Abstract
Ammonia is one of the major aquatic environmental pollutants that can bring detrimental effects to the growth and survival of aquatic organisms. However, the molecular mechanisms of ammonia toxicity and ammonia excretion in marine invertebrates especially mollusks are still poorly understood. Cephalopods are exclusively ammonotelic with high protein metabolism and ammonia excretion rate, making this taxonomic group an ideal specimen to explore the ammonia detoxification mechanism. In this study, comparative transcriptomes were employed to investigate the transcriptional changes of O. minor in responses to acute ammonia exposure. A total of 63,237 unigenes with an average length of 811 bp were discovered and 25,708 unigenes were successfully annotated. The transcription of 1845 genes were significantly changed after ammonia stress, including 315 up-regulated genes and 1530 down-regulated genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on differentially expressed genes (DEGs) revealed that 44 GO terms and 55 KEGG pathways were over-represented. Notably, a large number of genes involved in immune defense, citric acid (TCA) cycle, oxidative phosphorylation and amino acid metabolisms were significantly down-regulated, indicating the decelerated energy production and amino acid rate in response to acute ammonia stress. These results provide new insights into the potential molecular mechanism of ammonia detoxification on transcriptomic level and will facilitate further mechanism studies on mollusks.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Octopus minor is one of the most important commercial cephalopods in China and it is a promising species for aquaculture because of its benthic lifestyle, short life span and rapid growth rate (Zheng et al. 2014). In north China only, the overall amount of catching is more than 10,000 tons per year. High price and great demand require the development of O. minor industry. Although restrictions such as low fecundity, long embryonic development period and feeding problems still exist, the artificial culturing of O. minor has been working since 2009, and some progress has been made (Liu 2013; Qian et al. 2013; Zheng et al. 2014).
As short-lived aquatic livestock, cephalopods are quite vulnerable to environmental chaies and even slight changes in water quality might result in death (Pierce et al. 2006). Ammonia is one of the major indexes in the aquatic environment and can be toxic to aquatic animals, meanwhile, other parameters such as high temperature, pH, as well as low salinity can increase the amount of ammonia in seawater (Bower and Bidwell 1978; Colt and Armstrong 2009). Specifically, the amount of ammonia excretion in cephalopods is about 2–3 times higher than that of in fish in the same bodyweight (Lee 1994). Moreover, octopuses produce much more ammonia than other species within this family and can be extensively affected by the ammonia excretion (Vaz-Pires et al. 2004). All those make octopus a terrific model for ammonia toxicity mechanism studies.
Un-ionized ammonia in culturing water can enter the hemolymph through the gills and accumulate in the hemolymph, leading to the toxicity effects on aquatic animals (Bower and Bidwell 1978; Colt and Armstrong 2009). The mechanism of ammonia toxicity in aquatic animal is not clear, but many researches indicated that high ammonia can cause functional disorder and even influence the growth and survival (Chen and Kou 1992; Colt and Armstrong 2009; Smart 1978; Tomasso 1994). Generally, ammonia toxicity majorly includes the following aspects: (1) the histologic damages such as gill hyperplasia, liver and kidney degeneration (Benli et al. 2008; Peyghan and Takamy 2002), (2) physiological and biochemical changes such as disturbance of electrochemical gradients, blood pH decreasing, ATP consumption (Arillo et al. 1981; Smart 1978; Tomasso 1994; Vedel et al. 1998), (3) damages to the nervous system and immune system and hence susceptible to bacterial, fungal and parasites (Evans et al. 2006; Hurvitz et al. 1997; Vedel et al. 1998). In octopus, exposing to acute ammonia can lead to increases in skin mucus secretion, squirt of ink, prey consumption and changes in chromatophore, and can even influence the survival of octopus paralarvae (Feyjoo et al. 2011).
Immune response to ammonia stimulation varies in different species. Mollusks like cephalopods that lack the specific immune system, immune protecting mechanisms are mainly comprised of humoral factors and circulating hemocytes (Ford 1992). Hemocytes are fundamental in transporting and absorbing of nutrient substance, mediating and effecting the cellular defense to pathogen invasion and environmental stress, repairing the tissue damage caused by mechanical and biological trauma (Castellanos-Martinez et al. 2014a, b; Cheng 1977; Ford 1992; Perez and Fontanetti 2011). Cephalopod hemocytes are crucial in immune defense and are capable to encapsulate and neutralize the nonself materials and perform the function like phagocytosis (Ford 1992; Grimaldi et al. 2013). The number and the type of hemocytes can reflect the wellbeing of cephalopods and even can serve as the immune-marker of water pollution in some mollusks (Gestal et al. 2015; Mayrand et al. 2005; Perez and Fontanetti 2011). Apart from the immune responses against the ammonia stress, some recent researches also reported the metabolism and structural changes in accordance with the high ammonia accumulation, including cytoskeleton remodeling, disturbance of amino acids metabolism, nucleotide metabolism, lipid metabolism and changes in energy production (Wang et al. 2017; Xiao et al. 2019).
In recent years, high throughput sequencing data from crustaceans provide us new insights into molecular mechanisms of ammonia excretion, nevertheless in mollusks only limited information is available and molecular responses to ammonia toxicity and detoxification mechanisms in this phylum remain unclear (reviewed in Weihrauch and Allen 2018). Hence, in this study, we performed the hemocytes transcriptome analysis in O. minor to investigate the genes and metabolic pathways involved in the responses to ammonia stress. To our knowledge, this work exhibited the first RNA-seq data for cephalopods in responses to ammonia stress, and we hope it will improve our understanding of ammonia detoxification mechanisms in mollusks.
Materials and methods
Octopus minor cultivation and ammonia challenge experiment
Before the challenge experiment, we conducted a pre-experiment to explore the half-lethal dose of O. minor in 48 h. The result revealed the 48-h half-lethal concentration (LC50) of total ammonia in O. minor is 200.365 mg/L, which contains un-ionized ammonia (NH3) of 4.62 mg/L (Xu and Zheng 2018). A total number of 30 samples of mixed sex weighing from 86–214 g were selected and were acclimated for one week in tanks. All the tanks are in the same size (46 cm × 56 cm × 50 cm) and each tank was filled with about 50 L seawater (46 cm × 56 cm × 20 cm). All the samples were divided into two groups randomly, the control group with normal culturing (OM-C) and the experimental group with ammonia challenge (OM-E), and each group contained 3 biological repeats and each repeat included five individuals. During the acclimation period, all the samples were fed with live crabs (Asian shore crab: Hemigrapsus sanguineus). The seawater was changed twice a day and each time only half of the seawater (about 25 L) was renewed. Because the habitat environment of O. minor is commonly dark, the tank was covered with black plastic. Sand-filtered seawater was used in this study and the seawater conditions were shown as following: pH at 7.8 ± 0.2, temperature at 22 ± 1 °C, salinity at 28 ppt and oxygen higher than 6 mg/L.
After acclimation, the OM-E group were treated with 200.365 mg/L total ammonia for 8 h. All samples were anesthetized with 7.5% magnesium chloride before hemolymph extraction to follow the ethical procedures and reduce suffering (Andrews et al. 2013; Messenger et al. 1985). A dorsal incision was made on the mantle to collect the hemolymph from the cephalic aorta using the disposable syringe. About 1.5 mL of hemolymph were collected from each sample and then centrifuged at 3000 × g, 4 °C for 15 min. After centrifugation, only hemocytes on the bottom of the tube were kept and were re-suspended using 1 mL Trizol reagent (Invitrogen) and stored at −80 °C until the RNA extraction.
RNA isolation, mRNA library construction and sequencing
Total RNA of hemocytes from all 30 individuals was extracted using Trizol Reagent according to the manufacture’s protocol. In short, for the hemocytes suspended in Trizol reagent, 0.2 ml of chloroform was added (per 1 ml of Trizol reagent) and was incubated in room temperature for 3 min then centrifugated at 12,000 × g for 15 min at 4 °C. The upper aqueous phase was transferred into a new tube and mixed with 0.5 mL isopropyl alcohol for RNA precipitation (incubate for 10 min and centrifuge at 12,000 × g for 10 min at 4 °C). Then, the supernatant was removed and the gel-like pellet raw RNA was washed with 1 mL 75% ethanol followed by centrifugation at 7500 × g for 5 min at 4 °C (remove ethanol and repeat twice). Finally, air-dry RNA pellet and dissolve RNA in DEPC-treated water. The quality of RNA was detected by NanoDrop 2000 spectrophotometer and 1% agarose gels. RNA of five individuals from the same biological repeats were pooled to construct the mRNA libraries. The pooled RNA samples were purified though Oligo (dT) beads and then cleaved into short fragments using fragmentation buffer under high temperature. Those short fragments were reverse transcribed into cDNA with random primers and then the second-strand cDNA was synthesized. The synthesized cDNA was ligated to the sequencing adapters after purification and end repair step. The ligation products of 200–300 bp were selected by 2% agarose gel and were amplified by PCR. Then the products were sequenced using Illumine HiSeqTM 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
The raw data were submitted to the NCBI (accession number: SAMN08684187–SAMN08684192).
De novo transcriptome assembly and unigene annotation
The raw reads were filtered by removing the adapters, low quality reads and ploy-N to obtain high-quality clean reads for the downstream analysis. The de novo assembly was performed in the Trinity (Grabherr et al. 2011). Blastn was used to aligned the unigenes in the non-redundant nucleotide databases and Blastp were used to hit against the NCBI non-redundant protein (Nr), Swiss-Prot, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and COG/KOG databases with the E value threshold of 1e − 5. Protein functional annotations were obtained following the best alignment results.
Identification of differentially expressed genes and enrichment analysis
Before differential expression analysis, we filtered the contigs that matched bacteria and other possible contaminating species like Dicyema misakiense. Then the filtered data was inputed into DESeq2 (Love et al. 2014) to detect differentially expressed genes (DEGs). Taking together all six samples, genes with the sum of row reads less than 60 were filtered and the left genes were kept and normalized by estimateSizeFactors function in DESeq2. Genes with a fold change ≥ 2 and adjusted p values < 0.05 were identified as significant DEGs. Then the DEGs were subjected to enrichment analysis in topGO, using the Fisher test (Alexa, Rahnenfuhrer 2010). The enriched GO terms were visualized in REVIGO (http://revigo.irb.hr/) and CirGO (Kuznetsova et al. 2019). KEGG pathways analysis was also conducted to identify enriched metabolic pathways in DEGs by using KOBAS with the FDR ≤ 0.05 (Xie et al. 2011).
SSR and SNP detection
The potential SSRs in the whole transcriptome were detected by MIcroSAtellite (Thiel et al. 2003). The minimum repeats of di-, tri, tetra-, penta- and hexa-nucleotide were set as six, five, four, four, four, respectively. The SNPs were detected by GATK3 software (Van der Auwera et al. 2013). And SNP/InDel correlation analysis was performed using ANNOVAR (Wang et al. 2010).
Gene expression validation
To validate the sequencing data, seven DEGs including three up-regulated and four down-regulated were selected to conduct the qRT-PCR in both sets. The reference genes EF1-α and RPS18b were chosen as internal controls according to our previous research (Xu and Zheng 2018). The primer sequences of seven DEGs and the internal controls were shown in Table S1. A total amount of 1 μg RNA was reverse transcribed to the first-strand cDNA and a final volume of 20 µL cDNA was obtained by PrimerScript RT reagent kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. qRT-PCR was performed in LightCycler Roche480 and the final PCR reaction volume was 10 µL, including 1 µL cDNA, 5 µL of 2 × SYBR EVE PCR Mix (abm), 0.3 µL of each primer(10 µM) and 3.4 µL distilled water. The reaction conditions included denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 s, 55 °C for 30 s, 72 °C for 30 s. The relative expression levels of DEGs were calculated by comparative CT methods (Schmittgen et al. 2008).
Results
De novo assembly and annotation
Two libraries (OM-C & OM-E) were built and 376,166,676 raw reads were generated through sequencing (Table 1). After filtering, we have obtained 367,396,914 clean reads, including 174,442,374 in OM-C and 192,954,540 in OM-E. The libraries contain 54.11G clean bases and the mean values of Q20 and Q30 were 98.15% and 95.94%, respectively (Table S2). A total of 63,237 unigenes were discovered after de novo assembly. The GC content of the transcripts was 37.18% and the N50 length was 1291 bp. The length of unigenes ranged from 201 to 32,899 bp and the average length of unigenes was 811 bp (Fig. S1).
Among all the assembled unigenes, 25,708 were successfully annotated in at least one database and 10,777 were annotated in all databases described above. However, the other 37,529 unigenes did not hit in any database. Nr database annotated the highest number (25,597) of unigenes, accounting for 40.48% of all the unigenes, followed by 16,606 (26.26%) annotated in the Swissport database and 15,095 (23.87%) matched to the KOG database (Fig. S2). Among all the successfully annotated unigenes in the NR database, 13,919 unigenes (54.38%) showed strong homology to Octopus bimaculoides and the top ten hit species distribution based on Nr databases were shown in Fig. S3. KOG annotation is also an effective tool in identifying orthology and paralog proteins in eukaryotes. In this research, a total number of 15,095 unigenes were successfully annotated to 25 functional categories in the KOG database and 237 unigenes were involved in defense mechanisms (Fig. S4a), implying these genes might responsible for the immune defense in O. minor. In the KEGG database, 6526 unigenes were assigned to 234 known pathways, which can be classified into five categories and 32 subcategories (Fig. S4b). Among all the known 234 pathways, metabolic pathways assigned the highest number of unigenes (1932), accounting for 30.83% followed by translation pathways (17.44%) and folding, sorting and degradation (13.68%). Moreover, a total of 70 unigenes, involving 16 pathways were identified in the immune system subcategory. GO annotation revealed 7 unigenes involved in the immune system process, 166 unigenes involved in response to stimulus and 197 unigenes involved in biological regulation (Fig. S4c), which may provide useful information in responses to ammonia stress in O. minor.
Differentially expressed genes and functional enrichment analysis
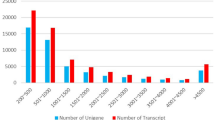
After filtering the contaminated sequences and low-quality counts sequences, 47,255 unigenes were included for differential expression analysis. In total, 1845 DEGs including 315 up-regulated genes and 1530 down-regulated genes were discovered (Fig. 1). Among all the 1845 DEGs, only 1349 DEGs were annotated in the public database, accounting for 73.12%. According to the annotations, some of these DEGs were involved in immune responses, energy metabolism, amino acid metabolism, ion transporter (Table 2). The functions of DEGs were further evaluated by GO and KEGG enrichment analysis. According to the GO enrichment analysis, a total of 44 GO terms were significantly enriched. Notably, the second-highest proportion of GO classification was ion transmembrane transport (13.8%), including inorganic cation/ion transmembrane transport and hydrogen ion transmembrane transport and meanwhile other GO terms involving proton transport were also enriched such as energy coupled proton transmembrane transport against electrochemical gradient (Fig. 2 and Table S3), which indicated that cation/ion transmembrane transport might play a central function in ammonia detoxification. For DEGs in KEGG pathway analysis, fifty-five metabolic pathways were significantly enriched, including metabolic pathways, oxidative phosphorylation, proteasome, citrate cycle, biosynthesis of amino acids and phagosome (Table 3). In general, these pathways can be represented by two major biological process: (1) metabolism to maintain the living state of the cells and organism and (2) innate immune defense represented by endocytosis, phagosome and lysosome, etc.
To validate the expression levels of unigenes in RNA-seq, we performed a qRT-PCR in 7 unigenes, including 3 up-regulated and four down-regulated unigenes. The results indicated that the expression pattern of unigenes in RNA-seq and quantitative results were highly consistent, which further confirmed the reliability of data (Fig. S5).
SSR and SNP analysis
SSR and SNP are two major molecular markers that are widely used in population genetic research and marker-associated breeding studies (Chistiakov et al. 2006; Dawson et al. 2013; Hubert et al. 2010). However, to date, only very limited SSR makers are available for O. minor (Gao et al. 2016; Zuo et al. 2011). Here, the potential SSR and SNP markers were examined through transcriptome analysis. The results revealed that the total number of identified SSRs is 25,026, which including 16,883 di-nucleotide, 6,680 tri-nucleotide, 1379 tetra-nucleotide, 76 penta-nucleotide and 8 hexa-nucleotide (Fig. S6a). AC/GT, AAT/ATT, ACAT/ATGT were the primary types in di-nucleotide, tri-nucleotide and tetra-nucleotide repeats, respectively. According to the transcriptome scan, a total of 268, 672 potential SNPs with the quality greater than 50 were detected, distributing in 40,112 unigenes. The number of transition and transversion were 175,775 and 92,897, respectively (Fig. S6b).
Discussion
Responses to acute ammonia stress
Marine organisms have employed diverse ammonia excretion strategies to survive in a various environment like seawater, freshwater, water film surrounding sand particles or mud. The published genome of O. minor also implied that the large gene redundancy and gene expansion in this species may relate to the adaptation of harsh environment fluctuation like diurnal temperature changes, steep salinity and pH gradients oxygen availability (Kim et al. 2018). Ammonia is one of the most common and detrimental stimulation in an aquaculture environment that can cause the immune system damage and even the death of octopods (Feyjoo et al. 2011). In recent years, several papers tried to explore the molecular mechanism of detoxification of ammonia in cephalopods (Hu et al. 2017; Peng et al. 2016). However, our understanding of the mechanism of ammonia tolerance and detoxification metabolism is still limited. In this study, we performed comparative transcriptome analysis in O. minor to provide for the first time the key genes and possible pathways involved in ammonia responses in O. minor.
In this study, we found several immune-related genes that were significantly down-regulated after ammonia stress, such as peptidoglycan-recognition protein SC2 (pgrp-SC2), TNF receptor-associated factor 2 (traf2), malectin, and endoplasmic reticulum lectin 1 (erlec1), interleukin 17-like protein (Table 2). PGRP-SC2 is a member of PGRP family that can specifically recognize the peptidoglycan (PGN) of bacteria and is involved in innate immune in bivalves (Ni et al. 2007). TRAF 2 belongs to the TRAF superfamily that plays a key role in signal transduction, inflammatory and apoptosis in mammals. However, researchers found that TRAF2 may also involve in host defense against immune challenges in mollusks (Qu et al. 2017). Malectin and ERLEC1 all belong to the endoplasmic reticulum lectins, particularly, malectin was found to be involved in ER-stress and innate immunity in scallops (Wang et al. 2019). Similar down-regulation of immune-related genes under ammonia stress was also observed in shrimps (Li et al. 2018; Lu et al. 2016; Wang et al. 2017). In addition, several pathways like lysosome, phagosome and endocytosis pathways that highly related to immune defense were significantly enriched (Table 3). The lysosome is a membrane-bound dynamic organelle that is responsible for degradation and recycling of intracellular and extracellular materials by phagocytosis and autophagy and endocytosis, and thus essential for innate immune responses to prevent pathogen and eliminate non-self molecules (Gao et al. 2017; Luzio et al. 2007). Phagocytosis especially hemocyte phagocytosis constitutes the major part of immune responses in marine mollusks (Zannella et al. 2017). All the above indicated the damages on the immune defense system in O. minor after ammonia stress, which also further explained why aquatic organisms in high ammonia accumulation are susceptible to pathogens (Evans et al. 2006; Hurvitz et al. 1997).
Researches in other organisms found that ammonia accumulation would induce mitochondria dysfunction and energy metabolism disturbances (Niknahad et al. 2017; Ott et al. 2005). In this study, two energy-producing pathways—oxidative phosphorylation and Citrate cycle (TCA cycle) pathways and the GO term—energy coupled proton transmembrane transport, against electrochemical gradient (GO:0015988) were significantly enriched. The transport of protons across a membrane to generate an electrochemical gradient (proton-motive force) that provides energy for the synthesis of ATP or GTP. Oxidative phosphorylation and TCA cycle are also closely related. Previous studies in marine organisms revealed that oxidative phosphorylation was highly involved in the responses to environmental stress (Tomanek 2015; Wang et al. 2017). The nicotinamide adenine dinucleotide (NADH) generated by the TCA cycle would transfer to the Oxidative phosphorylation, leading the ADP convert into ATP. Several main genes in mitochondrial membrane respiratory chain like NADH dehydrogenase and succinate dehydrogenase and genes in the TCA cycle like 2-oxoglutarate dehydrogenase and malate dehydrogenase were significantly down-regulated, indicating the rate of energy production in mitochondria might decline after ammonia stress.
Notably, a large amount of amino acid metabolism pathways was enriched in our study, including biosynthesis of amino acids, arginine and proline metabolism, valine, leucine and isoleucine degradation, cysteine and methionine metabolism, alanine, aspartate and glutamate metabolism, glycine, serine and threonine metabolism, glutathione metabolism. Amino acid metabolism changes were also observed in other marine organisms in response to cold stress, osmotic stress as well as ammonia stress (Nie et al. 2017; Xiao et al. 2019; Zhou et al. 2011). In these amino acids, some are important precursors in TCA cycles like valine, aspartate, glutamate, threonine, while others like alanine, serine and threonine are precursors of pyruvate which is the end-product of glycolysis and is also essential for ATP generation in respiration chain. Furthermore, amino acid metabolism is one of the major ways for nitrogenous degradation and thus is deemed as a strategy in aquatic organisms to avoid ammonia toxicity (Chen 2000; Xiao et al. 2019).
Mechanism of ammonia metabolism in O. minor
In this study, we used RNA-seq to investigate possible ammonia excretion mechanisms in O. minor. As shown in Fig. 3, high accumulation of ammonia would down-regulate the key enzyme-coded genes, in our case for example isocritate dehydrogenase, succinate dehydrogenase, malate dehydrogenase and thus inhibit the TCA cycle, which was also observed in Litopenaeus vannamei (Xiao et al. 2019). According to previous researches in other species, one detoxification pathway is to transform ammonia to glutamine. However, because of the down-regulation of the TCA cycle, this pathway is less possible for ammonia detoxification in our case. Another possible way is to convert ammonia to urea or uric acids. Researches in cephalopods indicated that they can produce guanine, urea or uric acids (Wells 1978; Hoeger et al. 1987; Boucher-Rodoni and Mangold 1989). Here, we found that the ornithine aminotransferase gene is down-regulated, which means a higher rate of urea cycle. However, other key genes in urea cycle were not differentially expressed and the key genes involved in producing uric acids were not detected in our RNA-seq data. Therefore, the excretion way through urea cycle or producing uric acids in O. minor needs further studies. Finally, we suspected that another possible way of ammonia detoxification under acute ammonia stress is through ammonia transporters. Perfused gills experiment in Octopus vulgaris revealed that four ammonia transporters (Na+/K+-ATPase, V-type H+-ATPase, Na+/H+-exchanger 3 and Rhesus protein) located in gill epithelial cell and blood vessels may be responsible for the ammonia homeostasis (Hu et al. 2017). K+ ions and hydrated NH4+ and have similar sizes and thus K+ can be substituted by NH4+ to excrete into the environment through Na+/NH4+ exchanger (Hu et al. 2017; Weihrauch and Allen 2018). V-type H+-ATPase could induce an “acid-trapping” mechanism to convert NH3 into NH4+. In shallow-water octopods (O. vulgaris), the major ammonia excretion organs are gill, renal appendages and the ammonia level in blood was significantly lower after the passage of the excretory organs (Hu et al. 2017). However, in this study, only hemolymph was analysis and the transcription of V-type H+-ATPase (VHA) and Na+/K+-ATPase (NKA) were significantly down-regulated after ammonia stress, indicating lower levels of NH4+ entrapment in hemolymph and a lower rate of NH4+ export. This situation could because both transporters are ATP-dependent, which means a decline in energy production may result in down-regulation of these two genes, and the harmful effects of ammonia stress may also influence the physiology of octopus. Hence, further experiments about blood ammonia levels and transcriptional regulation of ammonia transporters in excretory organs are necessary to reveal the role of ammonia transporters in ammonia detoxification.
Conclusion
In this study, we performed for the first time the comparative transcriptome analysis in Cephalopoda (O. minor) under acute ammonia stress. We found that most of the annotated DEGs were involved in immune responses, amino acids metabolism, energy production. According to the differential expression analysis and enrichment analysis, we speculated that high ammonia accumulation in the environment would reduce the ability of immune defense in O. minor, inhibit the TCA cycle and slow down the rate of ATP production. Possible ammonia excretion ways were proposed and further experiments would be necessary to verify these hypotheses. This study provided the basic information about ammonia toxicity and ammonia excretion in O. minor, meanwhile the genes and pathways found in this study will facilitate further studies of molecular mechanisms of ammonia excretion in mollusk.
References
Alexa A, Rahnenfuhrer J (2010) topGO: enrichment analysis for gene ontology. R package version 2.36.0
Andrews PLR, Darmaillacq AS, Dennison N et al. (2013) The identification and management of pain, suffering and distress in cephalopods, including anaesthesia, analgesia and humane killing. J Exp Mar Biol Ecol 447:46–64. https://doi.org/10.1016/j.jembe.2013.02.010
Arillo A, Margiocco C, Melodia F et al. (1981) Ammonia toxicity mechanism in fish: studies on rainbow trout (Salmo gairdneri Rich). Ecotoxicol Environ Saf 5:316–328. https://doi.org/10.1016/0147-6513(81)90006-3
Benli ACK, Koksal G, Ozkul A et al. (2008) Sublethal ammonia exposure of Nile tilapia (Oreochromis niloticus L.): effects on gill, liver and kidney histology. Chemosphere 72:1355–1358. https://doi.org/10.1016/j.chemosphere.2008.04.037
Boucher-Rodoni R, Mangold K (1989) Respiration and nitrogen excretion by the squid Loligo forbes. Mar Biol 103(3):333–338. https://doi.org/10.1007/BF00397267
Bower CE, Bidwell JP (1978) Ionization of ammonia in seawater: effects of temperature, pH, and salinity. J Fish Res Board Can 35:1012–1016. https://doi.org/10.1139/f78-165
Castellanos-Martinez S, Arteta D, Catarino S et al. (2014a) De novo transcriptome sequencing of the Octopus vulgaris hemocytes using Illumina RNA-Seq technology: response to the infection by the gastrointestinal parasite Aggregata octopiana. PLoS ONE 9:e107873. https://doi.org/10.1371/journal.pone.0107873
Castellanos-Martinez S, Prado-Alvarez M, Lobo-da-Cunha A et al. (2014b) Morphologic, cytometric and functional characterization of the common octopus (Octopus vulgaris) hemocytes. Dev Comp Immunol 44:50–58. https://doi.org/10.1016/j.dci.2013.11.013
Chen J (2000) Study on the free amino acid levels in the hemolymph, gill, hepatopancreas and muscle of Penaeus monodon exposed to elevated ambient ammonia. Aquatic Toxicol 50:27–37. https://doi.org/10.1016/s0166-445x(99)00095-8
Chen JC, Kou YZ (1992) Effects of ammonia on growth and molting of Penaeus japonicus juveniles. Aquaculture 104:249–260. https://doi.org/10.1016/0044-8486(92)90207-2
Cheng TC (1977) Biochemical and ultrastructural evidence for the double role of phagocytosis in molluscs: Defense and nutrition. In: Bulla LA, Cheng TC (eds) Comparative pathobiology, vol 3. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-7299-2_2
Chistiakov DA, Hellemans B, Volckaert FAM (2006) Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255:1–29. https://doi.org/10.1016/j.aquaculture.2005.11.031
Colt JE, Armstrong DA (2009) Nitrogen toxicity to crustaceans, fish, and molluscs. In: Allen LJ, Kinney EC (eds.) Proceedings of the bio-engineering symposium for fish culture. Northeast Society of Conservation Engineers, Bethesda, pp. 34–47
Dawson DA, Ball AD, Spurgin LG et al. (2013) High-utility conserved avian microsatellite markers enable parentage and population studies across a wide range of species. BMC Genom 14. https://doi.org/10.1186/1471-2164-14-176
Evans JJ, Pasnik DJ, Brill GC et al. (2006) Un-ionized ammonia exposure in Nile tilapia: toxicity, stress response, and susceptibility to Streptococcus agalactiae. N Am J Aquac 68:23–33. https://doi.org/10.1577/A05-032.1
Feyjoo P, Riera R, Felipe BC, Skalli A, Almansa E (2011) Tolerance response to ammonia and nitrite in hatchlings paralarvae of Octopus vulgaris and its toxic effects on prey consumption rate and chromatophores activity. Aquac Int 19:193–204
Ford LA (1992) Host defense mechanisms of cephalopods. Annu Rev Fish Dis 2:25–41. https://doi.org/10.1016/0959-8030(92)90054-2
Gao XL, Zheng XD, Bo QK et al. (2016) Population genetics of the common long-armed octopus Octopus minor (Sasaki, 1920) (Cephalopoda: Octopoda) in Chinese waters based on microsatellite analysis. Biochem Syst Ecol 66:129–136. https://doi.org/10.1016/j.bse.2016.03.014
Gao Y, Chen Y, Zhan S et al. (2017) Comprehensive proteome analysis of lysosomes reveals the diverse function of macrophages in immune responses. Oncotarget 8:7420–7440. https://doi.org/10.18632/oncotarget.14558
Gestal C, Castellanos-Martínez S et al. (2015) Understanding the cephalopod immune system based on functional and molecular evidence. Fish Shellfish Immunol 46:120–130. https://doi.org/10.1016/j.fsi.2015.05.005
Grabherr MG, Haas BJ, Yassour M et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Grimaldi AM, Belcari P, Pagano E et al. (2013) Immune responses of Octopus vulgaris (Mollusca: Cephalopoda) exposed to titanium dioxide nanoparticles. J Exp Mar Biol Ecol 447:123–127. https://doi.org/10.1016/j.jembe.2013.02.018
Hoeger U, Mommsen TP, O’Dor R et al. (1987) Oxygen uptake and nitrogen excretion in two cephalopods, octopus and squid. Comp Biochem Physiol Part A: Physiol 87(1):63–67. https://doi.org/10.1016/0300-9629(87)90426-9
Hu MY, Sung PH, Guh YJ et al. (2017) Perfused gills reveal fundamental principles of pH regulation and ammonia homeostasis in the Cephalopod Octopus vulgaris. Front Physiol 8:162. https://doi.org/10.3389/fphys.2017.00162
Hubert S, Higgins B, Borza T et al. (2010) Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua). BMC Genom 11(1):191. https://doi.org/10.1186/1471-2164-11-191
Hurvitz A, Bercovier H, VanRijn J (1997) Effect of ammonia on the survival and the immune response of rainbow trout (Oncorhynchus mykiss, Walbaum) vaccinated against Streptococcus iniae. Fish Shellfish Immunol 7:45–53. https://doi.org/10.1006/fsim.1996.0062
Kim BM, Kang S, Ahn DH et al. (2018) The genome of common long-arm octopus Octopus minor. GigaScience 7(11):giy119. https://doi.org/10.1093/gigascience/giy119
Kuznetsova I, Lugmayr A, Siira SJ et al. (2019) CirGO: an alternative circular way of visualising gene ontology terms. BMC Bioinform 20:84. https://doi.org/10.1186/s12859-019-2671-2
Lee PG (1994) Nutrition of cephalopods: fueling the system. Mar Freshw Behav Physiol 25:35–51. https://doi.org/10.1080/10236249409378906
Li Y, Zhou F, Huang J et al. (2018) Transcriptome reveals involvement of immune defense, oxidative imbalance, and apoptosis in ammonia-stress response of the black tiger shrimp (Penaeus monodon). Fish Shellfish Immunol 83:162–170. https://doi.org/10.1016/j.fsi.2018.09.026
Liu C (2013) Studies on culture of the life cycle of Octopus minor (Sasaki, 1920). Fisheries College, Ocean University of China, pp. 7–33. (Chinese in English abstract)
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Lu X, Kong J, Luan S et al. (2016) Transcriptome analysis of the hepatopancreas in the Pacific white shrimp (Litopenaeus vannamei) under acute ammonia stress. PLoS ONE 11:e0164396. https://doi.org/10.1371/journal.pone.0164396
Luzio JP, Pryor PR, Bright NA (2007) Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632. https://doi.org/10.1038/nrm2217
Mayrand E, St‐Jean SD, Courtenay SC (2005) Haemocyte responses of blue mussels (Mytilus edulis L.) transferred from a contaminated site to a reference site: can the immune system recuperate? Aquac Res 36:962–971. https://doi.org/10.1111/j.1365-2109.2005.01302.x
Messenger JB, Nixon M, Ryan KP (1985) Magnesium chloride as ananaesthetic for cephalopods. Comp Biochem Physiol C 82:203–205. https://doi.org/10.1016/0742-8413(85)90230-0
Ni D, Song L, Wu L et al. (2007) Molecular cloning and mRNA expression of peptidoglycan recognition protein (PGRP) gene in bay scallop (Argopecten irradians, Lamarck 1819). Dev Comp Immunol 31:548–558. https://doi.org/10.1016/j.dci.2006.09.001
Nie H, Jiang L, Chen P et al. (2017) High throughput sequencing of RNA transcriptomes in Ruditapes philippinarum identifies genes involved in osmotic stress response. Sci Rep 7:4953. https://doi.org/10.1038/s41598-017-05397-8
Niknahad H, Jamshidzadeh A, Heidari R et al. (2017) Ammonia-induced mitochondrial dysfunction and energy metabolism disturbances in isolated brain and liver mitochondria, and the effect of taurine administration: relevance to hepatic encephalopathy treatment. Clin Exp Hepatol 3:141–151. https://doi.org/10.5114/ceh.2017.68833
Ott P, Clemmesen O, Larsen FS (2005) Cerebral metabolic disturbances in the brain during acute liver failure: from hyperammonemia to energy failure and proteolysis. Neurochem Int 47:13–18. https://doi.org/10.1016/j.neuint.2005.04.002
Peng RB, Le KX, Wang PS et al. (2016) Detoxification pathways in response to environmental ammonia exposure of the cuttlefish, Sepia pharaonis: glutamine and urea formation. J World Aquac Soc 48:342–352. https://doi.org/10.1111/jwas.12341
Perez DG, Fontanetti CS (2011) Hemocitical responses to environmental stress in invertebrates: a review. Environ Monit Assess 177:437–447. https://doi.org/10.1007/s10661-010-1645-7
Peyghan R, Takamy GA (2002) Histopathological, serum enzyme, cholesterol and urea changes in experimental acute toxicity of ammonia in common carp Cyprinus carpio and use of natural zeolite for prevention. Aquac Int 10:317–325. https://doi.org/10.1023/A:1022408529458
Pierce G, Valavanis V, Pereira J et al. (2006) Fishing for cephalopods. ICES CIEM Newsl43:24–28
Qian Y, Zheng X, Liu C et al. (2013) Studies on the reproductive habit and embryonic development of Octopus minor under the artificial conditions. Oceanol Limnol Sin 44:165–170. https://doi.org/10.11693/hyhz201301024024. Chinese in English abstract
Qu F, Xiang Z, Zhou Y et al. (2017) A molluscan TNF receptor-associated factor 2 (TRAF2) was involved in host defense against immune challenges. Fish Shellfish Immunol 71:105–115. https://doi.org/10.1016/j.fsi.2017.09.076
Schmittgen TD, Livak KJ et al. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Smart GR (1978) Investigations of the toxic mechanisms of ammonia to fish-gas exchange in rainbow trout (Salmo gairdneri) exposed to acutely lethal concentrations. J Fish Biol 12:93–104. https://doi.org/10.1111/j.1095-8649.1978.tb04155.x
Thiel T, Michalek W, Varshney RK et al. (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422. https://doi.org/10.1007/s00122-002-1031-0
Tomasso JR (1994) Toxicity of nitrogenous wastes to aquaculture animals. Rev Fish Sci 2:291–314. https://doi.org/10.1080/10641269409388560
Tomanek L (2015) Proteomic responses to environmentally induced oxidative stress. J Exp Biol 218:1867–1879. https://doi.org/10.1242/jeb.116475
Van der Auwera GA, Carneiro MO, Hartl C et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform 43:11 10 11–33. https://doi.org/10.1002/0471250953.bi1110s43
Vaz-Pires P, Seixas P, Barbosa A (2004) Aquaculture potential of the common octopus (Octopus vulgaris Cuvier, 1797): a review. Aquaculture 238:221–238. https://doi.org/10.1016/j.aquaculture.2004.05.018
Vedel NE, Korsgaard B, Jensen FB (1998) Isolated and combined exposure to ammonia and nitrite in rainbow trout (Oncorhynchus mykiss): effects on electrolyte status, blood respiratory properties and brain glutamine/glutamate concentrations. Aquat Toxicol 41:325–342. https://doi.org/10.1016/S0166-445X(97)00071-4
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Wang MQ, Wang BJ, Liu M et al. (2019) The first identification of a malectin gene (CfMal) in scallop Chlamys farreri: sequence features and expression profiles. Invertebr Surviv J 16:25–33. https://doi.org/10.25431/1824-307X/isj.v0i0.25-33
Wang W, Yang S, Wang C et al. (2017) Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immunol 71:319–328. https://doi.org/10.1016/j.fsi.2017.10.028
Weihrauch D, Allen GJP (2018) Ammonia excretion in aquatic invertebrates: new insights and questions. J Exp Biol 221(2):jeb169219. https://doi.org/10.1242/jeb.169219
Wells MJ (1978) Octopus: physiology and behaviour of an advanced invertebrate. University Printing House, Cambridge
Xie C, Mao X, Huang J et al. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–322. https://doi.org/10.1093/nar/gkr483
Xiao J, Li QY, Tu JP et al. (2019) Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotoxicol Environ Saf 180:491–500. https://doi.org/10.1016/j.ecoenv.2019.05.029
Xu R, Zheng X (2018) Selection of reference genes for quantitative real-time PCR in Octopus minor (Cephalopoda: Octopoda) under acute ammonia stress. Environ Toxicol Pharmacol 60:76–81. https://doi.org/10.1016/j.etap.2018.04.010
Zannella C, Mosca F, Mariani F et al. (2017) Microbial diseases of bivalve mollusks: infections, immunology and antimicrobial defense. Mar Drugs 15(6):182. https://doi.org/10.3390/md15060182
Zheng, X, Qian, Y, Liu, C et al. (2014) Octopus minor. In: Iglesias J, Fuentes L, Villanueva R (eds), Cephalopod culture. Springer, New York, pp. 415–426
Zhou M, Wang AL, Xian JA (2011) Variation of free amino acid and carbohydrate concentrations in white shrimp, Litopenaeus vannamei: effects of continuous cold stress. Aquac Int 317:182–186. https://doi.org/10.1016/j.aquaculture.2011.04.033
Zuo ZR, Zheng XD, Yuan Y, Li Q (2011) Development and characterization of 12 polymorphic microsatellite loci in Octopus minor (Sasaki, 1920). Conserv Genet Resour 3:489–491. https://doi.org/10.1007/s12686-011-9386-7
Acknowledgements
The authors thank Mr. CAI Bing and Mr. CAI Hui from Mashan Fishery Company for their help in supplying O. minor samples and experiment sites. This work was supported by research grants from National Science Foundation of China (No. 31672257) and the Fundamental Research Funds for the Central Universities (No. 201822022).
Author information
Authors and Affiliations
Contributions
R.X. and X.D.Z conceived and designed the study. R.X. collected samples and performed the experimental work and data analysis. R.X. wrote the paper and X.D.Z revised the paper. All the authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards in Cephalopods (reviewed in Andrews et al. 2013).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, R., Zheng, X. Hemocytes transcriptomes reveal metabolism changes and detoxification mechanisms in response to ammonia stress in Octopus minor. Ecotoxicology 29, 1441–1452 (2020). https://doi.org/10.1007/s10646-020-02279-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-020-02279-0