e., the cysteine amino acid is separated from the other cysteine by 2–4 other amino acid residues, and these amino acids are separated from the two histidines by another 12 amino acid residues. The first and the third zinc finger discovered here exactly fit these characteristics; however, in the second zinc finger, 8 instead of 12 amino acid residues separated the extremities of the motif. The three motifs are located at the positions 44–67, 80–90 and 110–130 of the putative protein. Similar to what was identified in the PacC protein of C. acutatum (You et al. 2007) and C. gloeosporioides, at position 219–249 a nuclear localization signal was found, whose consensus sequence is (K/R)2–X10–12–(K/R)3 (Nigg 1997). In the central portion of the protein, glycine- and glutamine-rich regions were found, as observed in other organisms, such as A. nidulans and C. acutatum. In the carboxy-terminal region at positions 429 and 575, two consensus sequences were found, YPNL and IPIL, which participate in the recognition and binding of the PalA protein and in the resultant signaling cascade, respectively. These two sequences flank another well-conserved region, the protease signaling box, which is located at position 446–467. This is the position where the first proteolytic cleavage takes place during the activation of the PacC protein, which is cleaved by PalB. The presence of all of these conserved domains in the deduced PacCl protein strongly indicates that it is homologous to the PacC/Rim101 proteins and that C. lindemuthianum is therefore able to regulate pH-mediated gene expression the same way as other fungi.
The pacCl transcript was detected at all pH values tested, from acidic to alkaline conditions. The quantitative analysis showed that pacCl expression is pH-dependent, increasing at more alkaline pH levels. The same transcription profile is observed for the pacC gene of A. nidulans, in which PacC binds to cis elements in its own promoter region, self-activating its transcription in more alkaline pH (Tilburn et al. 1995). The increase in the expression of pacC/RIM101 genes at elevated extracellular pH appears to be common to many organisms, such as in S. sclerotiorum (Rollins 2003), F. oxysporum (Caracuel et al. 2003a), F. verticillioides (Flaherty et al. 2003), C. albicans (Ramon et al. 1999), Y. lipolytica (Lambert et al. 1997), and U. maydis (Aréchiga-Carvajal and Ruiz-Herrera 2005). Moreover, qRT-PCR demonstrated the expression of pacCl during the infection cycle of C. lindemuthianum in the biotrophic and, especially, in the necrotrophic phase.
The first physiological evidence that the pacCl gene was inactivated came from the growth evaluation of the wild-type and mutant lines cultured under different pH conditions. The Mutpac2 mutant showed a strong growth reduction in more alkaline pH in comparison with UPS9 isolates (Fig. 2). Decreased growth in mutants with pacC/RIM101 loss of function is frequently observed when they are inoculated in environments with elevated pH (Peñalva and Arst. 2002). In an alkaline pH, these mutants show phenotypes that mimic growth in an acidic pH, resulting in the expression of genes that, in the wild-type, are preferentially expressed in acidic environments, as well as in the reduction of the expression of genes that, in the wild-type, are expressed preferentially in alkaline environments. Most likely, the genes required for C. lindemuthianum adaptation in alkaline environments and whose expression is activated by the PacCl transcription factor at these pH values are not expressed in Mutpac2. These results clearly show that PacCl is necessary for the normal growth of C. lindemuthianum in alkaline pH, and they support the hypothesis that in the Mutpac2 transformant the pacC1 gene is inactivated, leading to loss of its function. The pacC/RIM101-null mutants show other phenotypes in addition to the inhibition of growth at elevated pH values. In S. sclerotiorum, PacC is needed for the development and normal maturation of sclerotia (Rollins 2003). The mutant line grows normally at lower pH but is completely inhibited at higher pH levels. Moreover, the accumulation of oxalic acid is drastically reduced and the pg1 gene derepressed at higher pH values (which repress this gene in wild-type lines). Morphological changes, increased sensitivity to lytic enzymes, and polysaccharide secretion are characteristics that had previously only been reported in the pacC mutant phytopathogen U. maydis (Aréchiga-Carvajal and Ruiz-Herrera 2005). This mutant is also sensitive to osmotic stress, caused by the increased Li+ and Na+ in the culture medium, as observed with pacC +/− mutants of F. oxysporum (Caracuel et al. 2003b). Altered conidiogenesis is reported for the fungi A. nidulans (Tilburn et al. 1995) and A. niger (MacCabe et al. 1996) when pacC is disrupted.
As shown in Fig. 3, the Mutpac2 line can be considered non-pathogenic due to its inability to macerate the plant tissue. Although it creates light-brown lesions, the infectious process leading to the development of anthracnose does not progress, and this line cannot complete its cycle of asexual reproduction in plants lacking lesions with exudates. C. lindemuthianum conidia can be found in bean tissue lesions, in which exudation occurs during the maceration process (Pellier et al. 2003).
During the colonization of the host tissue, the phytopathogen must maintain precise control over the timing of expression of its genes, efficiently adapting to the tissue ambient conditions to complete its life cycle. The limited availability of nutrients in plant tissue is one of the main signals that control the expression of genes involved in the pathogenicity of several pathogenic microorganisms (Divon and Fluhr 2007). Genes encoding general regulators of nitrogen and/or carbon absorption and utilization are important for the pathogenicity of phytopathogens, as they allow gene expression to adjust depending on nitrogen and carbon limitations in the host tissue. The clnr1 gene from C. lindemuthianum encodes a transcription factor that enables this phytopathogen to use different nitrogen sources, and its transcription is necessary for pathogenicity. This was demonstrated by the inability of the clnr1 − mutant to develop into the necrotrophic phase during the infection of susceptible cultivars of common bean (Pellier et al. 2003), as well as by the phenotype of the Mutpac2 line obtained in this work. A second gene, clta1, encodes another transcriptional activator in C. lindemuthianum that is necessary for biotrophic-to-necrotrophic phase transition (Dufresne et al. 2000). Clta1 shows homology to the GAL4 family of proteins. Thon et al. (2002) demonstrated that disruption of the CPR1 gene from C. graminicola results in a significant reduction in virulence. According to these authors, this gene encodes a component of a Signal Peptidase Complex (SPC), responsible for the signal peptide cleavage of proteins targeted to the endoplasmic reticulum. The authors believe, therefore, that the phenotype can be due the reduction in the transport of secretory proteins during the transition from biotrophy to a necrotrophic lifestyle, especially enzymes involved in degradation of plant cell wall polysaccharides. More recent work (Bhadauria et al. 2013), there is a novel effector gene, CtNUDIX, from C. truncatum that is exclusively expressed during the late biotrophic phase. It encodes a protein containing a signal peptide and a Nudix hydrolase domain, able to elicit the hypersensitive response (HR) pathway of defence against plant pathogens in tobacco. The ambient pH modulates the pathogenicity of many plant and insect pathogens (Prusky and Yakoby 2003; Akimitsu et al. 2004) and human pathogens, such as C. albicans (De Bernardis et al. 1998). Ambient pH controls the expression of virulence factors, such as cell wall hydrolytic enzymes and proteases, and even yeast filamentation. The phytopathogens can encounter both temporal and spatial pH variations during the process of host colonization. In this case, the transcription factor PacC/Rim101 could play a fundamental role in the regulation of genes necessary for efficient infection, thereby determining the virulence and/or pathogenicity of these organisms. In S. sclerotiorum, PacC functions as a virulence factor (Rollins 2003), similar to its orthologs in A. nidulans (Bignell et al. 2005) and C. albicans (Davis et al. 2000). Clearly, the role of PacC in virulence/pathogenicity varies according to the species in question. In the phytopathogen F. oxysporum, pacC mutants are more virulent than the wild-type, suggesting that PacC behaves as a negative virulence regulator (Caracuel et al. 2003a). Interestingly, the pacC mutant lines of U. maydis were as virulent as the wild-type when inoculated in maize (Aréchiga-Carvajal and Ruiz-Herrera 2005). Thus, an organism’s pathogenicity is characterized by its capacity to efficiently colonize the host and complete its life cycle. The results shown in Fig. 4 allow us to propose that pacCl is, in fact, involved in C. lindemuthianum pathogenicity.
PacC is a transcription factor involved in the activation or repression of a number of genes important in pathogenicity in filamentous fungi. To determine the effect of inactivating the pacCl gene on the expression of the gene encoding the C. lindemuthianum lipase enzyme, lipase activity was evaluated in the wild-type and Mutpac2 isolates. Lipase activity was detected in the wild-type culture, but not in the supernatant of the Mutpac2 culture (Fig. 5). Detecting lipase activity by the halo degradation method in different species of Colletotrichum has shown that lipase activity depends on pH. In general, lipase activity is absent at acidic pH and is prominent in the extracellular environment at alkaline pH (Maccheroni et al. 2004). In F. graminearum, as well as in other phytopathogenic fungi, the extracellular lipase is a necessary virulence factor for the infection of cereals (Voigt et al. 2005). The results in C. lindemuthianum indicate that Mutpac2 is deficient in the production or secretion of extracellular lipases. It is probable that PacCl is involved in the regulation of lipase synthesis in C. lindemuthianum. Taken together, our results indicate that the pacCl gene, which encodes a pH-responsive transcriptional regulator, is an important determinant of the pathogenicity of C. lindemuthianum towards its host, the common bean (P. vulgaris).
References
Akimitsu, K., Isshiki, A., Ohtani, K., Yamamoto, H., Eshel, D., & Prusky, D. (2004). Sugars and pH: a clue to the regulation of fungal cell wall-degrading enzymes in plants. Physiological and Molecular Plant Pathology, 65, 271–275.
Ansari, K. I., Palacios, N., Araya, C., Langin, T., Egan, D., & Doohan, F. M. (2004). Pathogenic and genetic variability among Colletotrichum lindemuthianum isolates of different geographic origins. Plant Pathology, 53, 635–642.
Aréchiga-Carvajal, A. T., & Ruiz-Herrera, J. (2005). The RIM101/pacC homologue from basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryotic Cell, 4, 999–1008.
Arst, H. N., Jr., & Peñalva, M. A. (2003). Recognizing gene regulation by ambient pH. Fungal Genetics and Biology, 40, 1–3.
Bailey, J. A., & Jeger, M. J. (1992). Colletotrichum: Biology, pathology and control. Wallingford, Oxon: CAB International.
Ballance, D. J. (1986). Important sequences for gene expression in filamentous fungi. Yeast, 2, 229–236.
Barcellos, Q. L., Souza, E. A., & Damasceno e Silva, K. J. (2011). Vegetative compatibility and genetic analysis of Colletotrichum lindemuthianum isolates from Brazil. Genetics and Molecular Research, 10, 230–242.
Benton, W. D., & Davis, R. W. (1977). Screening of Xgt recombinant clones by hybridization to single plaques in situ. Science, 196, 180–183.
Bhadauria, V., Banniza, S., Vandenberg, A., Selvaraj, G., & Wei, Y. (2013). Overexpression of a novel biotrophy-specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryotic Cell, 12, 2–11.
Bignell, E., Negrete-Urtasun, S., Calcagno, A. M., Haynes, K., Arst, H. N., Jr., & Rogers, T. (2005). The Aspergillus pH-responsive transcription factor PacC regulates virulence. Molecular Microbiology, 55, 1072–1084.
Blanchin-Roland, S., Cordero-Otero, R., & Gaillardin, C. (1994). Two upstream activation sequences control the expression of the XPR2 gene in the yeast Yarrowia lipolytica. Molecular Cell. Biology, 14, 327–338.
Bonett, L. P., Schewe, I., & Silva, L. I. (2008). Variability of Colletotrichum lindemuthianum in common bean in western Paraná. Scientia Agrária, 9, 207–210.
Brown, A. J. P., & Gow, N. A. R. (1999). Regulatory networks controlling Candida albicans morphogenesis. Trends in Microbiology, 7, 333–338.
Caddick, M. X., Brownlee, A. G., & Arst, H. N. (1986). Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Molecular and General Genetics, 203, 346–353.
Caracuel, Z., Roncero, M. I. G., Espeso, E. A., González-Verdejo, C. I., García-Maceira, F. I., & Pietro, A. (2003a). The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Molecular Microbiology, 48, 765–779.
Caracuel, Z., Casanova, C., Roncero, M. I. G., Di Pietro, A., & Ramos, J. (2003b). pH response transcription factor PacC controls salt stress tolerance and expression of the P-type Na+-ATPase Ena1 in Fusarium oxysporum. Eukaryotic Cell, 2, 1246–1252.
Casela, C. R., & Frederiksen, R. A. (1994). Pathogenic variation in monoconidial culture from a single lesion and monoconodial subcultures of the sorghum anthracnose fungus Colletotrichum graminicola. Tropica Plant Pathology, 19, 149–153.
Castro-Prado, M. A., Querol, C. B., Sant’Anna, J. R., Miyamoto, C. T., Franco, C. C., Mangolin, C. A., & Machado, M. F. (2007). Vegetative compatibility and parasexual segregation in Colletotrichum lindemuthianum, a fungal pathogen of the common bean. Genetics and Molecular Research, 6, 634–642.
Damasceno e Silva, K. J., Souza, E. A., & Ishikawa, F. H. (2007). Characterization of Colletotrichum lindemuthianum isolates from the State of Minas Gerais, Brasil. Journal of Phytopathology, 155, 241–247.
Davis, D., Edwards, J. E., Jr., Mitchell, A. P., & Ibrahim, A. S. (2000). Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infection and Immunity, 68, 5953–5959.
De Bernardis, F., Muhlschlegel, F. A., Cassone, A., & Fonzi, W. A. (1998). The pH of the host niche controls gene expression in and virulence of Candida albicans. Infection and Immunity, 66, 3317–3325.
Denison, S. H. (2000). Review: pH regulation of gene expression in fungi. Fungal Genetics and Biology, 29, 61–71.
Divon, H. H., & Fluhr, R. (2007). Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiology Letters, 266, 65–74.
Dufresne, M., Bailey, J. A., Michel, D., & Langin, T. (1998). clk1, a serine/threonine protein kinase-encoding gene, is involved in pathogenicity of Colletotrichum lindemuthianum on common bean. Molecular Plant-Microbe Interaction, 11, 99–108.
Dufresne, M., Perfect, S., Pellier, A. L., Bailey, J. A., & Langin, T. (2000). GAL4-like Protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell, 12, 1579–1589.
Elva, T., Aréchiga-Carvajal, Ruiz-Herrera, J. (2005). The HIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukariot. Cell. 4, 999–1008.
Felle, H. H., Herrmann, A., Hanstein, S., Hückelhoven, R., & Kogel, K.-H. (2004). Apoplastic pH signaling in barley leaves attackedby the powdery mildew fungus Blumeria graminis f. sp. hordei. Molecular Plant-Microbe Interactions, 17, 118–123.
Flaherty, J. E., Pirttilä, A. M., Bluhm, B. H., & Woloshuk, C. P. (2003). PAC1, a pH-regulatory gene from Fusarium verticillioides. Applied Environmental Microbiology, 69, 5222–5227.
Franco, C. C., Sant’Anna, J. R., Rosada, L. J., Kaneshima, E. N., Stangarlin, J. R., & Castro-Prado, M. A. (2011). Vegetative compatibility groups and parasexual segregation in Colletotrichum acutatum isolates infecting different hosts. Phytopathology, 101, 923–928.
Gurr, S. J., Unkles, S. E., & Kinghorn, J. R. (1987). The structure and organization of nuclear genes of filamentous fungi. In J. R. Kinghorn (Ed.), Gene structure in eukaryotic microbes (pp. 93–139). Oxford: IRL Press.
Horton, P., Park, K., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., & Nakai, K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Research, 35, 585–587.
Inoue, H., Nojima, H., & Okayama, H. (1990). High efficiency transformation of Escherichia coli with plasmids. Gene, 96, 23–28.
King, B.R., Guda, C. (2007). ngLOC: an n-gram-based Bayesian method for estimating the subcellular proteomes of eukaryotes. Genome Biol. 8, R68.
Kozak, M. (1984). Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Research, 12, 857–872.
Kramer-Haimovich, H., Servi, E., Katan, T., Rollins, J., Okon, Y., & Prusky, D. (2006). Effect of ammonia production by Colletotrichum gloesporioides on pelB activation, pectate lyase secretion and fruit pathogenicity. Applied and Environmental Microbiology, 72, 1034–1039.
Lamb, T. M., Xu, W., Diamond, A., & Mitchel, A. P. (2001). Alkaline response genes of Saccharomyces cereviseae and their relationship to the RIM101 pathway. Journal of Biological Chemistry, 276, 1850–1856.
Lambert, M., Blanchin-Roland, S., Le Louedec, F., Lepingle, A., & Gaillardin, C. (1997). Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Molecular Cell. Biology, 17, 3966–3976.
Larionov, A., Krause, A., & Miller, W. (2005). A standard curve based method for relative real time PCR data processing. BMC Bioinf., 6, 62.
MacCabe, A. P., Van den Hombergh, J. P. T. W., Visser, J., Tilburn, J., & Arst, H. N., Jr. (1996). Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Molecular and General Genetics, 250, 367–374.
Maccheroni, W., Jr., Araújo, W. L., & Azevedo, J. L. (2004). Ambient pH-regulated enzyme secretion in endophytic and pathogenic isolates of the fungal genus Colletotrichum. Scientific Agriculture, 61, 298–302.
Mahuku, G. S., & Riascos, J. J. (2004). Virulence and molecular diversity within Colletotrichum lindemuthianum isolates from Andean and Mesoamerican bean varieties and regions. European Journal Plant Pathology, 110, 253–263.
Merhej, J., Richard-Forget, F., & Barreau, C. (2011). The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genetics and Biology, 48, 275–284.
Moreno-Mateos, M. A., Delgado-Jarana, J., Codón, A. C., & Benítez, T. (2007). pH and Pac1 control development and antifungal activity in Trichoderma harzianum. Fungal Genetics and Biology, 44, 1355–1367.
Nigg, E. A. (1997). Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787.
Ochman, H., Gerber, A. S., & Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics, 120, 621–623.
Parisot, D., Dufresne, M., Veneault, C., Laugé, R., & Langin, T. (2002). cla1, a gene encoding a copper-transporting ATPase involved in the process of infection by the phytopathogenic fungus Colletotrichum lindemuthianum. Molecular Genetics and Genomics, 268, 139–151.
Pellier, A. L., Laugé, R., Veneault-Fourrey, C., & Langin, T. (2003). CLNR1, the AREA/NIT2-like global nitrogen regulador of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Molecular Microbiology, 48, 639–355.
Peñalva, M. A., & Arst, H. N., Jr. (2002). Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiology Molecular Biology R., 66, 426–446.
Peñalva, M. A., & Arst, H. N., Jr. (2004). Recent avances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annual Review of Microbiology, 58, 425–451.
Peñalva, M. A., Tilburn, J., Bignell, E., & Arst, H. N., Jr. (2008). Ambient pH gene regulation in fungi: making connections. Trends in Microbiology, 16, 291–300.
Perfect, S. E., Hughes, H. B., O’Connell, R. J., & Green, J. (1999). Colletotrichum: a model genus for studies on pathology and fungal-plant interaction. Fungal Genetics and Biology, 27, 186–198.
Prusky, D., & Yakoby, N. (2003). Pathogenic fungi: leading or led by ambient pH? Molecular Plant Pathology, 4, 509–516.
Ramon, A. M., Porta, A., & Fonzi, W. A. (1999). Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-Related transcription factor encoded by PRR2. Journal of Bacteriology, 181, 7524–7530.
Roca, M. G., Davide, L. C., & Mendes-Costa, M. C. (2003). Conidial anastomosis tubes in Colletotrichum. Fungal Genetics and Biology, 40, 138–145.
Rodriguez-Guera, R.M.T., Ramirez-Rueda, De La Vega, O.M., Simpson, J. (2003). Variation in genotype, pathotype and anastomosis groups of Colletotrichum lindemuthianum isolates from Mexico. Plant Pathol. 52, 228–235.
Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Molecular Plant-Microbe Interactions, 16, 785–795.
Rollins, J. A., & Dickman, M. (2001). pH signaling in Sclerotinia sclerotiorum: identification of a pacC/HIM101 homolog. Applied and Environmental Microbiology, 67, 75–81.
Rosada, L. J., Franco, C. C., Sant’Anna, J. R., Kaneshima, E. N., Gonçalves-Vidigal, M. C., & Castro-Prado, M. A. (2010). Parasexuality in Race 65 Colletotrichum lindemuthianum Isolates. Journal of Eukaryotic Microbiology, 57, 383–384.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Santos, L. V., Queiroz, M. V., Ferreira, M. S., Soares, M. A., Barros, E. G., Araújo, E. F., & Langin, T. (2012). Development of new molecular markers for the Colletotrichum genus using RetroCl1 sequences. World J. Microb. Biot., 28, 1087–1095.
Specht, C. A., DiRusso, C. C., Novotny, C. P., & Ullrich, R. C. (1982). A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Analytical Biochemistry, 119, 158–163.
Su, S. S., & Mitchell, A. P. (1993). Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Research, 21, 3789–3797.
Suárez, T., & Peñalva, M. A. (1996). Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Molecular Microbiology, 20, 529–540.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA6: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular and Biological Evolution, 24, 1596–1599.
Thon, M. R., Nuckles, E. M., Takach, J. E., & Vaillancourt, L. J. (2002). CPR1: a gene encoding a putative signal peptidase that functions in tathogenicity of Colletotrichum graminicola to maize. Molecular Plant-Microbe Interactions, 15, 120–128.
Tilburn, J., Sarkar, S., Widdick, D. A., Espeso, E. A., Orejas, M., Mungroo, J., Peñalva, M. A., & Arst, H. N., Jr. (1995). The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO Journal, 14, 779–790.
Voigt, C. A., Schafer, W., & Salomon, S. (2005). A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant Journal, 42, 364–375.
Wang, Q., & Szaniszlo, P. J. (2009). Roles of the pH signaling transcription factor PacC in Wangiella (Exophiala) dermatitidis. Fungal Genetics and Biology, 46, 657–666.
You, B., Choquer, M., & Chung, K. (2007). The Colletotrichum acutatum gene encoding a putative pH-responsive transcription regulator is a key virulence determinant during fungal pathogenesis on citrus. Molecular Plant-Microbe Interactions, 20, 1149–1160.
Zou, C., Tu, H., Liu, X., Tao, N., & Zhang, K. (2010). PacC in the nematophagous fungus Clonostachys rosea controls virulence to nematodes. Environmental Microbiology, 12, 1868–1877.
Acknowledgments
The authors would like to thank the Research Foundation of the State of Minas Gerais (FAPEMIG), the Brazilian Federal Agency for the Support and Evaluation of Graduate Education (CAPES), and the National Council for Scientific and Technological Development (CNPq) for their financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic Supplementary Material A1
pacCl inactivation analysis. (a) Determining the copy number of pacCl gene by DNA hybridization. (b) The pBKSpac::hph2 plasmid used in the transformation. The clear rectangle represents the pacCl cDNA fragment. The trans-primer (gray triangle) was inserted into the region between the annealing sequences of the oligonucleotides Opac1 and Opac2; it amplified a 1,079-bp fragment of the pBKSpac::hph2 and 1221-bp of the genomic DNA. (c) PCR results using the group of oligonucleotide pair Opac1/Opac2 or Ohph3/Ohph4 and DNA from lines Mutpac2, Tr1, Tr2, Tr3, and Tr4 of C. lindemuthianum; M: 100-bp marker. (d) Molecular characterization of the pacCl − mutant by DNA hybridization using as a probe a 1.8-kb fragment of the pacCl gene and the empty pBluescriptSK+. (M) - Lambda bacteriophage DNA digested with Hind III; (Mut): mutant line Mutpac2; (WT): wild-type line. (JPEG 2059 kb)
Rights and permissions
About this article
Cite this article
Soares, M.A., Nogueira, G.B., Bazzolli, D.M.S. et al. PacCl, a pH-responsive transcriptional regulator, is essential in the pathogenicity of Colletotrichum lindemuthianum, a causal agent of anthracnose in bean plants. Eur J Plant Pathol 140, 769–785 (2014). https://doi.org/10.1007/s10658-014-0508-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-014-0508-4