Abstract
Lasiodiplodia theobromae (Pat.) Griff. & Maubl, Neofusicoccum parvum Pennycook & Samuels, N. mangiferae Syd. & P. Syd., and Fusicoccum aesculi Corda, all anamorphs of Botryosphaeriaceae species, are the causal agents of mango stem-end rot and fruit rot in Taiwan. Identification of these fungal species based on morphology has not been easy due to their extensive plasticity for some of the morphological characters. To aid reliable identification of Botryosphaeriaceae species associated with mango fruits, four pairs of species-specific primers were designed according to sequences of the ribosomal internal transcribed spacers (ITS), and a rapid method was established based on nested multiplex polymerase chain reaction (PCR) in this study. To perform the analysis, PCR was first run with ITS1 and ITS4 as the primers, followed by a second PCR with the addition of all four sets of species-specific primers. With this method, a low limit of 100 fg-1 pg of purified fungal DNA was detectable. It could also successfully detect L. theobromae, N. parvum, N. mangiferae and F. aesculi in total DNA extracted from inoculated mango fruits. This assay provides a rapid and sensitive method for the identification of Botryosphaeriaceae species and diagnosis of mango fruit rot and stem-end rot as well.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mango (Mangifera indica L.) is an economically important fruit crop in Taiwan. Tainan, Kaohsiung and Pingtung in southern Taiwan are the main areas for mango fruit production. According to Agricultural Statistics Yearbook which was published by Council of Agriculture, Taiwan (http://www.coa.gov.tw/view.php), the area for cultivation of mango trees was approximately 16,796 ha in 2010, which resulted in a yield of 135 kilotons of mango fruits, with an annual crop valued over 157 million US dollars.
Stem-end rot and fruit rot, the major postharvest diseases of mango, are caused by a complex of fungal pathogens, including several species of the Botryosphaeriaceae (Ploetz et al. 1994; Slippers et al. 2005). Recently, Lasiodiplodia theobromae (Pat.) Griff. & Maubl (teleomorph Botryosphaeria rhodina), Neofusicoccum parvum Pennycook & Samuels (teleomorph B. parva), N. mangiferae Syd. & P. Syd. (teleomorph Botryosphaeria sp.), and Fusicoccum aesculi Corda (teleomorph B. dothidea) were identified as the major pathogens causing stem-end rot and fruit rot of mango in Taiwan (Ni et al. 2010; unpublished data). These pathogens have also been associated with mango stem-end rot and fruit rot in Australia, South Africa, and Brazil (de Oliveira Costa et al. 2010; Jacobs 2002; Slippers et al. 2005).
Botryosphaeria is a species-rich genus, and has been associated with various plant disease symptoms such as pistachio shoot blight (Michailides et al. 2002), eucalyptus canker and shoot blight (Yu et al. 2009), mango die-back (Javier-Alva et al. 2009), avocado branch canker (McDonald et al. 2009), avocado dieback (Zea-Bonilla et al. 2007), grapevine cankers (Urbez-Torres 2006), mango stem-end rot and fruit rot (Jacobs 2002; Ni et al. 2010; Slippers et al. 2005), and avocado fruit rot (Ni et al. 2009). These fungal species are easily distinguished from most other fungi by their grey to black, aerial mycelium, and grey to indigo-grey or -black pigment that is visible from the reverse side of Petri dishes (Slippers and Wingfield 2007). For further description of Botryosphaeria spp., the morphological characters of the anamorphs have been widely applied (Denman et al. 2000). These characters include size, shape, colour, septation, wall thickness, and texture of conidia, as well as cultural aspects such as colony morphology and effects of temperature on fungal growth (Alves et al. 2007). However, identification of these species through phenotypic characteristics is complicated by the extensive plasticity of some morphological characters. Taken for example, the size range of conidia of different species may overlap. Furthermore, age and state of maturity may also affect the pigmentation and septation of conidia (Alves et al. 2007). Therefore, tools that can provide accurate, reproducible, and rapid identification of Botryosphaeriaceae species are in urgent need for those not familiar with these fungi.
Over the past decade, DNA-based techniques have been applied to the identification and detection of Botryosphaeria spp. (Crous et al. 2006; Denman et al. 2000; Luchi et al. 2009; 2011; Phillips et al. 2005; Smith and Stanosz 2001). Specifically, sequence data from internal transcribed spacers (ITS) of the nuclear ribosomal DNA (rDNA) have been used to analyze intraspecific and interspecific relationships of Botryosphaeria spp. (Alves et al. 2007; Denman et al. 2000; Jacobs and Rehner 1998; Slippers et al. 2005; Smith and Stanosz 2001; Zhou and Stanosz 2001). The ITS1 and ITS2 regions within the nuclear ribosomal gene clusters are particularly attractive loci for the development of polymerase chain reaction (PCR)-based detection assays, because they are readily accessible using universal primers (White et al. 1990), and often exhibit sufficient interspecific sequence divergence for the design of species-specific primers. The identification of fungal pathogens based on PCR using species-specific primers is now widely used, especially for economically important plant pathogens such as quarantine-listed fungi or those that are difficult to isolate or cause symptomless infections (Chen et al. 2006; Côté et al. 2004; Hamelin et al. 1996; Mishra et al. 2003; Peres et al. 2007). The objectives of the present study were to develop specific primers for the identification of anamorphs of Botryosphaeriaceae species found on mango and establish a nested multiplex PCR method for the detection of L. theobromae, N. parvum, N. mangiferae, and F. aesculi on mango fruits.
Material and methods
Fungal cultures and fungal DNA isolation
Botryosphaeriaceae isolates used in this study were collected from mango fruits in southern Taiwan, and maintained on slants of potato dextrose agar (PDA) (Merck KGaA, Darmstadt, Germany) at 8°C until use. For routine culture, the isolates were grown on PDA at room temperature.
For preparation of DNA, each fungal isolate was grown in a 50-ml centrifuge tube containing 15 ml potato dextrose broth (PDB) (Difco Laboratories, Detroit, MI, USA) at 25°C for 5 days in darkness. The harvested mycelium was rinsed with sterile water and then lyophilized overnight by using a Freeze dryer (FD24-36P, Kingmech. Taiwan). Total genomic DNA of each isolate was extracted with the DNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. The concentration of DNA was quantified by the use of NanoDrop (Thermo Fisher Scientific, Wilmington, MA, USA). Purified genomic DNA was stored at −20°C until use.
Design of species-specific primers
The ribosomal internal transcribed spacers (ITS) of 38 Botryosphaeriaceae isolates were amplified from mycelial DNA using the universal primers ITS1 and ITS4 (White et al. 1990), and their sequences were determined (Table 1). The obtained ITS sequences and other ITS sequences of Botryosphaeriaceae species retrieved from the databases of NCBI (http://www.ncbi.nlm.nih.gov/) were analyzed by multiple sequence alignment using Vector NTI Advance 9 (Invitrogen, Carlsbad, CA, USA). Primers were then designed based on the sequence variability among the four Botryosphaeriaceae species (Table 2).
Assessment of primer specificity by PCR
The specificity of designed primers was tested using DNA of L. theobromae, N. parvum, N. mangiferae, or F. aesculi as the template. In addition, DNA from other fungal isolates commonly found on mango trees and fruits, including Phomopsis sp., Colletotrichum gloeosporioides, Fusarium sp., Pestalotiopsis sp., Alternaria sp., Guignardia sp., Cladosporium sp., and Botrytis sp. were also analyzed. PCR was carried out using a Bio-Rad iCycler Thermal cycler (Hercules, California, USA), with each reaction containing 1x PCR buffer (10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.1 % (w/v) gelatin, 1 % Triton X-100), 100 μM of each dNTP, 0.2 μM of each primer pair (Table 2: Lt397F/Lt397R for L. theobromae; Bd318F/Np479R for N. mangiferae; Np479F/Np479R for N. parvum; Bd318F/Bd318R for F. aesculi), 0.4 U of Prozyme DNA polymerase (Protech Technology Enterprise, Taipei, Taiwan), and 30 ng of template DNA in a total volume of 25 μl. The amplification program included an initial step of 3 min at 94°C, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and elongation at 72°C for 2 min. A final extension was performed at 72°C for 10 min. The amplified products were analyzed by 1.5 % agarose gel electrophoresis.
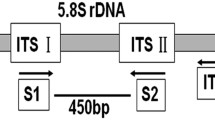
Nested multiplex PCR and assessment of its sensitivity
For nested multiplex PCR, fungal DNA was first subjected to amplification by PCR using the universal primers ITS1 and ITS4 (White et al. 1990). The reaction mixture (25 μl) contained 1x PCR buffer (10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.1 % (w/v) gelatin, 1 % Triton X-100), 100 μM of each dNTP, 0.2 μM of the primer pair, 0.4 U of Prozyme DNA polymerase (Protech Technology Enterprise), and 30 ng of fungal DNA. The amplification program included an initial step of 4 min at 94°C, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and elongation at 72°C for 30 s. A final extension was performed at 72°C for 10 min. Subsequently, the amplified products were diluted 30 folds and 1 μl was used as the template for a multiplex PCR as described in the previous section.
To analyze the sensitivity of the nested multiplex PCR, DNA of the four anamorphic species of Botryosphaeriaceae was diluted serially with distilled water to yield a final concentration ranging from 100 ng/μl to 10 fg/μl, which were then used as the templates for amplification by the aforementioned nested multiplex PCR, followed by analysis of the amplified products by 1.5 % agarose gel electrophoresis.
Inoculation of mango fruits and detection of fungal pathogens by nested multiplex PCR
For the inoculation experiment, L. theobromae (isolate B-878), N. parvum (isolate B-010), N. mangiferae (isolate B-763), and F. aesculi (isolate B-811) were grown on PDA for 7 days. Prior to inoculation, mango fruits (cv. Irwin) were first treated with hot water (60°C) for 20 s to prevent the break of latent infections caused by C. gloeosporioides. To inoculate the pathogen, mango fruits were wounded by pricking once with a sterile needle, and then covered with a mycelial agar block (5 mm in diameter) excised from the culture of the test fungal strain. In a parallel experiment, the wounded area was covered with an agar block without any mycelium to serve as a negative control. When the symptom of fruit rot appeared, a thin layer of the fruit peel surrounding the lesion was excised for DNA isolation by using a quick method that was modified from Wang et al. (1993). In brief, fruit peels (100 mg) were cut into pieces of approximately 1 cm2 and macerated with a homogenizer (Sample Preparation System, FastPrep-24, MP Biomedicals, CA, USA) in the presence of 200 μl of 0.5 N NaOH. Following centrifugation at 12,000 rpm for 5 min, the supernatant was collected and mixed thoroughly with 9 volumes of 0.1 M Tris buffer (pH 8.0). Aliquots of the mixture were then used as the template for nested multiplex PCR as described in the previous section.
Results
Design of pathogen-specific primers
To design primers for the specific detection of Botryosphaeriaceae species, complete sequences of the ITS regions of 38 fungal isolates were determined in this study (Table 1). All ITS sequences from fungal isolates of the same species were identical, despite that some of them were collected from different areas of Taiwan. Pairwise comparison of the sequences indicated that ITS of N. parvum was closely related to that of N. mangiferae, with a sequence identity of 97 %, followed by F. aesculi (93 % identity) and L. theobromae (87 % identity). ITS sequence of L. theobromae showed an identity of 86 % with that of F. aesculi. Multiple sequence alignment revealed the presence of sequence variations among different fungal pathogens (Fig. 1). Six primers were designed accordingly, which would make up four pairs, each specific for one pathogen: Lt397F/Lt397R for L. theobromae, Bd318F/Np479R for N. mangiferae, Np479F/Np479R for N. parvum, and Bd318F/Bd318R for F. aesculi, respectively (Table 2).
Nucleotide sequence alignment of the ribosomal internal transcribed spacer 1 (ITS1), 5.8 S rRNA, and ITS2 among Fusicoccum aesculi (Fa), Lasiodiplodia theobromae (Lt), Neofusicoccum mangiferae (Nm), and N. parvum (Np). Multiple sequence alignment was performed by using Vector NTI Advance 9 software. Sequences that are identical across all four species are shaded in grey. Positions of the primers for species-specific detection of Botryosphaeriaceae species are marked with rectangles
Specificity and sensitivity of the primers
As a preliminary test, PCR was performed with the addition of a single pair of primer and template DNA from the corresponding fungal species. Analysis by agarose gel electrophoresis indicated that, when DNA from indicated fungal species was used as the template, PCR primed with the corresponding primer pair amplified a single DNA product of the expected size, which is 397 bp in length for L. theobromae (Fig. 2, lanes 1–2), 378 bp for N. mangiferae (Fig. 2, lanes 3–4), 479 bp for N. parvum (Fig. 2, lanes 5–6), and 318 bp for F. aesculi (Fig. 2, lanes 7–8), respectively. Specificity of each primer pair was further verified by multiplex PCR with the addition of all six primers. Analysis by agarose gel electrophoresis indicated that, while amplified products of the expected size were obtained with DNA from L. theobromae, N. mangiferae, N. parvum, and F. aesculi as the template (Fig. 3, lanes 1–4), no fragment was detected from other fungal species isolated from mango fruits or branches (Fig. 3, lanes 5–9, 11, 12) except Guignardia sp., which gave rise to a weak band of 1.2 kb in length (Fig. 3, lane 10). Sensitivity of each primer pair was analyzed by using a serial dilution of purified fungal DNA as the template for PCR. Analysis of the amplified products by agarose gel electrophoresis indicated that the detection limit of PCR was approximately 10 pg for each of the primer pairs (data not shown).
Amplified products obtained by polymerase chain reaction with species-specific primer pairs. The amplification reaction was performed using DNA from Lasiodiplodia theobromae, Neofusicoccum mangiferae, N. parvum, or Fusicoccum aesculi as the template. Primers used in each reaction were: Lt397F/Lt397R (for L. theobromae, lanes 1–2), Bd318F/Np479R (for Neofusicoccum mangiferae, lanes 3–4), Np479F/Np479R (for N. parvum, lanes 5–6), and Bd318F/Bd318R (for Fusicoccum aesculi, lanes 7–8), respectively. M: 100-bp DNA ladder (Protech Technology Enterprise, Taipei, Taiwan)
Specificity test of the primers. Specificity of the primers was testified by performing multiplex polymerase chain reaction with the addition of all four sets of primers (Lt397F/Lt397R, Bd318F/Np479R, Np479F/Np479R, and Bd318F/Bd318R), using DNA from different fungal species as the template: Lasiodiplodia theobromae (lane 1), Neofusicoccum mangiferae (lane 2), N. parvum (lane 3), Fusicoccum aesculi (lane 4), Phomopsis sp. (lane 5), Colletotrichum gloeosporioides (lane 6), Fusarium sp. (lane 7), Pestalotiopsis sp. (lane 8), Alternaria sp. (lane 9), Guignardia sp. (lane 10), Cladosporium sp. (lane 11), and Botrytis sp. (lane 12). Lane13, no DNA template. M, 100-bp DNA ladder (Protech Technology Enterprise, Taipei, Taiwan)
To enhance the sensitivity, a method for detecting the pathogen by nested multiplex PCR was developed. PCR was first performed using ITS1 and ITS4 (White et al. 1990) as primers, the amplified product of DNA from F. aesculi, N. mangiferae, or N. parvum was 580 bp in length, while that obtained with L. theobromae was 540 bp in length (data not shown). The amplified products were then diluted and used as the template for a second PCR with the addition of all four pairs of primers, namely Bd318F/Bd318R, Lt397F/Lt397R, Bd318F/Np479R or Np479F/Np479R, gave rise to amplified products of the expected size (Fig. 4). Analysis of amplified products obtained from the second PCR indicated that, when the amount of purified DNA was reduced to 100 fg, PCR primed with Lt397F/Lt397R (for L. theobromae) and Bd318F/Bd318R (for F. aesculi) still generated DNA bands of significant intensity (Fig. 4, a and d). PCR primed with Bd318F/Np479R (for N. mangiferae) and Np479F/Np479R (for N. parvum), in contrast, needed a low limit of 1 pg of purified DNA for significant signals (Fig. 4, b and c).
Sensitivity of nested multiplex polymerase chain reaction. Serial dilutions of purified DNA from Lasiodiplodia theobromae (a), Neofusicoccum parvum (b), Neofusicoccum mangiferae (c), and Fusicoccum aesculi (d) were used as the template for the first PCR with ITS1 and ITS4 as the primer pair. The resultant amplified products were then diluted and used as the template for nested multiplex PCR with the addition of all four sets of species-specific primers (397 F/Lt397R, Np479F/Np479R, Bd318F/Np479R, and Bd/318 F/Bd318R). M: 100-bp DNA ladder (Protech Technology Enterprise, Taipei, Taiwan)
Detection of pathogens present on the inoculated mango fruits by nested multiplex PCR
To know if the nested PCR method is useful for the detection of fungal pathogens in the diseased tissues, mango fruits were inoculated with isolate of L. theobromae, F. aesculi, N. mangiferae, or N. parvum. Once the symptom of fruit rot appeared, DNA was isolated from the diseased tissue and analyzed by nested multiplex PCR. The results indicated that a single DNA band of the expected size was detected for each plant sample which had been inoculated with a specific fungal pathogen (Fig. 5, lanes 1–4). In contrast, no amplified product was detected when DNA from mock-inoculated mango fruits was used as the template (Fig. 5, lane 5). The results indicated that the nested multiplex PCR method established in this study may serve as an efficient tool for the detection and identification of Botryosphaeriaceae species which cause stem-end rot and fruit rot on mango.
Detection of the pathogen on artificially infected mango fruits by nested multiplex polymerase chain reaction. Following inoculation of the mango fruits with Lasiodiplodia theobromae (lane 1), Neofusicoccum parvum (lane 2), N. mangiferae (lane 3), or Fusicoccum aesculi (lane 4), DNA was isolated from the diseased tissues and analyzed by nested multiplex PCR. Primers used for the first PCR was ITS1 and ITS4 (White et al. 1990), and those for the second PCR included all four pairs of species-specific primers (Lt397F/Lt397R, Bd318F/Np479R, Np479F/Np479R, and Bd318F/Bd318R). Lane 5, DNA from mock-inoculated mango fruit; Lane 6, no DNA template. M, 100-bp DNA ladder (Protech Technology Enterprise, Taipei, Taiwan)
Discussion
L. theobromae, N. mangiferae, N. parvum, and F. aesculi have recently been recognized as major pathogens causing stem-end rot and fruit rot of mango in Taiwan (Ni et al. 2010 and unpublished data). These pathogenic fungi may simultaneously colonize on the mango fruits or inside the branch, and thus make accurate diagnosis of the disease more complicated. Traditional methods for the identification of fungal pathogens involve isolation of fungal strains, induction of sporulation, as well as examination of morphological characteristics under microscope. Identification of Botryosphaeriaceae species based on morphological characters could be even more difficult because the teleomorphs of most species were not found in the field (Denman et al. 2000; Jacobs and Rehner 1998). Moreover, colony and conidial characters of these species may vary considerably, which strongly hampered the differentiation of Botryosphaeriaceae species (Slippers et al. 2005).
Recently, several molecular methods have been developed for the identification of Botryosphaeriaceae species. Ma and Michailides (2002) designed a pair of group-specific PCR primers, BDI and BDII, based on ITS sequences, for the identification of Fusicoccum sp. from pistachio and other host plants in California. However, these primers were unable to differentiate other species of Botryosphaeriaceae pathogens assayed in the present study. Recently, fingerprinting methods such as MSP-PCR (microsatellite-primed polymerase chain reaction) and rep-PCR (repetitive-sequence based polymerase chain reaction) have been developed for rapid differentiation of Botryopshaeriaceae, including L. theobromae, N. parvum and F. aesculi (teleomorph: B. dothidea) (Alves et al. 2007). However, these methods are usually used to analyze highly variable regions of the genome, and thus serve as a better marker for taxonomic studies below the species level (Ordoñez and Kolmer 2009). Multiplex PCR is an advanced method in which one or more regions are simultaneously amplified in a single PCR reaction (Asano et al. 2010). In this study, we have successfully designed four pairs of primers which show good specificity towards a specific fungal pathogen, and no products were amplified by these primers for other fungal pathogens isolated from mango fruits and branches, including C. gloeosporioides, Phomopsis sp., Alternaria sp., Botrytis sp., and Cladosporium sp. With these primers, we developed a nested multiplex PCR to differentiate and detect L. theobromae, N. mangiferae, N. parvum, and F. aesculi on mango. This method can be of great value especially for those researchers who do not have enough experience with Botryosphaeriaceae species.
To enhance the sensitivity of detection, we first performed PCR with ITS1 and ITS4 as the primers in order to enrich fungal rDNA in the plant sample, and then used the amplified product as the template for multiplex PCR, which selectively amplified DNA of the target pathogen. By applying the nested multiplex PCR, the detection limit could reach a level of 1 pg-100 fg of DNA, which was 10–100 times more sensitive than with multiplex PCR alone. Increased sensitivity and specificity of nested PCR over conventional PCR was also noted in studies of other plant-pathogenic fungi (Ma et al. 2003; Pérez-Hernández et al. 2008; Tsai et al. 2006; Wang et al. 2009).
The preparation of total genomic DNA from peel of mangoes is critical for a successful PCR reaction. In this study, we extracted genomic DNA of Botryosphaeriaceae species from mango fruits by an alkaline (NaOH) solution method modified from Wang et al. (1993). With this method, DNA was obtained from mango peel within 30 min, and most importantly, it serves as a good template for amplification by the nested multiplex PCR. This method was also applied to successfully detect L. theobromae, N. mangiferae, F. aesculi, and N. parvum from artificially inoculated mango fruits. This ‘grinding and use’ protocol is convenient and efficient, especially in the examination of large numbers of mango fruits for the presence of potential fungal pathogens. Similar extraction protocols have been reported for the detection of Phytophthora on orchid (Tsai et al. 2006), Fusicoccum sp. (Ma and Michailides 2002), Colletotrichum lindemuthianum on bean (Wang, et al. 2008) and Cylindrocarpon liriodendri grapevine (Alaniz et al. 2009). Nonetheless, sufficient dilution of the DNA extract is necessary to eliminate the effect of potential inhibitors on PCR due to the presence of phenolic compounds on mango peels (Jitareerat and Wongs-Aree 2006). As well, the nested multiplex PCR method may be used to identify fungal isolates as long as there is a sufficient amount of fungal material (10 mg fresh weight), without the need to wait 2 weeks for the formation of mature pycnidia and pycnidiospores, which are required for identification based on morphology.
In conclusion, a highly specific and sensitive nested multiplex PCR method has been developed for Botryosphaeriaceae species which infects mango fruit. To our knowledge, this is the first report to develop a multiplex PCR which enables simultaneous identification of L. theobromae, N. mangiferae, N. parvum, and F. aesculi on mango in a single PCR amplification reaction. The species-specific primers designed here could thus contribute to improve the management of fruit rot and stem-end rot of mango due to reliable species identification. It also provides a rapid, effective, and validated detection protocol for one-day analysis of fruits. In the future, we could use the method described here to detect the existence of the aforementioned four pathogens in the field in order to understand the dynamics of their survival and their importance in disease development.
References
Alaniz, S., Armengol, J., Garcia-Jimenez, J., Abad-Campos, P., & Leon, M. (2009). A multiplex PCR system for the specific detection of Cylinddrocarpon liriodendri, C. macrodidymum, and C. pauciseptatum from grapevine. Plant Disease, 93, 821–825.
Alves, A., Phillips, A. J. L., Henriques, I., & Correia, I. A. (2007). Rapid differentiation of species of Botryosphaeriaceae by PCR fingerprinting. Research in Microbiology, 158, 112–121.
Asano, T., Senda, M., Suga, H., & Kageyama, K. (2010). Development of multiplex PCR to detect five Pythium species related to turfgrass diseases. Journal of Phytopathology, 158, 609–615.
Chen, L. S., Chu, C., Liu, C. D., Chen, R. S., & Tsay, J. G. (2006). PCR-based detection and differentiation of anthracnose pathogens, Colletotrichum gloeosporioides and C. truncatum, from vegetable soybean in Taiwan. Journal of Phytopathology, 154, 654–662.
Côté, M. J., Tardif, M. C., & Meldrum, A. J. (2004). Identification of Monilinia fructigena, M. fructicola, M. laxa, and M. polystroma on inoculated and naturally infected fruit using multiplex PCR. Plant Disease, 88, 1219–1225.
Crous, P. W., Slippers, B., Wingfield, M. J., Rheeder, J., Marasas, W. F. O., Philips, A. J. L., et al. (2006). Phylogenetics lineages in the Botryosphaeriaceae. Studies in Mycology, 55, 235–253.
de Oliveira Costa, V. S., Michereff, S. J., Martins, R. B., Câmara, M. P. S., Gava, C. A. T., & Mizubuti, E. S. G. (2010). Species of Botryosphaeriaceae associated on mango in Brazil. European Journal of Plant Pathology, 127, 509–519.
Denman, S., Crous, P. W., Taylor, J. E., Kang, J. C., Pascoe, I., & Wingfield, M. J. (2000). An overview of the taxonomic history of Botryosphaeria and re-evaluation of its anamorphs based on morphology and ITS rDNA phylogeny. Studies in Mycology, 45, 129–140.
Hamelin, R. C., Bérubé, P., Gignac, M., & Bourassa, M. (1996). Identification of root rot fungi in nursery seedling by nested multiplex PCR. Applied and Environmental Microbiology, 62, 4026–4031.
Jacobs, R. (2002). Characterization of Botryosphaeria species from mango in South Africa. M. Sc. thesis. University of Pretoria, Pretoria, South Africa. 162 pp.
Jacobs, K. A., & Rehner, S. A. (1998). Comparison of cultural and morphological characters and ITS sequences in anamorphs of Botryosphaeria and related taxa. Mycologia, 90, 601–610.
Javier-Alva, J., Gramaje, D., Alvarez, L. A., & Armengol, J. (2009). First report of Neofusicoccum parvum associated with dieback of mango trees in Peru. Plant Disease, 93, 426.
Jitareerat, P., & Wongs-Aree, C. (2006). Detection of quiescent infection of Colletotrichum gloeosporioides on green mango fruit by polymerase chain reaction. Acta Horticulture (ISHS), 712, 927–935.
Luchi, N., Pratesi, N., Simi, L., Pazzagli, M., Capretti, P., Scala, A., et al. (2011). High-resolution melting analysis: a new molecular approach for the early detection of Diplodia pinea in Austrian pine. Fungal Biology, 115, 715–723.
Luchi, N., Pinzani, P., Pazzagli, M., & Capretti, P. (2009). Detection of Botryosphaeriaceae species by real-time PCR. Phytopathologia Mediterranea, 48, 163.
Ma, Z., Luo, Y., & Michalides, T. J. (2003). Nested PCR assays for detection of Monilinia fructicola in stone fruit orchards and Botryosphaeria dothidea from pistachios in California. Journal of Phytopathology, 151, 312–322.
Ma, Z., & Michailides, T. J. (2002). A PCR-based technique for identification of Fusicoccum sp. from pistachio and various other hosts in California. Plant Disease, 86, 515–520.
McDonald, V., Lynch, S., & Eskalen, A. (2009). First report of Neofusicoccum australe, N. luteum, and N. parvum associated with avocado branch canker in California. Plant Disease, 93, 967.
Michailides, T. J., Morgan, D. P., & Felts, D. (2002). First report of Botryosphaeria rhodinea causing shoot blight of pistachio in California. Plant Disease, 86, 1273.
Mishra, P. K., Fox, R. T., & Culham, A. (2003). Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusaria. FEMS Microbiology Letter, 218, 329–332.
Ni, H. F., Liou, R. F., Hung, T. H., Chen, R. S., & Yang, H. R. (2009). First report of a fruit rot disease of avocado caused by Neofusicoccum mangiferae. Plant Disease, 93, 760.
Ni, H. F., Liou, R. F., Hung, T. H., Chen, R. S., & Yang, H. R. (2010). First report of fruit rot disease of mango caused by Botryosphaeria dothidea and Neofusicoccum mangiferae in Taiwan. Plant Disease, 94, 128.
Ordoñez, M. E., & Kolmer, J. A. (2009). Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in North America. Phytopathology, 99, 750–758.
Peres, N. A., Harakava, R., Carroll, G. C., Adaskaveg, J. E., & Timmer, L. W. (2007). Comparison of molecular procedures for detection and identification of Guignardia citricarpa and G. mangiferae. Plant Disease, 91, 525–531.
Pérez-Hernández, O., Nam, M. H., Gleason, M. L., & Kim, H. G. (2008). Development of nested polymerase chain reaction assay for detection of Colletotrichum acutatum on symptomless strawberry leaves. Plant Disease, 92, 1655–1661.
Phillips, A. J. L., Alves, A., Correia, A., & Luque, J. (2005). Two new species of Botryosphaeria with brown, 1-septate ascospores and Dothiroella anamorphs. Mycologia, 97, 513–529.
Ploetz, R. C., Zentmyer, G. A., Nishijima, W. T., Rohrbach, K. G., & Ohr, H. D. (Eds.). (1994). Compendium of tropical fruit diseases. St Paul: American Phytopathological Society Press.
Slippers, B., Johnson, G. I., & Crous, P. W. (2005). Phylogenetic and morphological re-evaluation of the Botryosphaeria species causing diseases of Mangifera indica. Mycologia, 97, 99–110.
Slippers, B., & Wingfield, M. J. (2007). Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews, 21, 90–106.
Smith, D. R., & Stanosz, G. R. (2001). Molecular and morphological differentiation of Botryosphaeria dothidea (anamorph Fusicoccum aesculi) from other fungi with Fusicoccum anamorphs. Mycologia, 93, 505–515.
Tsai, H. L., Huang, L. C., Ann, P. J., & Liou, R. F. (2006). Detection of orchid Phytophthora disease by nested PCR. Botanical Studies, 47, 379–387.
Urbez-Torres, J. R. (2006). Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Disease, 90, 1490–1503.
Wang, H., Qi, M., & Adrian, J. C. (1993). A simple method of preparing plant samples for PCR. Nucleic Acids Research, 21, 4153–4154.
Wang, X., Tang, C., Chen, J., Buchenauar, H., Zhao, J., Han, Q., et al. (2009). Detection of Puccinia striiformis in latently infected wheat leaves by nested polymerase chain reaction. Journal of Phytopathology, 157, 490–493.
Wang, W., Tang, J. H., & Wang, Y. C. (2008). Molecular detection of Colletotrichum lindemuthianum by duplex PCR. Journal of Phytopathology, 156, 431–437.
White, T. J., Bruns, T., Lee, S., & Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR Protocols: A Guide to Methods and Applications (pp. 315–322). San Diego: Academics Press.
Yu, L., Chen, X. L., Gao, L. L., Chen, H. R., & Huang, Q. (2009). First report of Botryosphaeria dothidea causing canker and shoot blight of Eucalyptus in China. Plant Disease, 93, 764.
Zea-Bonilla, T., Gonzalez-Sanchez, M. A., Martin-Sanchez, P. M., & Perez-Jimezez, R. M. (2007). Avocado dieback caused by Neofusicoccum parvum in the Andalucia region, Spain. Plant Disease, 91, 1052.
Zhou, S., & Stanosz, G. R. (2001). Relationships among Botryosphaeria species and associated anamorphic fungi inferred from the analyses of ITS and 5.8S rDNA sequences. Mycologia, 93, 516–527.
Acknowledgements
This project was support by the Council of Agriculture, Executive Yuan, R. O. C. We also gratefully acknowledge Ms. S. L. Hsu and S. Y. Lai for their help in the isolation of fungal pathogens, pathogenicity test, and sequence analysis.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ni, HF., Yang, HR., Chen, RS. et al. A nested multiplex PCR for species-specific identification and detection of Botryosphaeriaceae species on mango. Eur J Plant Pathol 133, 819–828 (2012). https://doi.org/10.1007/s10658-012-0003-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-012-0003-8