Abstract
In this study, we investigated the activities of β-1,3-glucanase and peroxidase enzymes in the leaves of pepper cultivar A3 infected with the incompatible strain PC and the compatible strain HX-9 of Phytophthora capsici. The activities of β-1,3-glucanase and peroxidase enzymes substantially increased in the incompatible interactions compared to the compatible interactions. We also analysed the expression patterns of four defence-related genes, including CABPR1, CABGLU, CAPO1 and CaRGA1, in the leaves and roots of pepper inoculated with different strains of P. capsici. All gene expression levels were higher in the leaves than in the roots. Markedly different expression patterns were observed between incompatible and compatible host-pathogen interactions. In the incompatible interactions, the expression levels of CABPR1, CABGLU and CAPO1 genes in leaves increased by a maximum of 17.2-, 13.2- and 20.5-fold at 24, 12 and 12 h, respectively, whereas the CaRGA1 gene expression level increased to a lesser degree, 6.0-fold at 24 h. However, in the compatible interactions, the expression levels of the four defence-related genes increased by a maximum of 11.2-, 8.6-, 7.9- and 2.0-fold at 48, 24, 48 and 72 h, respectively. Compared to the leaves, the expression levels of the four defence-related genes were much lower in the roots. The highest levels of mRNA were those of the CABPR1 gene, which increased 5.1-fold at 24 h in the incompatible and 3.2-fold at 48 h in the compatible interactions. The other three genes exhibited lower expression levels in the incompatible and compatible interactions. These results further confirmed that defence-related genes might be involved in the defence response of pepper to P. capsici attack.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytophthora capsici is a soil-borne pathogen that causes root rot, crown rot, foliar blight and fruit rot in nearly all cultivars of Capsicum annuum (Alcantara and Bosland 1994). Although P. capsici was originally thought to be host-specific to pepper, it was recently reported to interact with tomato, eggplant, cucurbits (cucumber, melon, pumpkin and others) and lima beans. The symptoms vary considerably according to the host, areas of infection and environmental conditions (Lamour et al. 2012). Recently, the incidence of P. capsici has increased, leading to severe economic losses in agricultural and horticultural crops (Ristaino and Johnston 1999). To date, no useful control methods have been developed, as chemical and biological controls are ineffective in preventing the spread of P. capsici to pepper plants (Oelke et al. 2003).
Plants have developed a range of defence mechanisms against such attacks, including both the elaboration of structural barriers and the induction of active host-specific responses that provide resistance (Singh et al. 2000). The active mechanisms imply the recognition of the pathogen by the host plant. Once the contact has been established, active defences are activated, and they consist of morphological barriers (cell wall thickening), secondary metabolites (phytoalexins), and defence-related proteins (Silvar et al. 2008). These defences can include the induction and activation of genes coding for enzymes of the phenylpropanoid pathway or for pathogenesis-related (PR) proteins, which have been implicated in active defence and play roles in restricting pathogen development and spread within the plant (Van Loon and Van Strien 1999). Defence-related enzymes include chitinase, β-1,3-glucanase and peroxidases, and these enzyme activities are known to be induced in many plants in response to infection with fungal pathogens and are also correlated with induced resistance (Saikia et al. 2005; Govindappa et al. 2010). β-1,3-glucanases are grouped in the PR-2 family of pathogenesis-related (PR) proteins. They also defend plants against fungal pathogens either alone or in association with chitinases and other antifungal proteins (Balasubramanian et al. 2012). Peroxidase is related to the resistance against diseases in plant-pathogen interactions. Peroxidase accumulates both in the extracellular environment and in the cell wall in cell suspension studies. This result might be related to lignin biosynthesis, which forms a mechanical barrier against pathogen infection (Egea et al. 2001). In a previous study, an increase in peroxidase activity was associated with CM334 resistance to P. capsici (Fernández-Pavia 1997) compared to susceptible plants. In incompatible plant-nematode interactions, peroxidases played a critical role in the generation of reactive oxygen species (ROS) associated with the hypersensitive reaction (HR) (Melillo et al. 2006). The change in plant metabolism is significantly different for susceptible and resistant plants (Esra et al. 2011). Those studies concerned with obtaining pepper varieties resistant to root rot have highlighted the necessity of examining and comparing the metabolisms of varieties in which resistance to P. capsici is different (Silvar et al. 2008). Unfortunately, the roles of many of the plant substances that play important roles in the physiological and biochemical mechanisms governing the defence against this disease are not fully understood (Egea et al. 1996; Requena et al. 2005). Recent plant microarray data have confirmed that the differences between susceptibility and resistance are associated with differences in the timing and magnitude of the induced response (Marate et al. 2004; Huang et al. 2005; Yang et al. 2007). However, little is known about the defence responses of the same host plant inoculated with different strains of P. capsici.
In a previous study, we isolated a Capsicum annuum blight resistance protein RGA1 from the leaves of the pepper cultivar CM334 inoculated with P. capsici. This gene was cloned on the basis of the conserved gene sequences of potato and was named CaRGA1 (GenBank: GQ386945.1). The functional role of the CaRGA1 gene in the defence response to pathogens was analysed by virus-induced gene silencing (VIGS) and over-expression (genetic transformation) in pepper. In pepper plants, CaRGA1 gene expression was strongly induced by the defence-related plant hormone methyl jasmonic acid. In addition, CaRGA1 gene expression was also strongly induced by biotic and abiotic stress treatments, including cold, high salinity and pathogen infection. A loss-of-function of the CaRGA1 gene in virus-induced gene silenced pepper plants led to an increased susceptibility to pathogen attack. In contrast, the over-expression of CaRGA1 in transgenic pepper plants conferred an enhanced tolerance to high salt stress, while also enhancing the resistance to pathogen infection. Taken together, these results provide evidence for the involvement of CaRGA1 in plant defence responses to pathogens and salt stress (data not shown). Along with these results, the CaRGA1 gene and another three reported defence-related genes were used to compare the defence responses between pepper and different strains of P. capsici.
In this study, the pepper cultivar A3 was used as the host plant because it is susceptible to ph3 (HX-9 strain) but resistant to ph1 (PC strain) and thus provides the opportunity to investigate both compatible and incompatible interactions in the same genetic background. The aim of the present work was to access the potential role of defence-related enzyme activity (β-1, 3-glucanase and peroxidase) in the defence response of pepper infected with incompatible and compatible isolates of P. capsici. We also aimed to examine the differences in expression of defence-related genes (CABPR1, CABGIU, CAPO1 and CaRGA1) in the leaves and roots of pepper plants infected with compatible and incompatible isolates of P. capsici. Our study will facilitate a better understanding of the plant response to P. capsici infection and will help develop strategies to improve plant defences through genetic engineering.
Materials and methods
Plant material and seedling culture conditions
The pepper cultivar A3 was used as host plant to inoculate with P. capsici. Seeds of cultivar A3 were supplied from Northwest A&F University (Yangling, China) and soaked in warm water (55 °C) for approximately 20 min to promote germination. The seeds were rinsed twice every 24 h and then placed on moist gauze in an incubator (28 °C, 60 % relative humidity in darkness). When the seeds were at least 80 % germinated, they were sown at a depth of 1.0 cm in 9-cm-deep plastic pots filled with growth medium consisting of grass charcoal and perlite at a ratio of 3:1. The seedlings were grown in a growth chamber at 25 °C with a 16 h light / 8 h dark photoperiod cycle until they were used for pathogen inoculation at the six-true-leaf stage.
Pathogen preparation and inoculation
The virulent strain HX-9 (ph3, compatible with A3) and the avirulent strain PC (ph1, incompatible with A3) were isolated from blighted pepper plants collected from a field in China and identified as P. capsici, as described by Zhang et al. (2009). The preparation of P. capsici inocula was previously described (Kim et al. 2008). The oomycete pathogen was grown on potato dextrose agar (PDA) medium in the dark at 28 °C for 7 days. The mycelia were scraped and incubated under fluorescent light for 2 days to promote sporangium formation. Zoospores were prepared by sporulating cultures with sterile distilled water and incubating at 4 °C for 1 h followed by 30 min at 28 °C to initiate zoospore release. The zoospores were collected by filtering through four layers of cheesecloth and numbers estimated using a haemocytometer. The concentration of the zoospore suspension was adjusted to 1 × 104 zoospores / ml with sterile water.
Pepper plants at the six-true-leaf stage were used for the P. capsici inoculation. The pots were saturated with sterile distilled water 24 h prior to the inoculation. The pepper plants were inoculated by adding 2.5 ml zoospore suspensions (1 × 104 zoospores / ml) to the soil in each pot. Each pot had two pepper seedlings. The inoculated pepper plants were kept in a growth chamber at 28 °C, 60 % relative humidity with a 16 h light / 8 h dark photoperiod cycle. The P. capsici-infected pepper leaves and roots were collected at 0, 6, 12, 24, 48, 72 and 96 h after inoculation and stored at −80 °C for later use.
Determination of root activity
The root activity was measured by a modified triphenyltetrazolium chloride (TTC) method from Ou et al. (2011). The P. capsici-infected roots at different time points after inoculation (including non-infected roots) were washed with sterile water for 10 min before the TTC tests. Their surfaces were dried carefully with absorbent papers. A root tip (approximately 0.3 g) was selected, placed in a small beaker and incubated with 10 ml 1:1 (v/v) mixture of 1 % TTC solution and 0.l M phosphate buffer (pH 7.0) at 37 °C for 1 h in the dark. After this incubation, 2.0 ml 1 M sulphuric acid was to added to inhibit the reaction. The root samples were rinsed twice with distilled water and ground with quartz sand in 3.0 ml ethyl acetate 2–3 times, in which the extracts of TTF (a derivative of TTC from the reduced reaction) were obtained and transferred to a 10-ml tube. The residue was rinsed with ethyl acetate. The rinsed solution was mixed with the earlier extracts and adjusted to a volume of 10 ml. The absorption of the diluted extraction was measured with a spectrophotometer at 485 nm. The control was similarly performed except the sulphuric acid was added first, the root sample second, and the mixture last. The reduced TTC amount was obtained from the standard curve and its intensity in the root tip was calculated as follows: TTC reduction intensity [mg g−1 h] = reduced TTC amount / FW.h (FW-fresh root mass; h-the incubation time). Each treatment was conducted in three independent experiments and each measurement was repeated three times.
β-1,3-glucanase (EC 3.2.1.39) enzymatic activities assay
The β-1,3-glucanase activity was measured by a colorimetric assay from Kauffmann et al. (1987) using laminarin as a substrate. The samples were collected from the third and fourth leaves at different time points after inoculation with P. capsici and stored at −80 °C. The lyophilised leaf powder (approximately 1.0 g) was ground in a mortar and homogenised with 5.0 ml pre-chilled extraction buffer (0.1 M sodium acetate buffer, pH 5.2). The homogenate was centrifuged at 12,000 rpm for 25 min. The supernatant fraction was used as a crude extract for the enzyme activity assays. All procedures were carried out at 4 °C. The substrate buffer was 0.1 M sodium acetate buffer (pH 5.2) containing laminarin (1.0 mg / ml buffer). The reaction mixture contained 0.9 ml substrate buffer and 0.1 ml enzyme solution (leaf extract). The reaction tubes were incubated at 37 °C for 1 h and measured with a spectrophotometer at 540 nm. The resulting reducing sugars were measured using the method of Nelson (1994). β-1,3-glucanase activities were expressed as μmol glucose equivalents released / g fresh weight tissue /1 h. Each treatment was conducted in three independent experiments and each measurement was repeated three times.
Peroxidase (EC 1.11.1.7) enzymatic activity assay
The activity of peroxidase (PX) was quantified using the technique of Beffa et al. (1990). The samples were collected from the third and fourth leaves at different time points after inoculation with P. capsici and stored at −80 °C. The lyophilised leaf powder (approximately 1.0 g) was ground in a mortar and homogenised with 5.0 ml pre-chilled extraction buffer (0.1 M potassium phosphate buffer (pH 7.5), 1 mM EDTA, and 4 % polyvinylpyrrolidone). The homogenate was centrifuged at 12,000 rpm for 25 min. The supernatant fraction was used as a crude extract for the enzyme activity assays. All procedures were carried out at 4 °C. The reaction mixtures contained 25 μl 50 mM H2O2, 5 μl 250 mM guaiacol, 195 μl 12.5 mM 3,3-dimethylglutaric acid (3,3-DGA)-NaOH (pH 6.0) and 25 μl enzyme extract. The optical density was recorded at 470 nm. The amount of enzyme required for the formation of 1 μmol tetraguaiacol / min at room temperature was defined as 1 unit (U) of PX activity. Each treatment was conducted in three independent experiments and each measurement was repeated three times.
RNA extraction and real-time PCR for defence-related gene expression analysis
The total RNA was isolated from the pepper leaves at different time points after pathogen inoculation. The RNA was extracted using the TRIZOL (Invitrogen, USA) RNA Kit according to the manufacturer’s instructions. The concentration of the total RNA was measured spectrophotometrically using a NanoDrop instrument (Thermo Scientific NanoDrop 2000C Technologies, Wilmington, USA), and the purity was assessed using the A260/280 and A260/230 ratios provided by NanoDrop. For quantitative real-time RT-PCR analysis, first strand cDNA was synthesised from 500 ng total RNA using a PrimeScriptTM Kit (TaKaRa, Bio Inc, China) following the manufacturer’s protocols. Quantitative real-time RT-PCR was performed with an iCycler iQTM Multicolor PCR Detection System (Bio-Rad, Hercules, USA) and carried out using SYBR® Premix Ex Taq™ II (TaKaRa, Bio Inc, China). The quantitative real-time RT-PCR analysis was performed on a 20 μl mixture containing 10.0 μl SYBR® Premix Ex Taq™ II, 2.0 μl diluted cDNA, and 0.8 μl forward and reverse primers. The quantitative real-time RT-PCR cycling conditions were as follows: 95 °C for 1 min and 45 cycles of 95 °C for 10 s, 52 °C for 30 s, and 72 °C for 30 s. The suitable annealing temperature is 52 °C, which was tested by PCR with the primer pairs of the other PR genes. The primers for real-time RT-PCR are shown in Table 1. As a reference gene, the expression of the pepper ubiquitin-conjugating protein CaUbi3 gene was used (Wan et al. 2011). The expression levels were reported as the mean values with standard errors. The entire experiment was repeated three times. Each real-time RT-PCR measurement was performed in three replicates.
Statistical analysis
The data presented are the means ± standard deviation measures for three replicates. The data from replicates of the same experiment were pooled together for a single ANOVA and differences between means of treatments were determined using the least significant difference (LSD). The statistical procedures were performed using Statistical Analysis System software (SAS Institute, version 8.2). The differences with P ≤ 0.05 were considered significant.
Results
Root activity in the roots of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici
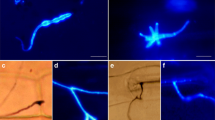
The disease symptoms were observed in the pepper cultivar A3 after inoculation with incompatible strain PC and compatible strain HX-9 of P. capsici at the six-true-leaf stage (Fig. 1a). The results indicate that the disease symptoms occurred in the compatible interaction but not in the incompatible interaction within 96 h of inoculation. To confirm the symptoms of disease, a 2,3,5-triphenyltetrazolium chloride (TTC) assay of a cell vitality was performed (Chen et al. 2006). The activities of the roots inoculated with different strains of P. capsici are shown in Fig. 1b. The activities of roots at different time points changed clearly between the incompatible and compatible interactions with P. capsici. The differences in the root activity were monitored at 6 h after inoculation both in the incompatible and compatible interactions (Fig. 1c). A significant difference was found from 12 to 96 h after pathogen inoculation, showing 2–3 times the activity in the incompatible than in the compatible interactions with P. capsici. By analysis of the root activity in two interactions, we concluded that there was a difference in the same host infected with different strains of P. capsici.
Phenotype in roots of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici. (a) Disease symptoms in pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici; (b) Phenotype in roots of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici; (c) Time courses of root activity in roots of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici. Values are the means ± standard deviation from three independent experiments. Means followed by bars denoted with different letters are significantly different at P ≤ 0.05, according to the least significant difference (LSD) test. Capital letters correspond to the comparison between incompatible and compatible interaction of P. capsici at the same time point. Small letters represent differences in the times of P. capsici inoculation in the same strain
β-1,3-glucanase activities in the leaves of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici
The β-1,3-glucanase activities were measured in the leaf extracts from plants inoculated with the incompatible strain PC and the compatible strain HX-9 of P. capsici (Fig. 2). The activity of β-1,3-glucanase increased significantly with increasing time after P. capsici infection compared to the control plants. As increase in the β-1,3-glucanase activity was detected at 6 h after pathogen inoculation with the maximum increase recorded at 12 h after pathogen inoculation in the incompatible interaction; however, there was a gradual increase in the induction of β-1,3-glucanase, reaching a peak at 24 h following injection in the compatible interaction. The accumulation of the β-1,3-glucanase activity was much higher in the incompatible interaction than the compatible interaction at different time points after inoculation with P. capsici.
Time courses of β-1,3-glucanase activities in leaves of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici. Values are the means ± standard deviation from three independent experiments. Means followed by bars denoted with different letters are significantly different at P ≤ 0.05, according to the least significant difference (LSD) test. Capital letters correspond to the comparison between incompatible and compatible interactions of P. capsici at the same time point. Small letters represent differences in the times of P. capsici inoculation in the same strain
Peroxidase activities in the leaves of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici
The activities of peroxidase in the leaves of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici are shown in Fig. 3. There were significant differences in the peroxidase activity between the two interactions, especially in the incompatible interaction with P. capsici inoculation. The peroxidase activity increased greatly from 6 to 72 h after inoculation and the maximum increase was recorded at 12 h after inoculation with the PC strain of P. capsici. In comparison the highest activity of peroxidase occurred late at 48 h in the compatible interaction with the HX-9 strain. Moreover, the increase in the compatible interaction with the HX-9 strain was much lower than in the incompatible interaction at the same time points in our study.
Time courses of peroxidase activities in leaves of pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici. Values are the means ± standard deviation from three independent experiments. Means followed by bars denoted with different letters are significantly different at P ≤ 0.05, according to the least significant difference (LSD) test. Capital letters correspond to the comparison between incompatible and compatible interactions of P. capsici at the same time point. Small letters represent differences in the times of P. capsici inoculation in the same strain
Expression of defence-related genes in the leaves and roots from pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici
To evaluate whether there is a correlation between the host plant and the strain of P. capsici, the expression patterns of some defence-related genes that may take part in defence responses was analysed by real-time PCR. The expression patterns of four defence-related genes encoding for the PR-1 protein (CABPR1), β-1,3-glucanase (CABGLU) and peroxidase (CAPO1), as well as CaRGA1, were examined in the leaves and roots inoculated with the incompatible and compatible strains of P. capsici. All the defence-related genes were up-regulated but presented differences in the expression patterns (Fig. 4).
Relative expression of defence-related genes in the leaves (left) and roots (right) from pepper cultivar A3 after inoculation with incompatible and compatible strains of P. capsici. (a) CABPR1; (b) CABGLU; (c) CAPO1 and (d) CaRGA1. Error bars presented means ± standard deviation from three replicates
The expression levels of CABPR1 were much higher in leaves than in the roots (Fig. 4a). The up-regulated expression was observed at 6 h after inoculation between in the leaves and roots. In the leaves, CABPR1 gene expression increased greatly from 6 to 24 h and recorded a peak 17.2-fold at 24 h in the incompatible interaction, whereas the highest level of CaPR1 gene expression was comparatively less, 11.2-fold at 48 h in the compatible interaction. CABPR1 gene expression then gradually declined from 48 to 96 h after inoculation. In addition, the levels of CABPR1 gene expression were much lower in the roots at the same time points. The maximum levels of the transcripts were 5.1-fold in the incompatible interaction and 3.2-fold in the compatible interaction.
The differences in CABGLU gene expression in the different interactions with P. capsici were clear (Fig. 4b). In the incompatible interaction, an accumulation of CABGLU mRNA was detected 6 h after inoculation and remained at the highest level of 13.2-fold at 12 h in the leaves. In the compatible interaction, the accumulation of CABGLU mRNA was much lower than in the incompatible interaction. The highest transcript level was detected in the compatible response 8.6-fold at 24 h in the leaves; however, the mRNA levels gradually declined from 24 to 96 h after inoculation. With regards to the roots, the levels of CABGLU gene expression were lower than in the leaves. In particular, the accumulation of CABGLU mRNA drastically increased at 12 h in the infected roots after inoculation with the avirulent strain, up to 3.1-fold compared to the basal expression levels. The CABGLU gene expression levels gradually decreased from 12 to 48 h, while they presented an increase 72 h after inoculation. In the compatible interaction, the level of CABGLU gene expression was slightly decreased at 6 h, continued to rise thereafter, and retained a peak of 2.0-fold at 24 h after inoculation with the virulent strain. From 24 to 96 h, the expression of CABGLU was decreased.
The expression levels of CAPO1 varied markedly in the leaves inoculated with incompatible and compatible strains of P. capsici (Fig. 4c). In the incompatible interaction, an up-regulation was observed 6 h after inoculation. At this time point, there was a slight increase in the compatible interaction. The levels of CAPO1 gene expression were the highest, 20.5-fold at 12 h and 7.9-fold at 48 h, in the incompatible and compatible interactions, respectively. From 48 to 96 h, CAPO1 gene expression greatly decreased in the compatible interaction. However, there was a slight increase at 48 h among the gradual decline from 12 to 96 h after inoculation with the avirulent strain. Compared to the leaves, the levels of CAPO1 gene expression were relatively lower in the roots. The highest transcript level of 1.3-fold was observed in the compatible interaction at 48 h, which was slightly lower than 2.0-fold at 24 h after inoculation in the incompatible interaction. From 48 to 96 h after inoculation, the levels of CAPO1 gene expression decreased. In the incompatible interactions, the up-regulated expression was detected at 6 h and reached to a peak (2.0-fold) at 24 h after inoculation, after which the levels of CAPO1 gene expression gradually decreased.
We also investigated the expression pattern of the resistance-related gene CaRGA1, which can be induced by P. capsici inoculation (Fig. 4d). Unlike the other three defence-related genes, the expression levels of CaRGA1 were strikingly lower in the leaves. In addition, there were no obvious differences in the accumulation of CaRGA1 mRNA between the leaves and roots. In the leaves, CaRGA1 mRNA began to accumulate at 12 h and reached a peak of 6.0-fold at 24 h after inoculation in the incompatible interaction. However, the compatible interaction exhibited a lower of accumulation of the CaRGA1 mRNA than that of the incompatible interaction, and the highest expression level of 2.0-fold occurred at 72 h after inoculation. In the roots, CaRGA1 gene transcript level gradually increased from 24 to 96 h and peaked (1.9-fold) at 48 h in the incompatible interaction. However, the induction of CaRGA1 gene expression began at 48 h and the highest accumulation level of 1.3-fold recorded at 72 h after inoculation with the compatible strain HX-9 of P. capsici.
Discussion
Plants are able to defend themselves against pathogens through the so-called “biochemical” defences, which normally include secondary metabolites (phytoanticipins and phytoalexins) and defence proteins. (Huang and Knopp 1998; Mithöfer et al. 2004). In the present study, we tested whether a different defence response was induced by inoculation with different strains of P. capsici in the same genetic background (pepper cultivar A3). Defence-related enzymes (β-1,3-glucanase and peroxidase) and four genes involved in plant defence were used markers to test the defence response of pepper to P. capsici infection. A huge number of comparisons were performed between the compatible and incompatible interactions with P. capsici. The results showed that there are differences between the compatible and incompatible interactions between pepper plants and P. capsici.
Defence-related enzymes include chitinase, β-1,3-glucanase and peroxidase, and the activities of these enzymes are known to be induced in many plants (Kook Hwang et al. 1997; Saikia et al. 2005; Govindappa et al. 2010; Chmielowska et al. 2010; Koç and Üstün 2012; Abd EI-Rahman et al. 2012). The root activity was measured and used as a cell vitality indicator using the 2,3,5-triphenyltetrazolium chloride (TTC) method (Chen et al. 2006). The results from our research indicated that there is a difference between the incompatible and compatible interactions. The root activity was much higher in the incompatible than in the compatible strains of P. capsici. The most evident difference in the root activity was observed 12 h after inoculation, being 3.3 greater times in the incompatible than in the compatible interactions (Fig. 1c). Yeom et al. (2011) reported that differences in the root activity were significant from 24 to 48 hpi in CM334 and in Chilsungcho. This difference could be due to the use of different isolates, the cultivars used, or environmental conditions. The resistance to P. capsici in C. annuum is genetically and physiologically complex (Quirin et al. 2005).
β-1,3-glucanases enzymes are believed to play key roles in combating fungal pathogens (Mauch et al. 1988). It has been proposed that they can act in at least two ways, one of which is directly offering some protection against pathogens through their capacity to hydrolyse the cell walls of fungal pathogens. The proposed hydrolysing role is supported by in vitro experiments in which the erosion of fungal walls caused by a combination of β-1,3-glucanases leads to the lysis of the hyphal tip and the inhibition of the growth of different fungi (Mauch et al. 1988). The other method is more indirect, by promoting the release of cell wall-derived materials that can elicit an active defence reaction (Boller 1993). The β-1,3-glucanase activity increased more rapidly in resistant melon cultivars than in susceptible cultivars following inoculation with cucurbit powdery mildew fungus (Rivera et al. 2002). Higher levels of glucanase activity have been reported in the Phytophthora-infected tissues of a tolerant variety of black pepper compared to two susceptible varieties (Jebakumar et al. 2001). The activity of β-1,3-glucanase increased significantly with increasing time after P. capsici infection compared to the control plants in our study (Fig. 2). It is in accordance with the results of Kim and Hwang (1994) and supports the hypothesis that this enzyme may be involved in the reaction against oomycete infection. Kim & Hwang concluded that the accumulation of β-1,3-glucanase was more pronounced in the incompatible pepper-P. capsici interaction, suggesting that β-1,3-glucanase may play a role in the expression of the effective resistance of pepper to invasion by P. capsici. We found that β-1,3-glucanase activity at 12 h after pathogen inoculation in the incompatible interaction, compared to at 24 h following inoculation in the compatible interaction (Fig. 2). However, other authors have not confirmed differences in the compatible and incompatible plant-pathogen reactions (Ahl Goy et al. 1992). These contrasting results may be due to differences in the type of tissue analysed, the method used to inoculate the pathogen, and the time span between the inoculation of P. capsici and the determination of enzymatic activities.
Peroxidase activity is related to the resistance against disease in plant-pathogen interactions (Ye et al. 1990; Graham and Graham 1991). The resistance to P. capsici in some genotypes of C. annuum has been associated with an increase in peroxidase activity (Fernández-Pavia 1997; Egea et al. 2001; Gayoso et al. 2004; Koç and Üstün 2012). For example, in cultivar Smith-5, which is resistant to P. capsici, the increment in peroxidase activity was related to cell wall thickening by lignin accumulation at 24 h after oomycete inoculation (Egea et al. 2001). Such a relationship could be due to the fact that peroxidases catalyse the last step of lignin biosynthesis by polymerising hydroxyl and methoxycinnamic alcohols (Boudet 2000). The increases in the peroxidase activity in the plant pathogen-interaction could be a consequence of an increase in the expression of the PX genes encoding it (Williamson and Hussey 1996; Vercauteren et al. 2001; Gheysen and Fenoll 2002). The earliest response in the leaves was generated 6 h after inoculation in our study, whereas the highest level of peroxidase activity was observed at 12 and 48 h after infection with incompatible and compatible stains of P. capsici, respectively (Fig. 3). A gradual increase in the time of the enzymatic activity of peroxidase in the infected leaves may account for the involvement of this enzyme in the defence mechanism. This enzyme generates a toxic environment and inhibits the pathogen from growing towards inner cells (Melo et al. 2006). The increase in peroxidase activity in pepper is considered to be a general reaction required in phenylpropanoid pathways. The stimulation of phenylpropanoid pathways, depending on the pathogen infection and hence the synthesis of antioxidant compounds and phenolics, is one of the most characteristic properties of this hypersensitive response (Gholizadeh et al. 2004).
Plants exhibit a variety of responses during pathogens infection or abiotic stresses, many of which involve the activation of host pathogenesis-related proteins (Taheri and Tarighi 2012). PR proteins are considered to be stress proteins directed to increase the harmful effects of cellular degradation products on the invading pathogen and prevent further ingress (Mérillon et al. 2012). It is well known that plants respond to attacks by pathogenic fungi by inducing the expression of the genes encoding PR proteins such as PR-1, β-1,3-glucanase, and peroxidase (Conrath et al. 2001). PR proteins are constitutively expressed in plants at low levels, but the expression of most PR proteins is turned on in response to pathogen attack. The induction of PR proteins is a consequence of the activation of plant defensive pathways, which limit the entry or further spread of the pathogen (Baker et al. 1997; Conrath et al. 2002).
However, despite efforts to understand the basis of the compatibility between pepper species and P. capsici, the mechanisms by which this pathogen blocks the defence response are not clear (Núñez-Pastrana et al. 2011). Real-time RT-PCR evaluation of the expression levels of the defence-related genes CABPR1, CABGLU, CAPO1 and CaRGA1 was performed to analysis the nature of the defence response of pepper cultivar A3 inoculated with different strains of P. capsici in our study. Defence-related genes can be activated in both resistant and susceptible plants in response to pathogen attack. However, they often are expressed more rapidly and to a greater extent in incompatible interactions in which a resistant plant is challenged with an avirulent pathogen (Dong et al. 1991). In order to better understand the defence responses to pathogen attack, PR proteins were chosen as positive markers for P. capsici infection (Silvar et al. 2008). The results demonstrated that the expression levels of defence-related genes were much higher in the leaves than in the roots (Fig. 4). This result is in accordance with Silvar et al. (2008), who reported that the highest mRNA levels were found in the leaves, followed by the stems and roots.
In the case of the PR1 protein, several biological functions have been proposed, but its precise role is still unknown. However, increasing evidence implicates the PR-1 protein in the resistance to fungal and oomycete infection. PR-1 proteins may be involved in cell wall thickening and may offer resistance to the spread of the pathogen in the apoplast (Eyal et al. 1992; Chmielowska et al. 2010). Our results revealed that CABPR1 gene expression was up-regulated in the compatible and incompatible interactions (Fig.4a). This result is in agreement with that of Lee et al. (2000), who reported that the accumulation of CABPR1 transcripts was greater and more rapid in an incompatible interaction of pepper with P. capsici. The over-expression of CABPR1 in plants other than pepper, such as tomato, tobacco or Arabidopsis, confers protection against different pathogens (Sarowar et al. 2005, 2006; Hong and Hwang 2005). These results and the findings presented here suggest a role for the basic PR-1 protein in restricting pathogen attack.
β-1,3-glucanase proteins are considered to be part of the defence response of plants to fungal pathogens, presumably by inhibiting growth through their hydrolytic capacity (Mauch et al. 1988). They may act in at least two different ways: directly by degrading the cell walls of the pathogen or indirectly by aiding in the generation of signal molecules that may function as elicitors of further defence mechanisms (Van Loon et al. 2006). Mauch et al. (1988) reported that β-1,3-glucanase activity was similarly induced by both virulent and avirulent strains of P. capsici, which was inconsistent with our statement. Additionally, some researchers have reported that the level and onset of β-glucanase expression is often positively correlated with the degree of resistance to the pathogen. Rivera et al. (2002) demonstrated that β-glucanase activity increased more rapidly in resistant melon cultivars than in susceptible cultivars following inoculation with cucurbit powdery mildew fungus. Similar results have been reported by Egea et al. (1999), who studied β-glucanase expression in resistant and susceptible pepper cultivars infected with Phytophthora capsici L. Our results corroborate these findings: 12 h after inoculation, there was a clear increase in the transcripts of CABGLU in the roots, and this induction was much higher in the incompatible than in the compatible interactions with P. capsici. These results indicated that β-glucanase may be involved in pathogenesis and the response to disease resistance.
Peroxidases, grouped in the PR9 family, are activated in response to pathogen attacks, and are especially related to resistance (Passardi et al. 2005). They can produce massive amounts of reactive oxygen species (oxidative burst) that are involved in plant cell signalling and also create a highly toxic environment for pathogens (Chmielowska et al. 2010). Do et al. (2003) demonstrated that the CAPO1 gene is more strongly induced in plants inoculated with an avirulent isolate than in those inoculated with virulent P. capsici. These conclusions were in consistent with our finding in this study. The reason for this phenomenon may be related to ROS-associated defence responses because peroxidases are closely correlated with H2O2 accumulation during the hypersensitive response in resistant cultivars (Do et al. 2003). Silvar et al. (2008) verified the hypothesis that CAPO1 plays an important role in the oxidative burst that occurs in the early resistance response to pathogen attack. A similar outcome has been presented in our work, where the expression level of CAPO1 was evident at 6 h after inoculation in the incompatible interaction, compared to a slight increase in the compatible interaction. Moreover, CAPO1 encodes for a basic peroxidase, and basic peroxidases have recently been related to the lignification process (Barceló et al. 2004). Chmielowska et al. (2010) have reported that CAPO1 gene expression in pepper stems treated with copper may be related to participation of this gene in the formation of defensive barriers. However, this relationship is a hypothesis and requires further research to confirm.
Although a large number of plant defence-related genes have been cloned, there has been very limited research on the expression levels of these genes in the host-pathogen interactions with different strains of P. capsici (Kramer et al. 2009). The Capsicum annuum blight resistance protein gene CaRGA1 was cloned from the leaves of pepper cultivar CM334 inoculated with P. capsica by using a homology-based approach in combination with thermal asymmetric interlaced (TAIL)-PCR and rapid amplification of cDNA ends (RACE). The gene is cloned on the basis of the conserved sequence of the potato (Solanum bulbocastanum L.) blight resistance protein RPI gene (AY426259) has been designated CaRGA1 (GenBank: GQ386945.1). The expression patterns of CaRGA1 under biotic and abiotic stresses were analysed by quantitative RT-PCR in the pepper cultivar CM334. The results from a previous study indicated that CaRGA1 is involved in the response to abiotic stresses and disease resistance (unpublished results). In this study, we analysed the expression pattern of CaRGA1 in the leaves and roots inoculated with different strains of P. capsici (Fig. 4d). There expression levels of CaRGA1 in the roots were lower from in the early stage of pathogen infection (6 to 24 h) but significantly increased in the later stage (48 to 96 h). The results suggested that CaRGA1 gene expression began later and was lower compared to the other three defence-related genes. Some research has indicated that the expression of some resistance genes can also be influenced by the host genetic background (Cao et al. 2007) and by other environmental factors that may be favourable to pathogen infection (Wang et al. 2001). These differences may be due to the diversity of gene expression and require further investigation in future studies.
In summary, the findings in our study contribute to a better understanding of the correlation between the host plant and pathogen. The defence-related enzymes of β-1,3-glucanase and peroxidase exhibited higher activities in incompatible than in the compatible interactions with different strains of P. capsici. In addition, we further verified by real-time RT-RCR that the expression of the defence-related genes CABPR1, CABGLU, CAPO1 and CaRGA1 were much higher in incompatible than in compatible interactions in the same background.
References
Abd EI-Rahman, S. S., Mazen, M. M., Heba, I. M., et al. (2012). Induction of defence related enzymes and phenolic compounds in lupin (Lupinus albus L.) and their effects on host resistance against Fusarium wilt. European Journal of Plant Pathology, 134, 105–116.
Ahl Goy, P., Felix, G., Matraux, J. P., et al. (1992). Resistance to disease in the hybrid Nicotiana glutinosa × Nicotiana clebney is associated with high constitutive levels of β-l, 3-glucanase, chitinase, peroxidase and polyphenoloxidase. Physiological and Molecular Plant Pathology, 41, 11–21.
Alcantara, T. P., & Bosland, P. W. (1994). An inexpensive disease screening technique for foliar blight of chile pepper seedlings. HortScience, 29, 1182–1183.
Baker, B., Zamsryski, P., Staskaweiz, B., & Dinesh-Kumar, S. P. (1997). Signaling in plant-microbe interactions. Science, 272, 726–733.
Balasubramanian, V., Vashisht, D., Cletus, J., et al. (2012). Plant β-1, 3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnology Letters, 34, 1983–1990.
Barceló, A. R., Gómez Ros, L. V., Gabaldón, C., et al. (2004). Basic peroxidases: the gateway for lignin evolution ? Phytochemistry Reviews, 3, 61–78.
Beffa, R., Martin, H. V., & Pilet, P. E. (1990). In vitro oxidation of indoleacetic acid by soluble auxin-oxidases and peroxidases from maize roots. Plant Physiology, 94, 485–491.
Boller, T. (1993). Antimicrobial functions of the plant hydrolases chitinase and β-1, 3-glucanase. In B. Fritig & M. Legrand (Eds.), Mechanisms of plant defence responses (pp. 391–400). Dordecht: Kluwer Academic Publishers.
Boudet, A. M. (2000). Lignins and lignification: selected issues. Plant Physiology and Biochemistry, 38, 81–96.
Cao, Y., Ding, X., Cai, M., et al. (2007). Expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics, 177, 523–533.
Chen, C. W., Yang, Y. W., Lur, H. S., et al. (2006). A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant & Cell Physiology, 47, 1–13.
Chmielowska, J., Veloso, J., Gutierrez, J., et al. (2010). Cross-protection of pepper plants stressed by copper against a vascular pathogen is accompanied by the induction of a defence response. Plant Science, 178, 176–182.
Conrath, U., Thulke, O., Katz, V., et al. (2001). Priming as a mechanism in induced systemic resistance of plants. European Journal of Plant Pathology, 107, 113–119.
Conrath, U., Pieterse, M. J., & Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends in Plant Science, 7(5), 21–26.
Do, H. M., Hong, J. K., Jung, H. W., et al. (2003). Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Molecular Plant-Microbe Interactactions, 16, 196–205.
Dong, X., Mindrinos, M., Davis, K. R., & Ausubel, F. M. (1991). Induction of Arabidopsis defence genes by virulent and avirulent pseudomonas syringae strain and by a cloned avirulence gene. The Plant Cell, 3, 61–72.
Egea, C., Alcázar, M. D., & Candela, M. E. (1996). Capsidiol: its role in the resistance of Capsicum annuum to Phytophthora capsici. Physiologia Plantarum, 98, 737–742.
Egea, C., Dickinson, M. J., Candela, M., & Candela, M. E. (1999). β-1, 3-glucanase isozymes and genes in resistant and susceptible pepper (Capsicum annuum) cultivars infected with Phytophthora capsici. Physiologia Plantarum, 107, 312–318.
Egea, C., Ahmed, A. S., Candela, M., et al. (2001). Elicitation of peroxidase activity and lignin biosynthesis in pepper suspension cells by Phytophthora capsici. Journal of Plant Physiology, 158, 151–158.
Esra, K., Üstüna, A. S., \( {\mathop {\text{I}}\limits^. } \)slekb, C., et al. (2011). Defense responses in leaves of resistant and susceptible pepper (Capsicum annuum L.) cultivars infected with different inoculum concentrations of Phytophthora capsici Leon. Scientia Horticulturae, 128, 434–442.
Eyal, Y., Sagge, O., & Fluhr, R. (1992). Dark induced accumulation of a basic PR-1 transcript and a light requirement for its induction by ethylene. Plant Molecular Biology, 19(4), 589–599.
Fernández-Pavia, S. (1997). Host-Pathogen interactions in the root rot resistant Phytophthora capsici /Capsicum annuum resistant CM-334 pathosystem. Ph. D. Dissertation. New Mexico State University.
Fung, R. W. M., Wang, C. Y., Smith, D. L. et al. (2004). MeSA and MeJA increase steady-state transcript levels of alternative oxidase and resistance against chilling injury in sweet peppers (Capsicum annuum L.). Plant Science, 166, 711–719.
Gayoso, C., Pomar, F., Merino, F., et al. (2004). Oxidative metabolism and phenolic compounds in Capsicum annuum L. var. annuum infected by Phytophthora capsici Leon. Sci Hortic, 102, 1–13.
Gayoso, C., Martinez de ilarduya, O., Pomar, F., et al. (2007). Assessment of real-time PCR as a method for determining the presence of Verticillium dahliae in different Solanaceae cultivars. European Journal of Plant Pathology, 118, 199–209.
Gheysen, G., & Fenoll, C. (2002). Gene expression in nematode feeding sites. Annual Review of Phytopathology, 40, 191–219.
Gholizadeh, A., Kumar, M., Balasubrahmanyam, A., Sharma, S., et al. (2004). Antioxidant activity of antiviral proteins from Celosia cristata. J Plant Biochem Biotechnol, 13, 13–18.
Govindappa, M., Lokesh, S., Rai, V. R., et al. (2010). Induction of systemic resistance and management of safflower Macrophomina phaseolina root rot disease by biocontrol agents. Arch Phytopathol Plant Protect, 43, 26–40. doi:10.1080/03235400701652227.
Graham, M. Y., & Graham, T. L. (1991). Rapid accutnulation of anionic peroxidases and phenolic polymers in soybean cotyledon tissues following treatment with Phytophthiira megasperma f. sp. ghcinea wall glucan. Plant Physiology, 97, 1445–1455.
Hong, J. K., & Hwang, B. K. (2005). Induction of enhanced disease resistance and oxidative stress tolerance by overexpression of pepper basic PR-1 gene in Arabidopsis. Physiologia Plantarum, 124, 267–277.
Huang, J. S., & Knopp, J. A. (1998). Involvement of nitric oxide in Ralstonia solanacearum-induced hypersensitive reaction in tobacco. In P. Prior, C. Allen, & J. Elphinstone (Eds.), Bacterial wilt disease: Molecular and ecological aspects (pp. 218–224). Berlin: INRA and Springer Editions.
Huang, Z., Yeakley, J. M., Garcia, E. W., et al. (2005). Salicylic acid-dependent expression of host genes in compatible Arabidopsis-virus interactions. Plant Physiology, 137, 1147–1159.
Jebakumar, R. S., Anandaraj, M., & Sarma, Y. R. (2001). Induction of PR-proteins and defence related enzymes in black pepper due to inoculation with phytophthora capsici. Indian Phytopathol, 54, 23–28.
Kauffmann, S., Legrand, M., Geoffroy, P., & Fritig, B. (1987). Biological function of pathogenesis-related proteins: four PR proteins of tobacco have β-1, 3-glucanase activity. EMBO Journal, 6, 3209–3212.
Kim, Y. J., & Hwang, B. K. (1994). Differential accumulation of β-1-3-glucanase and chitinase isoforms in pepper stems infected by compatible and incompatible isolates of Phytophthoracapsici. Physiological and Molecular Plant Pathology, 45, 195–209.
Kim, H. J., Nahm, S. H., Lee, H. R., et al. (2008). BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theoretical and Applied Genetics, 118, 15–27.
Koç, E., & Üstün, A. S. (2012). Influence of Phytophthora capsici L. inoculation on disease severity, necrosis length, peroxidase and catalase activity, and phenolic content of resistant and susceptible pepper (Capsicum annuum L.) plants. Turkish Journal of Biology, 36, 357–371.
Kook Hwang, B., Sunwoo, J. Y., Kim, B. S., et al. (1997). Accumulation of β-1, 3-glucanase and chitinase isoforms, and salicylic acid in the DL-b-amino-n-butyric acid-induced resistance response of pepper stems to Phytophthora capsici. Physiological and Molecular Plant Pathology, 51, 305–322.
Kramer, L. C., Choudoir, M. J., Wielgus, S. M., et al. (2009). Correlation between transcript abundance of the RB gene and the level of the RB-mediated late blight resistance in potato. Mol Plant-Microbe Interact, 22, 447–455.
Lamour, K. H., Stam, R., Jupe, J., et al. (2012). The oomycete broad-host-range pathogen Phytophthora capsici. Molecular Plant Pathology, 13, 329–337.
Lee, Y. K., Hippe-Sanwald, S., Jung, H. W., et al. (2000). In situ localization of chitinase Mrna and protein in compatible and incompatible interactions of pepper stems with Phytophthora capsici. Physiological and Molecular Plant Pathology, 57, 111–121.
Marate, R., Guan, Z., Anandalakshmi, R., et al. (2004). Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Molecular Biology, 55, 501–520.
Mauch, F., Mauch-Mani, B., & Boller, T. (1988). Antifungal hydrolases in pea tissue.II. Inhibition of fungal growth by combinations of chitinase and β-1, 3-glucanase. Plant Physiology, 88, 936–942.
Melillo, M. T., Leonetti, P., Bongiovanni, M., et al. (2006). Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. New Phytologist, 170, 501–512.
Melo, G. A., Shimzu, M. M., & Mazzafera, P. (2006). Polyphenoloxidase activity in coffee leaves and its role in resistance against the coffee leaf miner and coffee leaf rust. Phytochemistry, 67, 277–285.
Mérillon, J.M., Ramawat, K.G., et al. (2012). Pathogenesis Related Proteins in Plant Defence Response. Plant Defence: Biological Control, Springer Netherlands. 12, 379–403.
Mithöfer, A., Schulze, B., & Boland, W. (2004). Biotic and heavy metal stress response in plants: evidence for common signal. FEBS Letters, 566, 1–5.
Nelson, N. (1994). A photometric adaption of the Somogyi method for the degradation of glucose. Journal of Biological Chemistry, 153, 375–380.
Núñez-Pastrana, R., Arcos-Ortega, G. F., Souza-Perera, R. A., et al. (2011). Ethylene, but not salicylic acid or methyl jasmonate, induces a resistance response against Phytophthora capsici in Habanero pepper. European Journal of Plant Pathology, 131(4), 669–683.
Oelke, L. M., Bosland, P. W., & Steiner, R. (2003). Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. Journal of the American Society for Horticultural Science, 128, 213–218.
Ou, L. J., Dai, X., Zhang, Z. Q., et al. (2011). Responses of pepper to waterlogging stress. Photosynthetica, 49, 339–345.
Passardi, F., Cosio, C., Penel, C., et al. (2005). Peroxidases have more functions than army knife. Plant Cell Reports, 24, 255–265.
Quirin, E. A., Ogundiwin, E. A., Prince, J. P., et al. (2005). Development of sequence characterized amplified region (SCAR) primers for the detection of Phyto.5.2, a major QTL for resistance to Phytophthora capsici Leon. in pepper. Theoretical and Applied Genetics, 110, 605–612.
Requena, M. E., Egea-Gilabert, C., & Candela, M. E. (2005). Nitric oxide generation during the interaction with Phytophtora capsici of two Capsicum annuum varieties showing different degrees of sensitivity. Physiologia Plantarum, 124, 50–60.
Ristaino, J. B., & Johnston, S. A. (1999). Ecologically based approaches to management of Phytophthora blight on bell pepper. Plant Disease, 83, 1080–1089.
Rivera, M. E., Codina, J. C., Olea, F., et al. (2002). Differential expression of β-1, 3-glucanase in susceptible and resistant melon cultivars in response to infection by Sphaerotheca fusca. Physiological and Molecular Plant Pathology, 61, 257–265.
Saikia, R., Singh, B. P., Kumar, R., & Arora, D. K. (2005). Detection of pathogenesis related proteins chitinase and β-1,3 glucanase in induced chickpea. Current Science, 89, 659–663.
Sarowar, S., Kim, Y. J., Kim, E. N., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Reports, 24, 216–224.
Sarowar, S., Kim, Y. J., Kim, E. N., Kim, K. D., et al. (2006). Constitutive expression of two pathogenesis-related genes in tomato plants enhanced resistance to oomycete pathogen Phytophthora capsici. Plant Cell, Tissue Organ Culture, 86, 7–14.
Silvar, C., Merino, F., & Díaz, J. (2008). Differential activation of defence-related genes in different pepper cultivars infected with phytophthora capsici. Journal of Plant Physiology, 165, 1120–1124.
Singh, R., Adholeya, A., & Mukerji, K. G. (2000). Mycorrhiza in control of soil borne pathogens. In K. G. Mukerji, B. P. Chamola, & J. Singh (Eds.), Mycorrhizal biology (pp. 173–196). New York: Kluwer Academic Publishers.
Taheri, P. & Tarighi, S. (2012). The Role of Pathogenesis-Related Proteins in the Tomato-Rhizoctonia solani Interaction. Journal of Botany, article ID 137037, 6 pages.
Van Loon, L. C., & Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type protein. Physiology Molecular Plant Pathology, 55, 85–97.
Van Loon, L. C., Rep, M., & Pieterse, C. M. J. (2006). Significance of inducible defence-related proteins in infected plants. Annual Review of Phytopathology, 44, 135–162.
Vercauteren, I., Van Der Schueren, E., Van Montagu, M., et al. (2001). Arabidopsis thaliana genes expressed in the early compatible interaction with root-knot nematodes. Molecular Plant-Microbe Interactactions, 14, 288–299.
Wan, H. J., Yuan, W., Ruan, M. Y., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochemical and Biophysical Research Communications, 416, 24–30.
Wang, Z.-X., Yamanouchi, U., Katayose, Y., et al. (2001). Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and by chemical signals that trigger secondary plant defences. Plant Molecular Biology, 47, 653–661.
Williamson, V. M., & Hussey, R. S. (1996). Nematode pathogenesis and resistance in plants. The Plant Cell, 8, 1735–1745.
Yang, C., Guo, R., Jie, F., et al. (2007). Spatial analysis of Arabidopsis thaliana gene expression in response to Turnip mosaic virus infection. Molecular Plant-Microbe Interactactions, 20, 358–370.
Ye, X. S., Pan, S. Q., & Kuc, J. (1990). Activity, isoenzyme pattern and cellular localization of peroxidase as related to systemic resistance of tobaceo to blue mold (Peronospora tabacina) and to tobacco mosaic virus. Phytopathology, 80, 1295–1299.
Yeom, S. I., Baek, H. K., Oh, S. K., et al. (2011). Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Molecular Plant-Microbe Interactactions, 24, 671–684.
Zhang, Y. L., Gong, Z. H., Li, D. W., et al. (2009). Identification of Phytophthora capsici in Shaanxi Province and screening of the Fungicides against Phytophthora blight of pepper. Acta Agriculturae Boreali-occidentalis Sinica, 5, 336–340.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 31272163), “The Twelfth Five-Year” Plan of National Science and Technology in Rural Areas (No.2011BAD12B03) and the Shaanxi Provincial Science and Technology Coordinating Innovative Engineering Project (2012KTCL02-09). Language help was provided by American Journal Experts (AJE).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, JE., Li, DW., Zhang, YL. et al. Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici . Eur J Plant Pathol 136, 625–638 (2013). https://doi.org/10.1007/s10658-013-0193-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-013-0193-8